Abstract
Elevated seawater temperatures have contributed to the rise of coral disease mediated by bacterial pathogens, such as the globally distributed Vibrio coralliilyticus, which utilizes coral mucus as a chemical cue to locate stressed corals. However, the physiological events in the pathogens that follow their entry into the coral host environment remain unknown. Here, we present simultaneous measurements of the behavioral and transcriptional responses of V. coralliilyticus BAA-450 incubated in coral mucus. Video microscopy revealed a strong and rapid chemokinetic behavioral response by the pathogen, characterized by a two-fold increase in average swimming speed within 6 min of coral mucus exposure. RNA sequencing showed that this bacterial behavior was accompanied by an equally rapid differential expression of 53% of the genes in the V. coralliilyticus genome. Specifically, transcript abundance 10 min after mucus exposure showed upregulation of genes involved in quorum sensing, biofilm formation, and nutrient metabolism, and downregulation of flagella synthesis and chemotaxis genes. After 60 min, we observed upregulation of genes associated with virulence, including zinc metalloproteases responsible for causing coral tissue damage and algal symbiont photoinactivation, and secretion systems that may export toxins. Together, our results suggest that V. coralliilyticus employs a suite of behavioral and transcriptional responses to rapidly shift into a distinct infection mode within minutes of exposure to the coral microenvironment.
Similar content being viewed by others
Introduction
Coral reefs are declining worldwide due to rising sea surface temperatures and increasing prevalence of coral disease outbreaks [1,2,3]. Elevated sea surface temperatures cause physiological stress in corals [4] and provide distinct advantages for some coral pathogens [5]. One well-studied coral pathogen, Vibrio coralliilyticus BAA-450, displays tightly regulated temperature-dependent virulence against its coral host, Pocillopora damicornis. While this V. coralliilyticus strain is avirulent at temperatures below 24 °C, it is capable of attacking the coral symbiotic dinoflagellates [6, 7] and lysing coral tissue [8] at temperatures above 27 °C.
V. coralliilyticus displays two distinct behavioral adaptations enabling targeted infection of corals that are physiologically stressed and therefore more vulnerable to pathogenic invasion. First, the bacterial pathogen uses chemotaxis to target chemical signatures present in the mucus of stressed corals [9]. Second, V. coralliilyticus displays chemokinesis, which is the ability to change swimming speed in response to a change in chemical concentration, to potentially enable faster environmental exploration in the presence of its coral host mucus [9, 10]. While both chemotaxis and chemokinesis are behaviors associated with motility and chemical sensing, chemotaxis specifically refers to the ability to detect and follow chemical gradients (without necessarily any change in swimming speed), whereas chemokinesis refers to the ability to change swimming speed in response to an overall change in concentration in the environment (without any reference to whether cells follow gradients). In V. coralliilyticus, these two behaviors combine to enable efficient and rapid targeting of stressed corals. However, chemokinesis, in contrast to chemotaxis, has remained more rarely studied in bacteria and is almost entirely undescribed in the context of marine disease [11,12,13].
Coral mucus—in addition to triggering increased motility and chemotaxis which are behaviors necessary for infection by V. coralliilyticus [14, 15]—also represents the critical interface where pathogen activities can dictate the outcome of an infection. Corals secrete up to half of the carbon assimilated by their algal symbionts as mucus [16, 17] and its production represents a sizable energetic investment that is important for nutrient cycling across the entire reef system [18,19,20,21]. In addition, mucus provides corals with protection against desiccation and is an ancient and evolutionarily conserved first line of defense against pathogens [22]. During infection studies, corals have been observed to actively expel ingested pathogens by spewing out bacteria-laden mucus from the mouths of polyps [23, 24]. However, entry into host mucus may also signal to the bacterial pathogen that contact with a potential host is imminent. Thus, elucidating the behavioral and transcriptional responses of V. coralliilyticus in the context of its coral host environment, and in particular coral mucus, is important in elucidating the mechanisms underpinning coral infection.
Here, we present experiments in which we simultaneously used video microscopy and RNA sequencing to measure the behavioral and transcriptional responses of V. coralliilyticus upon a sudden exposure to mucus from its coral host. To study chemokinesis independently from chemotaxis, we conducted our experiments in the absence of chemical gradients. This represents the first investigation to couple behavioral and transcriptomic analyses to decipher the mechanisms promoting coral infection. We show that behavioral and transcriptional responses occur concomitantly over a surprisingly rapid timescale of only minutes, highlighting the agility of the pathogens in seizing what are likely to be limited windows of opportunity [25] to target and ultimately infect their host.
Materials and methods
Coral mucus collection
Five small colonies of the coral Pocillopora damicornis were collected from Heron Island (23.4423 °S, 151.9148 °E, Great Barrier Reef, Australia) in April 2015 and maintained in aquaria for 2 weeks at 25 °C ± 1 °C and 35 ppt salinity on a 12 h light–dark cycle. Mucus was collected with a sterile 1-ml syringe (Becton Dickinson, NJ, USA) from corals subjected to repeated 3-min air exposure, and snap-frozen in a sterile 50 ml tube (Falcon, Corning Life Sciences, MA, USA) in liquid nitrogen and maintained at −80 °C until use in experiments. Due to the large volume required (55 ml), mucus was collected and pooled from the five coral colonies over 3 consecutive days (separate snap-frozen tube per day). Directly before the experiments, a single pool of coral mucus was created by thawing in a room temperature water bath and pooling all samples into a sterile glass bottle.
Bacterial culture
Vibrio coralliilyticus type strain ATCC BAA-450 was acquired from the American Type Culture Collection (Manassas, VA, USA). A frozen stock (−80 °C, 25% glycerol) of V. coralliilyticus was streaked onto a marine broth 2216 (BD Difco, Franklin Lakes, NJ, USA) culture plate and incubated at room temperature for 24–48 h. For liquid culture inoculation, five colonies were resuspended in 20 µl filtered (0.2 µm) artificial seawater (FASW, 35 g/L sea salt; Instant Ocean, Blacksburg, VA, USA) and 5 µl of this suspension was inoculated into each of three sterile 250-ml Erlenmeyer flasks containing 80 ml 1% marine broth medium (99% FASW, v/v). The timing of inoculation of the triplicate liquid cultures was staggered to allow 17–18 h of growth (to OD600 0.04) before the start of each replicate experiment. Liquid cultures were incubated at 30 °C in an orbital shaker (250 rpm).
Preparation of filtered spent medium
Experimental controls consisted of V. coralliilyticus cells incubated in their own filtered spent medium, which was prepared immediately prior (≤10 min) to the start of each replicate experiment. Approximately 20 ml V. coralliilyticus overnight culture was aseptically filtered through a sterile 0.2-µm syringe filter (polyethersulfone membrane; VWR International) attached to a sterile 10-ml syringe, into a sterile 50-ml Falcon tube. Filters were changed every 10 ml of volume filtered to minimize clogging.
Experimental setup
Three biological replicate experiments were sequentially performed on the same day using three independent cultures of V. coralliilyticus exposed to their respective filtered spent media (control) or aliquots of the same pooled coral mucus (treatment) (Fig. 1). Aliquots of 25 ml of bacteria were placed into each of two RNase-free 50-ml Falcon tubes for the mucus and control treatments. Bacteria (before and after addition of mucus), coral mucus, and filtered spent media were maintained at 30 °C ± 1 °C in a water bath for the duration of the experiment to prevent temperature shock. All experiments were performed at 30 °C to capture conditions in which V. coralliilyticus is capable of infecting its coral host [8]. Incubation was initiated by the 1:1 (v/v at t = 0 min) addition of coral mucus or filtered spent medium to bacteria, directly followed by thorough mixing by repeated pipetting to eliminate any chemical gradients.
Experimental timeline showing time points for microscopy video acquisition and RNA-seq sampling. At t = 0 min (red arrow), coral mucus (treatment) or filtered spent medium (control) was added (1:1, v/v) to V. coralliilyticus cultured in 1% marine broth. The experiment was conducted at 30 °C, and repeated three times sequentially in a single day, using three different cultures of V. coralliilyticus.
Bacteria were repeatedly and manually sampled from populations incubated with mucus or filtered spent media. At each time point, 40 µl of the bacteria-containing solution was removed from the top of the incubation tube using a pipette and introduced into a straight microfluidic channel (60 mm × 4 mm, 90 µm depth), followed immediately by video acquisition. A single microfluidic device, fabricated using soft lithography [26] and permanently bonded to a glass microscope slide (75 mm × 25 mm × 1 mm; VWR International), contained two identical channels in parallel to accommodate the mucus and the control experimental conditions. A temperature-controlled microscope stage insert (Warner Instruments, Hamden, CT, USA) was used to maintain cells at 30 °C ± 1 °C during imaging. After each time point, cells were removed from the device and discarded using a pipette, before the introduction of freshly sampled cells at the next time point. A new microfluidic device was used for each replicate experiment.
Microscopy and video analysis
Experiments were imaged using phase-contrast microscopy on an inverted epifluorescence TE2000 microscope (Nikon, Tokyo, Japan), controlled through Nikon Elements software, with a 20× objective (S Plan Fluor ELWD ADM Ph1 20×, 0.45 NA; Nikon) and 1.5× optical magnification (combined objective magnification of 30×). An sCMOS camera (Andor Neo, 2560 × 2160 pixels, 6.5 µm/pixel; Andor, Belfast, Northern Ireland) was used to acquire videos (300 frames at 29.79 fps) from the center of the microfluidic channels, with the focal plane at channel mid-depth to avoid wall effects on motility.
Analysis of microscopy videos was performed in MATLAB (MathWorks, Natick, MA, USA) using an automated image segmentation and trajectory reconstruction software developed in-house (detailed methods in Supplementary Methods). Briefly, cells in each frame were identified using a pixel intensity threshold, and their swimming trajectories were reconstructed using their x, y-positions in sequential frames (Fig. 2a, b). To discriminate motile from non-motile bacteria, we determined the type of motion (ballistic or diffusive) of each bacterium by calculating the mean squared displacement (MSD) as a function of short time intervals (Δt), and quantifying the exponent α of this dependence (MSD ~ Δtα). Non-motile bacteria were identified as slow-moving cells (median instantaneous speed < 10 µm/s) displaying purely diffusive motion (α < 1). Motile bacteria were instead identified as those cells moving more rapidly (median instantaneous speed ≥ 10 µm/s) and having a higher value of the MSD exponent (α ≥ 1) (Supplementary Fig. 1). A sensitivity analysis showed that our results were robust against the selection of different threshold values (Supplementary Fig. 2). Amongst motile bacteria, the swimming speed of each cell was calculated by averaging the instantaneous speed over the duration of its trajectory, and the mean speed of the population was quantified by averaging over all trajectories of motile bacteria detected in each microscopy video (mean ± s.d. of number of trajectories in videos, n = 1298 ± 312 pre-addition, n = 753 ± 230 post-addition; Supplementary Fig. 3), which represented a single time point in mucus or control conditions (Fig. 2).
Experiments were conducted at 30 °C. a, b Swimming tracks of V. coralliilyticus before (−3 min, a) and after (10 min, b) addition of coral mucus. Swimming tracks of 70 motile cells were randomly selected for each panel. Black circles mark the start, and colors indicate the mean swimming speed, of each track. Colored circles (3× zoom of gray box provided for visibility) represent frames of a microscopy video (0.03 s between frames). c, d Probability distributions of swimming speeds of motile cells, before (−3 min, c) and after (10 min, d) addition of coral mucus (orange) or filtered spent medium (gray). e Average swimming speeds of motile cells before and after 1:1 addition (v/v at t = 0 min) of coral mucus (orange) or filtered spent medium (black, control). Averages (filled circles) and standard deviation (shaded regions) were calculated using data obtained from three replicate experiments (○, *, □). Data were interpolated to match time points across replicates. RNA-seq samples were taken at −5, 10, and 60 min (green circles). Data from replicate 3 are presented in panels (a–d).
RNA sampling, isolation, sequencing, and sequence alignment
To characterize changes in gene expression accompanying shifts in swimming behavior, RNA sequencing (RNA-seq) was performed on the same V. coralliilyticus populations from which samples for tracking by video microscopy were obtained. Incubation tubes containing bacteria were swirled vigorously to mix before taking samples of mucus and control cells at three time points during each replicate experiment (Fig. 1). Sample volumes of 8 ml before mucus or filtered spent medium (control) addition (t = 0 min), and 16 ml after addition (t = 10 min and t = 60 min; larger volume to account for biomass dilution) were each filtered through a 0.22-µm Sterivex-GP filter unit (polyethersulfone membrane; Millipore) attached to a sterile 60-ml syringe to capture bacteria onto the membrane. Subsequently, the filtrate end of the Sterivex filter cartridge was briefly flamed and pinched to close, and 2 ml of RNAlater (Thermo Fisher Scientific, Waltham, MA, USA) was added with a pipette through the other end of the cartridge to immerse the cell-containing filter membrane in the RNA-stabilizing solution. Sample-containing filter cartridges were incubated at 4 °C for 24 h, then preserved at −80 °C until RNA extraction. A sample of coral mucus (~2 ml) was also preserved, and its RNA extraction confirmed that little to no RNA was present in coral mucus.
Total RNA extraction was performed as described previously [27] (detailed description in Supplementary Methods). Depletion of rRNA (using Ribo-Zero rRNA Removal Kit for bacteria (Epicentre Biotechnologies)) and subsequent mRNA sequencing (using an Illumina HiSeq2500 sequencer), assembly, and alignment were performed by the Joint Genome Institute (Los Alamos, NM, USA) (detailed description in Supplementary Methods). Due to RNA degradation, the mucus-treatment sample at 10 min from replicate 2 could not be sequenced. Thus, 17 samples in total were sequenced (Sequence Read Archive accession PRJNA707316). Raw reads from each library were aligned to the reference genome (V. coralliilyticus ATCC BAA-450, NCBI Taxon ID 675814). As a result, 99.48% (5022 genes) of the filtered FASTQ reads mapped to the V. coralliilyticus reference genome. GenBank protein IDs (prefix “EEX”) were obtained from V. coralliilyticus BAA-450 genome assembly ASM17613v1 and matched by locus tags (prefix “VIC_”) of each gene.
Differential expression analysis
The DESeq2 package [28] (v1.26.0) in R was used for differential expression analyses. The DESeq2 algorithm uses negative binomial generalized linear models to test for differential abundances in raw count data, and controls for differences in sequencing depth between libraries by estimating size and dispersion factors. Adjusted p values were calculated in DESeq2 using the Benjamini-Hochberg procedure, which accounts for multiple comparisons. Statistically significant gene expression differences were assessed using the Wald test with a false discovery rate (FDR) cutoff at adjusted p < 0.01. No fold-change cutoff was applied. Time-point-matched, pairwise mucus–control comparisons were performed. All instances of gene differential expression given in the text are statistically significant (unless otherwise noted), and the fold-difference values provided in each case are relative to the control at the same time point.
For principal component analysis (PCA) and sample-to-sample distance calculation, raw count data were transformed using the variance stabilizing transformation (VST) method [29] to remove the dependence of the variance on the count mean, especially when count means are low. VST uses the experiment-wide trend of variance over mean in order to transform the data to remove the experiment-wide trend. Transformed values are on the log2 scale.
Gene set enrichment analysis (GSEA)
Gene set enrichment analysis was performed using the GSEA software [30, 31] v4.0.3 to identify differential representation of metabolic pathways in transcripts sequenced in mucus or control conditions. Only genes with significant differential expression determined by DESeq2 were included in the GSEA analyses (GSEAPreranked protocol). Gene sets were defined by KEGG PATHWAY, and significance was determined at an FDR q value cutoff of 0.25. Genes were ranked by log2 fold-change values. Further details are described in Supplementary Methods.
Results
V. coralliilyticus exhibits strong chemokinesis within minutes of exposure to coral mucus
Video microscopy revealed a strong and rapid behavioral response by V. coralliilyticus cells following exposure to Pocillopora damicornis coral mucus (Fig. 2) at 30 °C, a temperature at which this pathogen is capable of infecting its coral host [8]. The viscosity of coral mucus in our experiments (0.750 cP) was similar to that of filtered artificial seawater (0.731 cP) at room temperature (Supplementary Methods). Thus, bacterial responses measured in this study were assumed to be mostly due to chemical components of coral mucus, although temperature-dependent viscosity changes of coral mucus may influence bacterial swimming [32, 33].
V. coralliilyticus responded to coral mucus with strong chemokinesis. Video microscopy revealed that the average swimming speed of V. coralliilyticus cells increased from 48.0 ± 3.4 µm/s (mean ± s.d.), measured 5 min before addition of coral mucus, to 81.0 ± 9.6 µm/s within 2 min of coral mucus addition (the first time point at which speed was measured post mucus addition; Fig. 2e). The maximum swimming speed (95.5 ± 4.6 µm/s) was reached at 6 min post-addition, representing a two-fold increase compared to the pre-addition state (Fig. 2e). After reaching their maximum speed, the average swimming speed of bacteria in coral mucus gradually decreased over time, but remained significantly higher than the swimming speeds of pre-addition and control cells for the entire experimental duration (65 min; two-tailed t tests, p < 0.01). When samples were collected for RNA-seq, at 10- and 60-min post-addition of mucus, swimming speeds were 90.1 ± 2.9 µm/s and 64.5 ± 0.9 µm/s, or 1.9× or 1.3× compared to baseline, respectively (Fig. 2e).
Chemokinesis by V. coralliilyticus is a temperature-dependent response and appears to be driven by the influx of nutrients and other signaling molecules. First, coral mucus–induced chemokinesis was attenuated in an experiment conducted at a lower temperature (18.7 °C), in which the observed speed enhancement was only 1.1-fold at 7 min post mucus addition (Supplementary Fig. 4). Second, in a separate experiment conducted at 30 °C, rich medium and coral mucus both led to a doubling of swimming speed (2.3-fold increase for rich medium, 2.2-fold increase for coral mucus) within 5 min (Supplementary Fig. 5), suggesting that, at least partially, chemokinesis is a response to the nutrient influx upon addition of mucus. Third, control cells, exposed to filtered spent medium in place of coral mucus, displayed a weak and short-lived increase in swimming speed that peaked at 1.3-fold the pre-addition speed after 8 min and returned to baseline within 15 min (Fig. 2e), which may have been caused by oxygenation of the spent medium during filtration (Supplementary Discussion). Taken together, our results suggest that the swimming speed of V. coralliilyticus is highly sensitive to chemical changes in the environment, and that at the virulence-inducing temperature of 30 °C, chemokinesis in response to coral mucus is likely due to the influx of nutrients, and is especially rapid, larger in magnitude, and longer in duration compared to any other conditions tested.
Probability distributions of the swimming speeds of motile cells before and after mucus addition showed that the observed increase in average swimming speed was caused by a shift of the entire motile V. coralliilyticus population toward a faster swimming regime (Fig. 2c, d, Supplementary Fig. 6), rather than the emergence of behaviorally distinct subpopulations. In contrast, only a subset of motile control cells increased swimming speeds upon the addition of filtered spent media (Supplementary Fig. 6). Furthermore, ~75% of the population was motile over the whole experimental duration in both conditions (exposed to mucus or spent medium; Supplementary Fig. 7). These results suggest that speed enhancement within the motile fraction of the population underlies the increase in average swimming speed observed in coral mucus, and that non-motile cells mostly maintain their non-swimming state upon mucus addition.
Within minutes of coral mucus exposure, V. coralliilyticus initiates a transcriptional response
Sequencing and alignment of mRNA libraries to the V. coralliilyticus BAA-450 reference genome, which contains 5078 protein-coding sequences [34], resulted in 5020 genes with non-zero total read count (Supplementary Tables 1, 2). Coral mucus led to a genome-wide response, with significant upregulation of 1379 genes (27% of detected genes) after 10 min, and 1159 genes (23%) after 60 min, relative to controls at the same time points (Fig. 3a). In addition, coral mucus exposure was associated with significant downregulation of 1326 genes (26%) after 10 min and 1076 genes (21%) after 60 min (Fig. 3a). Of all significantly differentially expressed genes, 1521 genes (30%) were shared between 10 and 60 min (Supplementary Table 3). These results suggest that exposure to coral mucus leads to large shifts in the gene expression profile of V. coralliilyticus.
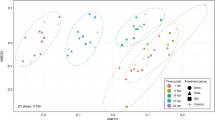
a Number of genes that were significantly upregulated or downregulated in coral mucus at each time point (adjusted p < 0.01). b Principal component analysis (PCA) plot showing all RNA-seq samples in the 2D plane spanned by the first two principal components. PCA was performed using raw read count data after variance stabilizing transformation. c, d Gene set enrichment analyses (GSEA) on mucus vs. control at 10 min (c) and 60 min (d). Gene sets (KEGG pathways, bar labels) that were significantly upregulated or downregulated in coral mucus (FDR q < 0.25) are shown, and their bars are ordered top to bottom by FDR q values (smallest to largest; i.e., most significant to least significant) within each expression category (upregulated in mucus, blue/orange, or downregulated in mucus, gray). Number of significantly differentially expressed genes (adjusted p < 0.01) in mucus that were included in the GSEA (bold), and the total number of genes in the V. coralliilyticus genome assigned to each KEGG pathway, are shown. Orange bars represent gene sets that were upregulated in mucus at both 10 and 60 min.
The changes in V. coralliilyticus gene expression following exposure to coral mucus occurred rapidly and on the same timescales as the chemokinetic responses. Cells exposed to mucus (10 min) displayed the largest transcriptional shift in the principal component analysis (PCA) space (first two principal components captured 88.3% of the variance; Fig. 3b). Replicate libraries possessed small sample-to-sample distances (Supplementary Fig. 8) and clustered tightly together in the PCA space (Fig. 3b), indicating little inter-replicate variance. Control and mucus cells at 10 min shared similar transcriptional shifts along the second component (PC2, 15.9%, Fig. 3b), suggesting that the transcriptional changes in control cells were also present in mucus cells. After 60 min of mucus incubation, gene expression occupied a distinct space on PC2 in comparison with other time points, indicating a potential physiological switch that requires the expression of a different group of genes compared to the early time point (10 min). Taken together, our results reveal a rapid (within 10 min) transcriptional response of the pathogen to coral mucus that mirrors the timescales of its behavioral changes.
Given the strong increase in swimming speed observed by video microscopy within the first 10 min of mucus exposure (Fig. 2), we searched for potential mechanisms underpinning the chemokinesis behavior in the transcriptome (Supplementary Tables 4, 5). Our results from differential expression analysis (DESeq2) revealed the upregulation of all six genes of the Na+-translocating NADH:ubiquinone oxidoreductase (Na+-NQR) enzyme in coral mucus compared to controls at the 10 min time point only (Fig. 4a, Supplementary Table 6). The Na+-NQR enzyme participates in the respiratory electron transport chain and is responsible for generating a sodium motive force that drives flagellar rotation in Vibrios [35, 36]. Although the rapid onset (within 2 min) of chemokinesis suggests that this behavior is not entirely a result of changes in gene expression (bacterial protein production typically takes 10 min or longer [37]), the increase in Na+-NQR enzyme production may have enabled the sustained chemokinesis observed over the duration of our experiments.
Log2-transformed fold difference between mucus and control (log2 FD) and their adjusted p values were determined using DESeq2. a Na+-NQR genes nqrA–F were identified through homology with nqr genes of Vibrio alginolyticus (Supplementary Table 6). b Vibrio biofilm genes in the vps and rbm gene clusters were identified through homology with V. cholerae genes (Supplementary Table 7). Nqr genes (nqrA–F) (a) and vps genes (vpsU, vpsA–P, vpsR, vpsT) (b) are labeled with their respective suffix letters. Lengths of arrows are proportional to protein size (scale bar; aa = amino acids). c–e Log2 FD of quorum-sensing autoinducer synthase genes (c), Vibrio master transcription regulators (d), and toxins (e) at 10 and 60 min. Homology was found with genes in other Vibrios (Supplementary Table 8). Fold difference before addition was assumed to be 1:1 (mucus:control, log2 FD = 0). Error bars represent standard error estimates for the log2 FD values. f Heatmap showing differential expression of secretion system genes identified through KEGG pathway assignments (Supplementary Table 10). The V. coralliilyticus genome has two sets of type 3 secretion genes [34]. CPI-1, Coralliilyticus Pathogenicity Island-1. Colors (a, b, f) indicate log2 FD values. Asterisks (a, b, f), black outlines of arrows (a–b), and filled circles (c–e) mark genes with significant differential expression (adjusted p < 0.05). Open circles, not significant.
Surprisingly, chemotaxis genes and flagella genes were downregulated in coral mucus compared to controls at 10 min (Fig. 3c, Supplementary Fig. 9), despite the strong increase in swimming speed observed by video microscopy at the same time point (Fig. 2). Gene Set Enrichment Analysis (GSEA) revealed that chemotaxis (KEGG 02030) and flagellar assembly (KEGG 02040) gene sets were downregulated within 10 min of incubation with coral mucus (Fig. 3c, Supplementary Fig. 9). All 17 chemotaxis che genes (cheA, cheB, cheD, cheR, cheV, cheW, cheY, cheZ) were downregulated, except cheX (EEX30576) (Supplementary Fig. 9). The majority of the 50 methyl-accepting chemotaxis protein (MCP) genes encoded in the V. coralliilyticus genome were differentially expressed, with 26 downregulated and only 15 upregulated (Supplementary Fig. 9). None of the 74 flagellar assembly genes (KEGG 02040 plus four additional genes found manually) were upregulated in coral mucus at 10 min, with all 31 differentially expressed genes downregulated (adjusted p < 0.05; Supplementary Fig. 9). Taken together, these patterns of gene expression led us to speculate that coral mucus, while inducing a rapid and strong response of chemokinesis, may also provide a cue for the transition from a motile to a non-motile (e.g., surface-associated) lifestyle.
V. coralliilyticus growth was observed in coral mucus (Supplementary Fig. 10), which, similar to other animal mucus, is rich in sugars, lipids, peptides, amino acids (especially serine and threonine), and sulfur compounds [38,39,40,41,42]. Metabolic pathways involved in the catabolism of natural constituents of mucus were enriched in transcripts in mucus-treated cells at 10 min (Fig. 3c). Enriched pathways included metabolism of starch and sucrose (KEGG 00500), glycerophospholipid (KEGG 00564), glycine, serine and threonine (KEGG 00260), sulfur compounds (KEGG 00920), and fatty acids (KEGG 00071) (Fig. 3c, Supplementary Discussion). On the other hand, V. coralliilyticus transcriptomes after mucus treatment (10 min) were depleted in functions involved in the assimilation of inorganic nitrogen (nitrate and nitrite reductases in KEGG 00910; Fig. 3c), which may suggest a metabolic switch toward the utilization of mucus-derived organic nitrogen (e.g., amino acids and peptides, [39]). Almost all ribosomal protein genes (KEGG 03010) and aminoacyl-tRNA synthetases (KEGG 00970) were upregulated in coral mucus compared to controls at 10 min (Fig. 3c, Supplementary Fig. 11), which is indicative of elevated metabolism. Similarly, markers of cell growth, ftsZ and rpoD (EEX34708 and EEX34866; Supplementary Fig. 12) were upregulated in V. coralliilyticus cells exposed to coral mucus. This suite of changes suggests that upon exposure to the nutrient-rich coral mucus, V. coralliilyticus rapidly (within 10 min) and substantially alters its transcriptome to increase metabolism, protein production, and growth.
Upregulation of functions associated with quorum sensing (KEGG 02024) and biofilm formation (KEGG 05111) was observed in V. coralliilyticus cells exposed to coral mucus. Upregulated genes included those involved in quorum-sensing signal molecule production (luxM, luxS) and regulation (aphA, vcpR), as well as biofilm-related polysaccharide production (vps and rbm genes) (Fig. 4b–d, Supplementary Tables 7, 8). The V. coralliilyticus genes responsible for the production of different quorum-sensing autoinducer molecules (AI-1, AI-2, CAI-1) [34] displayed diverse but significant responses, with luxM (AI-1; EEX31502) upregulated and cqsA (CAI-1; EEX33462) downregulated in mucus at both 10 and 60 min relative to controls at the same time points (Fig. 4c). In contrast, luxS (AI-2; EEX35562) was first downregulated at 10 min (0.35×) and subsequently upregulated at 60 min (1.8×) in mucus relative to controls (Fig. 4c). Furthermore, the quorum-sensing master transcription regulators, aphA (EEX30687) and vcpR (EEX34823; homologous to luxR of V. harveyi and hapR of V. cholerae [43]), were up- and downregulated, respectively, which is consistent with their reciprocal behavior in which AphA represses vcpR expression [44, 45] (Fig. 4d). At 10 min in coral mucus, aphA was upregulated (2.8×) while vcpR was downregulated (0.5×) compared to controls (Fig. 4d). In Vibrios, quorum sensing is tightly coupled with biofilm formation [44, 46,47,48]. Indeed, the majority of the 18 vps (Vibrio polysaccharide) and 5 rbm (rugosity and biofilm structure modulator) genes, which are essential for biofilm formation in V. cholerae [49,50,51,52], were upregulated in coral mucus at 10 min (Fig. 4b). These early-onset expression changes of specific genes involved in quorum sensing and biofilm formation were followed by the significant upregulation of their associated pathways (39 quorum sensing genes in KEGG 02024; 37 biofilm formation genes in KEGG 05111) at 60 min (Fig. 3d). Taken together, these results suggest that upon exposure to coral mucus, V. coralliilyticus initiates a gene expression program for biofilm formation that may be involved in host colonization.
Virulence factors characteristic of Vibrio pathogens were amongst the most strongly and significantly upregulated genes in V. coralliilyticus incubated in coral mucus (Supplementary Figs. 13, 14). The master regulator of virulence, ToxR (EEX35320), and its associated stabilizer [53, 54], ToxS (EEX35319), were upregulated in coral mucus compared to controls at both the 10- and 60-min time points (Fig. 4d). Concurrently, several toxin genes were upregulated in coral mucus compared to controls at both time points (Fig. 4e). The important V. coralliilyticus virulence factor, VcpB zinc metalloprotease (EEX32371), was one of the most strongly and significantly upregulated genes in coral mucus at both 10 min (8.6×; Supplementary Fig. 13) and 60 min (53.5×; Supplementary Fig. 14). In addition, the VchA hemolysin (EEX31069) and the associated putative chaperone VchB (EEX31068), which are homologs of the primary virulence factors for Vibrio vulnificus [55, 56], were significantly upregulated in coral mucus at 60 min (3.9× and 11.8×, respectively) (Fig. 4e, Supplementary Fig. 14). Furthermore, the upregulation of other zinc metalloproteases (Supplementary Fig. 15) suggests the existence of multiple, as yet uncharacterized, zinc metalloproteases available to V. coralliilyticus (Supplementary Discussion, Supplementary Table 9). Together, these virulence factors and toxins may be responsible for the tissue lysis of corals previously observed during V. coralliilyticus BAA-450 infection at elevated temperatures [8].
The bacterial secretion system (KEGG 03070) and protein export (KEGG 03060) gene sets were significantly upregulated in coral mucus at 60 min (Fig. 3d), suggesting elevated secretion of proteins. In particular, types 2, 6 (T2SS, T6SS) and Sec secretion system genes were collectively upregulated in coral mucus (Fig. 4f, Supplementary Table 10). Indeed, vipB (EEX32048), which encodes an essential component of T6SS [57], and sec genes were amongst the most significantly and highly upregulated genes in coral mucus at 60 min (Supplementary Fig. 14, Supplementary Table 5). In addition, the upregulation of β-lactam resistance (KEGG 01501; Fig. 3d), as well as several of the multidrug-resistance efflux pump (vex) genes (Supplementary Fig. 16), may confer resistance against antibiotic compounds produced by commensal bacteria within coral mucus [58,59,60,61,62,63]. Taken together, these results suggest that exposure to coral mucus induces V. coralliilyticus to upregulate toxin production, secretion and antibiotic resistance genes, which may be important for host damage and for defense and competition against the commensal microbiome during host colonization.
Discussion
We have reported a rapid behavioral and transcriptional response of V. coralliilyticus to coral mucus exposure, which led to a two-fold increase in swimming speed and significant differential expression of 53% of the genes in the genome within 10 min. Our findings identify coral mucus as a potential chemical signal that induces pathogens to prepare for host colonization and infection. These responses are in line with the behavioral and physiological versatility characteristic of marine copiotrophic bacteria, which are often adapted to boom and bust lifestyles [64, 65]. Yet the extent and the rapidity of the responses observed here suggest that temporally precise orchestration of behavior and gene expression is important for coral host colonization by V. coralliilyticus.
Chemokinesis in response to exposure to coral mucus is potentially a strategy for V. coralliilyticus to seize a limited window of opportunity to reach the coral surface. By increasing swimming speed, bacteria also enhance their chemotactic velocity, leading to a decrease in the time required to follow a chemical gradient to its source. This was previously shown for V. coralliilyticus using microfluidic gradient experiments [9, 10] and appears to be a more general feature of Vibrios, having also been observed in V. alginolyticus chemotaxing toward amino acids [13]. While swimming fast is expensive in the typically dilute ocean environment [66], energy is no longer limiting once nutrient-rich mucus is available. Instead, what is limiting is the window of time during which bacteria can exploit that mucus signal to reach the host. Not only can ambient water currents transport bacteria past the coral surface, but intense vortical flows produced by the corals themselves through cilia on their surfaces—moving at speeds much greater than bacterial swimming speeds—can result in rapid alternation of transport toward and away from the coral surface [25]. In this hydrodynamic environment, the colonization of a host by a bacterial pathogen is a challenging behavioral feat, where the opportunity to home in and attach to the coral surface may only last minutes or even less. The rapid behavioral response we reported here is consistent with this dynamic environment. In particular, the strong chemokinesis—where bacteria doubled their speed—is consistent with the need to reduce the time required to migrate to the coral surface once the detection of mucus indicates the presence of a coral. Furthermore, we observed that chemokinesis in response to coral mucus was almost entirely absent at a temperature at which V. coralliilyticus is avirulent (18.7 °C), which is consistent with the temperature dependence of chemokinesis observed in our previous study [10]. Thus, we propose that chemokinesis is a virulence trait that is important for successful host colonization by bacterial pathogens in the dynamic host surface environment.
Entry into coral mucus represents a dramatic change in nutrient exposure for V. coralliilyticus compared to the oligotrophic reef waters. Accordingly, V. coralliilyticus rapidly upregulated metabolic pathways of nutrients that are present in coral mucus, which may fuel the energetically expensive chemokinesis trait, as well as protein production (ribosome and tRNA biosynthesis) and cell growth (ftsZ and rpoD) genes that may enable rapid proliferation and confer a competitive advantage to the pathogens as they invade the coral host microbiome [67, 68]. Chemokinesis upon homogeneous addition of nutrients has been observed in other bacteria including Rhodobacter sphaeroides [69], Escherichia coli [70] and Azospirillum brasilense [71], and it has been speculated that this swimming speed enhancement is mediated by increasing the proton motive force that is responsible for flagellar rotation [12, 71]. In line with this, V. coralliilyticus exposed to coral mucus increased the expression, on a similar timescale as the chemokinesis behavior, of genes encoding the Na+-NQR enzyme, suggesting that regulation of periplasmic sodium levels may help control swimming speed. Thus, we hypothesize that the metabolism of mucus substrate stimulates Na+-NQR activity, which in turn enables sustained chemokinesis. Additional experimental work is required to test this hypothesis.
Despite the ~2× increase in swimming speed observed through video microscopy, flagellar genes were downregulated at the early RNA-seq time point (10 min). The swimming phenotype may thus persist using the existing polar flagellum, while downregulation of flagellar genes may be a strategy to prevent further replenishment of the flagellar apparatus during the transition to a non-motile phase, evidenced by the concurrent upregulation of biofilm genes. This observation has a parallel in the removal and downregulation of flagella observed in pathogens within the human mucosa, where it is speculated to be a strategy to escape immunological detection by the host, since flagella are strong inducers of pro-inflammatory signaling [72]. While corals possess innate and adaptive-like immunity [73], whether a similar dynamic occurs on the coral surface is currently not known.
Following only 10 min of exposure to coral mucus, the master regulator of Vibrio virulence, ToxR, and its associated protein ToxS, were upregulated. ToxR is known to be essential for coral infection by V. coralliilyticus [74, 75], and in other Vibrio pathogens the ToxR regulatory system coordinates the transcription of colonization, motility, and virulence genes in response to environmental conditions [76, 77]. These downstream effects of ToxR were indeed observed in our RNA-seq results. The temporal modulation of quorum-sensing autoinducer molecule (AI-1, AI-2, CAI-1) producers as seen in our RNA-seq data may be a strategy to coordinate metabolic and lifestyle transitions at the population level, as has been observed in Vibrio harveyi [78]. One such lifestyle transition may be biofilm formation, which is tightly regulated by quorum sensing in Vibrio pathogens [47, 79]. Indeed, we observed the upregulation of biofilm-related vps and rbm gene clusters in coral mucus at 10 min. Furthermore, we observed the upregulation of important Vibrio toxins, VcpB zinc metalloprotease, and VchA and VchB hemolysins, in coral mucus at both 10- and 60-min time points. Similarly, several secretion systems were upregulated, including the Sec-dependent and type 2 secretion systems, which are together responsible for extracellular secretion of a broad range of proteins, including toxins and degradative enzymes involved in the pathogenesis of many Gram-negative bacteria [80,81,82]. Type 6 secretion systems have been observed to be responsible for the injection of toxic effector proteins into bacterial cells in antagonistic interactions [83,84,85]. Taken together, the upregulation of toxR and toxS, as well as their downstream gene expression effects, suggest that coral mucus serves as an environmental signal for V. coralliilyticus to activate host colonization and virulence gene expression programs.
The VcpB zinc metalloprotease is a key virulence factor of V. coralliilyticus that causes photoinactivation of coral endosymbionts and coral tissue lesions [7], and its rank as one of the most highly and significantly upregulated genes in our RNA-seq dataset suggests that the bacterium rapidly responded to coral mucus as a cue to initiate its virulence program. However, the second zinc metalloprotease that has been implicated in V. coralliilyticus infections of corals, VcpA (EEX33179) [8], was downregulated in our experiment. The two zinc metalloproteases (VcpA and VcpB) may thus play redundant roles in V. coralliilyticus infections and may be important in different environmental contexts (Supplementary Discussion).
Our results underscore the rapidity of behavioral and transcriptional changes that occur in a coral pathogen upon entry into the host environment (Fig. 5). These changes in swimming and gene expression patterns paint a clear sequence of events immediately preceding infection—although further validation with direct phenotypic evidence is required. Upon exposure to coral mucus, the coral pathogen V. coralliilyticus (known to chemotax towards coral mucus [9]) increases swimming speed by up to two-fold within minutes, a response that, in the natural environment, would lead to faster chemotaxis and a halving of the time required for the pathogen to track the coral surface from which the mucus signal originates. This capacity to rapidly chemotax into the coral surface microenvironment is important because of the short window of opportunity available to pathogens in the hydrodynamic environment surrounding corals. Simultaneously, transcriptional changes indicate that mucus exposure immediately prompts V. coralliilyticus to increase nutrient metabolism and prepare for host colonization and damage. The downregulation of motility genes, puzzling at first in view of the strong chemokinetic response, is in fact consistent with the upregulation of quorum sensing and biofilm formation genes, together suggesting a “final dash” to the coral surface enabled by enhanced swimming speed, followed by a rapid transition to a non-motile, coral surface-associated lifestyle. The upregulation of metabolism, growth, and antibiotic resistance genes suggests that the pathogen takes advantage of mucus as an energy source, and prepares to colonize the coral surface and compete with commensal bacteria. The upregulation of host damage genes and secretion systems responsible for toxin export suggests preparation for the infection process itself.
Precise temporal control of pathogenesis is a hallmark of Vibrio pathogens [86, 87], which are capable of rapidly modulating their lifestyle between free-swimming and biofilm phases in response to their environment, in particular temperature changes [34, 86, 88]. The frequency of acute temperature-rise in reef waters is increasing [89], giving additional opportunities for temperature-dependent bacterial pathogens, such as V. coralliilyticus, to infect corals [90, 91]. In this context, understanding the mechanisms underlying the earliest stages of bacterial infections is critical in anticipating future disease outbreaks and curbing coral mortality to protect the ecosystems that they support.
Our results suggest that exposure to coral mucus triggers a suite of behavioral and transcriptional responses in V. coralliilyticus leading up to infection. Within 2 min of coral mucus exposure, V. coralliilyticus induces strong chemokinesis, which, coupled with chemotaxis [9, 10], allows the pathogens to reach the coral surface faster. Also early upon coral mucus exposure, upregulation of genes for metabolism of mucus components, biofilm formation, quorum sensing, and antibiotic resistance, and downregulation of flagella- and chemotaxis-related genes, enable host colonization and competition with commensal bacteria. Toxin genes (zinc metalloproteases and hemolysins; yellow stars) and secretion system genes are upregulated in coral mucus, which may lead to host tissue and symbiont damage. Solid arrows indicate bacterial responses for which we have direct observational evidence; dotted arrows indicate hypothesized phenomena based on our RNA-seq data. Figure adapted from Garren et al., 2014 [9].
Data availability
The data that support the findings of this study are available from the corresponding author on request (total data size ~2 TB). Raw, filtered sequencing data reported in this paper have been deposited in the Sequence Read Archive (accession PRJNA707316).
Code availability
All computer code (in MATLAB and R) developed for this study is available from the corresponding author on request.
References
De’Ath G, Fabricius KE, Sweatman H, Puotinen M. The 27-year decline of coral cover on the Great Barrier Reef and its causes. Proc Natl Acad Sci U.S.A. 2012;109:17995–9.
Randall CJ, van Woesik R. Contemporary white-band disease in Caribbean corals driven by climate change. Nat Clim Chang. 2015;5:375–9.
Maynard J, van Hooidonk R, Eakin CM, Puotinen M, Garren M, Williams G, et al. Projections of climate conditions that increase coral disease susceptibility and pathogen abundance and virulence. Nat Clim Chang. 2015;5:688–95.
Cziesielski MJ, Schmidt-Roach S, Aranda M. The past, present, and future of coral heat stress studies. Ecol Evol. 2019;9:10055–66.
Bourne D, Iida Y, Uthicke S, Smith-Keune C. Changes in coral-associated microbial communities during a bleaching event. ISME J. 2008;2:350–63.
van de Water JAJM, Chaib De Mares M, Dixon GB, Raina JB, Willis BL, Bourne DG, et al. Antimicrobial and stress responses to increased temperature and bacterial pathogen challenge in the holobiont of a reef-building coral. Mol Ecol. 2018;27:1065–80.
Sussman M, Mieog JC, Doyle J, Victor S, Willis BL, Bourne DG. Vibrio zinc-metalloprotease causes photoinactivation of coral endosymbionts and coral tissue lesions. PLoS ONE. 2009;4:1–14.
Ben-Haim Y, Zicherman-Keren M, Rosenberg E. Temperature-regulated bleaching and lysis of the coral Pocillopora damicornis by the novel pathogen Vibrio coralliilyticus. Appl Environ Microbiol. 2003;69:4236–41.
Garren M, Son K, Raina J-B, Rusconi R, Menolascina F, Shapiro OH, et al. A bacterial pathogen uses dimethylsulfoniopropionate as a cue to target heat-stressed corals. ISME J. 2014;8:999–1007.
Garren M, Son K, Tout J, Seymour JR, Stocker R. Temperature-induced behavioral switches in a bacterial coral pathogen. ISME J. 2016;10:1363–72.
Barbara GM, Mitchell JG. Marine bacterial organisation around point-like sources of amino acids. FEMS Microbiol Ecol. 2003;43:99–109.
Seymour JR, Marcos, Stocker R. Resource patch formation and exploitation throughout the marine microbial food web. Am Nat. 2009;173:E15–29.
Son K, Menolascina F, Stocker R. Speed-dependent chemotactic precision in marine bacteria. Proc Natl Acad Sci U.S.A. 2016;113:8624–9.
Meron D, Efrony R, Johnson WR, Schaefer AL, Morris PJ, Rosenberg E, et al. Role of Flagella in virulence of the coral pathogen Vibrio coralliilyticus. Appl Environ Microbiol. 2009;75:5704–7.
Ushijima B, Häse CC. Influence of chemotaxis and swimming patterns on the virulence of the coral pathogen Vibrio coralliilyticus. J Bacteriol. 2018;200:1–16.
Crossland CJ, Barnes DJ, Borowitzka MA. Diurnal lipid and mucus production in the staghorn coral Acropora acuminata. Mar Biol. 1980;60:81–90.
Davies PS. The role of zooxanthellae in the nutritional energy requirements of Pocillopora eydouxi. Coral Reefs. 1984;2:181–6.
Rix L, de Goeij JM, Mueller CE, Struck U, Middelburg JJ, van Duyl FC, et al. Coral mucus fuels the sponge loop in warm-and cold-water coral reef ecosystems. Sci Rep. 2016;6:1–11.
Naumann MS, Haas A, Struck U, Mayr C, El-Zibdah M, Wild C. Organic matter release by dominant hermatypic corals of the Northern Red Sea. Coral Reefs. 2010;29:649–59.
Wild C, Huettel M, Klueter A, Kremb SG, Rasheed MYM, Jørgensen BB. Coral mucus functions as an energy carrier and particle trap in the reef ecosystem. Nature. 2004;428:66–70.
Bythell JC, Wild C. Biology and ecology of coral mucus release. J Exp Mar Bio Ecol. 2011;408:88–93.
Bakshani CR, Morales-Garcia AL, Althaus M, Wilcox MD, Pearson JP, Bythell JC, et al. Evolutionary conservation of the antimicrobial function of mucus: a first defence against infection. NPJ Biofilms Microbiomes. 2018;14:1–12.
Gibbin E, Gavish A, Krueger T, Kramarsky-Winter E, Shapiro O, Guiet R, et al. Vibrio coralliilyticus infection triggers a behavioural response and perturbs nutritional exchange and tissue integrity in a symbiotic coral. ISME J. 2019;13:989–1003.
Gavish AR, Shapiro OH, Kramarsky-Winter E, Vardi A. Microscale tracking of coral-vibrio interactions. ISME Communications. 2021;1:1–18.
Shapiro OH, Fernandez VI, Garren M, Guasto JS, Debaillon-Vesque FP, Kramarsky-Winter E, et al. Vortical ciliary flows actively enhance mass transport in reef corals. Proc Natl Acad Sci U.S.A. 2014;111:13391–6.
Seymour JR, Ahmed T, Stocker R. A microfluidic chemotaxis assay to study microbial behavior in diffusing nutrient patches. Limnol Oceanogr Methods. 2008;6:477–88.
Penn K, Wang J, Fernando SC, Thompson JR. Secondary metabolite gene expression and interplay of bacterial functions in a tropical freshwater cyanobacterial bloom. ISME J. 2014;8:1866–78.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:1–21.
Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:1–12.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U.S.A. 2005;102:15545–50.
Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehar J, et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–73.
Schneider WR, Doetsch RN. Effect of viscosity on bacterial motility. J Bacteriol. 1974;117:696–701.
Martinez VA, Schwarz-Linek J, Reufer M, Wilson LG, Morozov AN, Poon WCK. Flagellated bacterial motility in polymer solutions. Proc Natl Acad Sci U.S.A. 2014;111:17771–6.
Kimes NE, Grim CJ, Johnson WR, Hasan NA, Tall BD, Kothary MH, et al. Temperature regulation of virulence factors in the pathogen Vibrio coralliilyticus. ISME J. 2012;6:835–46.
Kojima S, Yamamoto K, Kawagishi I, Homma M. The polar flagellar motor of Vibrio cholerae is driven by an Na+ motive force. J Bacteriol. 1999;181:1927–30.
Sowa Y, Hotta H, Homma M, Ishijima A. Torque-speed relationship of the Na+-driven flagellar motor of Vibrio alginolyticus. J Mol Biol. 2003;327:1043–51.
Milo R, Phillips R. Cell biology by the numbers. 1st ed. New York, NY: Garland Science; 2016.
Crossland CJ. In situ release of mucus and DOC-lipid from the corals Acropora variabilis and Stylophora pistillata in different light regimes. Coral Reefs. 1987;6:35–42.
Wild C, Woyt H, Huettel M. Influence of coral mucus on nutrient fluxes in carbonate sands. Mar Ecol Prog Ser. 2005;287:87–98.
Ducklow HW, Mitchell R. Composition of mucus released by coral reef coelenterates. Limnol Oceanogr. 1979;24:706–14.
Meikle P, Richards GN, Yellowlees D. Structural determination of the oligosaccharide side chains from a glycoprotein isolated from the mucus of the coral Acropora formosa. J Biol Chem. 1987;262:16941–7.
Coddeville B, Maes E, Ferrier-Pagès C, Guerardel Y. Glycan profiling of gel forming mucus layer from the scleractinian symbiotic coral Oculina arbuscula. Biomacromolecules. 2011;12:2064–73.
Hasegawa H, Häse CC. TetR-type transcriptional regulator VtpR functions as a global regulator in Vibrio tubiashii. Appl Environ Microbiol. 2009;75:7602–9.
Ball AS, Chaparian RR, van Kessel JC. Quorum sensing gene regulation by LuxR/HapR master regulators in Vibrios. J Bacteriol. 2017;199:1–13.
Rutherford ST, Van Kessel JC, Shao Y, Bassler BL. AphA and LuxR/HapR reciprocally control quorum sensing in vibrios. Genes Dev. 2011;25:397–408.
Hammer BK, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol Microbiol. 2003;50:101–4.
Waters CM, Lu W, Rabinowitz JD, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic Di-GMP levels and repression of vpsT. J Bacteriol. 2008;190:2527–36.
Burger AH. Quorum Sensing in the Hawai’ian Coral Pathogen Vibrio coralliilyticus strain OCN008. University of Hawaii at Manoa; 2017.
Yildiz FH, Schoolnik GK. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc Natl Acad Sci U.S.A. 1999;96:4028–33.
Fong JCN, Syed KA, Klose KE, Yildiz FH. Role of Vibrio polysaccharide (vps) genes in VPS production, biofilm formation and Vibrio cholerae pathogenesis. Microbiology. 2010;156:2757–69.
Fong JCN, Karplus K, Schoolnik GK, Yildiz FH. Identification and characterization of RbmA, a novel protein required for the development of rugose colony morphology and biofilm structure in Vibrio cholerae. J Bacteriol. 2006;188:1049–59.
Fong JCN, Yildiz FH. The rbmBCDEF gene cluster modulates development of rugose colony morphology and biofilm formation in Vibrio cholerae. J Bacteriol. 2007;189:2319–30.
DiRita VJ, Mekalanos JJ. Periplasmic interaction between two membrane regulatory proteins, ToxR and ToxS, results in signal transduction and transcriptional activation. Cell. 1991;64:29–37.
Almagro-Moreno S, Root MZ, Taylor RK. Role of ToxS in the proteolytic cascade of virulence regulator ToxR in Vibrio cholerae. Mol Microbiol. 2015;98:963–76.
Lee SE, Ryu PY, Kim SY, Kim YR, Koh JT, Kim OJ, et al. Production of Vibrio vulnificus hemolysin in vivo and its pathogenic significance. Biochem Biophys Res Commun. 2004;324:86–91.
Senoh M, Okita Y, Shinoda S, Miyoshi S. The crucial amino acid residue related to inactivation of Vibrio vulnificus hemolysin. Micro Pathog. 2008;44:78–83.
Bröms JE, Ishikawa T, Wai SN, Sjöstedt A. A functional VipA-VipB interaction is required for the type VI secretion system activity of Vibrio cholerae O1 strain A1552. BMC Microbiol. 2013;13:1–12.
Vizcaino MI, Johnson WR, Kimes NE, Williams K, Torralba M, Nelson KE, et al. Antimicrobial resistance of the coral pathogen Vibrio coralliilyticus and Caribbean sister phylotypes isolated from a diseased octocoral. Micro Ecol. 2010;59:646–57.
Ritchie KB. Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar Ecol Prog Ser. 2006;322:1–14.
Nissimov J, Rosenberg E, Munn CB. Antimicrobial properties of resident coral mucus bacteria of Oculina patagonica. FEMS Microbiol Lett. 2009;292:210–5.
Shnit-Orland M, Kushmaro A. Coral mucus-associated bacteria: a possible first line of defense. FEMS Microbiol Ecol. 2009;67:371–80.
Rypien KL, Ward JR, Azam F. Antagonistic interactions among coral-associated bacteria. Environ Microbiol. 2010;12:28–39.
Alagely A, Krediet CJ, Ritchie KB, Teplitski M. Signaling-mediated cross-talk modulates swarming and biofilm formation in a coral pathogen Serratia marcescens. ISME J. 2011;5:1609–20.
Stocker R, Seymour JR, Samadani A, Hunt DE, Polz MF. Rapid chemotactic response enables marine bacteria to exploit ephemeral microscale nutrient patches. Proc Natl Acad Sci U.S.A. 2008;105:4209–14.
Polz MF, Hunt DE, Preheim SP, Weinreich DM. Patterns and mechanisms of genetic and phenotypic differentiation in marine microbes. Philos Trans R Soc B Biol Sci. 2006;361:2009–21.
Taylor JR, Stocker R. Trade-offs of chemotactic foraging in turbulent water. Science. 2012;338:675–9.
Krediet CJ, Ritchie KB, Cohen M, Lipp EK, Patterson Sutherland K, Teplitski M. Utilization of mucus from the coral Acropora palmata by the pathogen Serratia marcescens and by environmental and coral commensal bacteria. Appl Environ Microbiol. 2009;75:3851–8.
Krediet CJ, Ritchie KB, Alagely A, Teplitski M. Members of native coral microbiota inhibit glycosidases and thwart colonization of coral mucus by an opportunistic pathogen. ISME J. 2013;7:980–90.
Packer HL, Armitage JP. The chemokinetic and chemotactic behavior of Rhodobacter sphaeroides: two independent responses. J Bacteriol. 1994;176:206–12.
Deepika D, Karmakar R, Tirumkudulu MS, Venkatesh KV. Variation in swimming speed of Escherichia coli in response to attractant. Arch Microbiol. 2015;197:211–22.
Zhulin IB, Armitage JP. Motility, chemokinesis, and methylation-independent chemotaxis in Azospirillum brasilense. J Bacteriol. 1993;175:952–8.
Ramos HC, Rumbo M, Sirard J-C. Bacterial flagellins: mediators of pathogenicity and host immune responses in mucosa. Trends Microbiol. 2004;12:509–17.
Reed KC, Muller EM, van Woesik R. Coral immunology and resistance to disease. Dis Aquat Organ. 2010;90:85–92.
Ushijima B, Videau P, Poscablo D, Stengel JW, Beurmann S, Burger AH, et al. Mutation of the toxR or mshA genes from Vibrio coralliilyticus strain OCN014 reduces infection of the coral Acropora cytherea. Environ Microbiol. 2016;18:4055–67.
Ushijima B, Richards GP, Watson MA, Schubiger CB, Häse CC. Factors affecting infection of corals and larval oysters by Vibrio coralliilyticus. PLoS ONE. 2018;13:e0199475.
Peterson KM, Mekalanos JJ. Characterization of the Vibrio cholerae ToxR regulon: identification of novel genes involved in intestinal colonization. Infect Immun. 1988;56:2822–9.
Provenzano D, Klose KE. Altered expression of the ToxR-regulated porins OmpU and OmpT diminishes Vibrio cholerae bile resistance, virulence factor expression, and intestinal colonization. Proc Natl Acad Sci U.S.A. 2000;97:10220–4.
Waters CM, Bassler BL. The Vibrio harveyi quorum-sensing system uses shared regulatory components to discriminate between multiple autoinducers. Genes Dev. 2006;20:2754–67.
Mukherjee S, Bassler BL. Bacterial quorum sensing in complex and dynamically changing environments. Nat Rev Microbiol. 2019;17:371–82.
Sikora AE, Zielke RA, Lawrence DA, Andrews PC, Sandkvist M. Proteomic analysis of the Vibrio cholerae type II secretome reveals new proteins, including three related serine proteases. J Biol Chem. 2011;286:16555–66.
Korotkov KV, Sandkvist M, Hol WGJ. The type II secretion system: biogenesis, molecular architecture and mechanism. Nat Rev Microbiol. 2012;10:336–51.
Stathopoulos C, Hendrixson DR, Thanassi DG, Hultgren SJ, St. Geme III JW, Curtiss III R. Secretion of virulence determinants by the general secretory pathway in Gram-negative pathogens: an evolving story. Microbes Infect. 2000;2:1061–72.
Hood RD, Singh P, Hsu FS, Güvener T, Carl MA, Trinidad RRS, et al. A Type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe. 2010;7:25–37.
Zheng J, Ho B, Mekalanos JJ. Genetic analysis of anti-amoebae and anti-bacterial activities of the Type VI secretion system in Vibrio cholerae. PLoS ONE. 2011;6:e23876.
MacIntyre DL, Miyata ST, Kitaoka M, Pukatzki S. The Vibrio cholerae type VI secretion system displays antimicrobial properties. Proc Natl Acad Sci U.S.A. 2010;107:19520–4.
Lee SH, Hava DL, Waldor MK, Camilli A. Regulation and temporal expression patterns of Vibrio cholerae virulence genes during infection. Cell. 1999;99:625–34.
Pennetzdorfer N, Lembke M, Pressler K, Matson JS, Reidl J, Schild S. Regulated proteolysis in Vibrio cholerae allowing rapid adaptation to stress conditions. Front Cell Infect Microbiol. 2019;9:1–9.
Liu R, Chen H, Zhang R, Zhou Z, Hou Z, Gao D, et al. Comparative transcriptome analysis of Vibrio splendidus JZ6 reveals the mechanism of its pathogenicity at low temperatures. Appl Environ Microbiol. 2016;82:2050–61.
Hughes TP, Anderson KD, Connolly SR, Heron SF, Kerry JT, Lough JM, et al. Spatial and temporal patterns of mass bleaching of corals in the Anthropocene. Science. 2018;359:80–3.
Vezzulli L, Previati M, Pruzzo C, Marchese A, Bourne DG, Cerrano C, et al. Vibrio infections triggering mass mortality events in a warming Mediterranean Sea. Environ Microbiol. 2010;12:2007–19.
Zaneveld JR, Burkepile DE, Shantz AA, Pritchard CE, McMinds R, Payet JP, et al. Overfishing and nutrient pollution interact with temperature to disrupt coral reefs down to microbial scales. Nat Commun. 2016;7:1–12.
Acknowledgements
This research was funded by a Gordon and Betty Moore Foundation Marine Microbiology Initiative Investigator Award (GBMF3783) and Symbiosis in Aquatic Systems Initiative Investigator Award (GBMF9197) to R.S., a U.S. Department of Energy Joint Genome Institute Emerging Technologies Opportunity grant (DE-AC02-05CH11231) to R.S., a grant from the Simons Foundation through the Principles of Microbial Ecosystems (PriME) collaboration (542395) to R.S., a National Science Foundation (NSF) Graduate Research Fellowship (Grant No. 1122374) to C.G, and an NSF Postdoctoral Research Fellowship in Biology (Grant No. DBI-1202865) to K.P. The authors thank Dr. K.M. Wheeler (MIT), Dr. B. Ushijima (Smithsonian Marine Station), C. McLean (MIT/WHOI), Dr. U. Pfreundt (ETH Zurich), Dr. C. Martinez-Pérez (ETH Zurich), Prof. C.S. Smillie (Broad Institute/MGH), and Eric Hill (NU Singapore) for discussions on RNA-seq analyses and interpretations; Dr. T. Kassis (MIT) for providing computational resources; Prof. J.S. Guasto (Tufts University) for insights and scripts on particle tracking; Prof. D.A. Lauffenburger (MIT) and Prof. K. Ribbeck (MIT) for scientific discussions; and Dr. R. Naisbit for editing this paper.
Funding
Open Access funding provided by ETH Zurich.
Author information
Authors and Affiliations
Contributions
CG, MG, KP, JRT, and RS designed the study. CG, MG, and KP performed experiments. JBR collected coral mucus. CG, VIF, and RS created the video analysis software. CG performed RNA-seq data analyses. JRS and JBR provided conceptual guidance. CG, JBR, and RS wrote the paper. All authors edited the paper before submission.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gao, C., Garren, M., Penn, K. et al. Coral mucus rapidly induces chemokinesis and genome-wide transcriptional shifts toward early pathogenesis in a bacterial coral pathogen. ISME J 15, 3668–3682 (2021). https://doi.org/10.1038/s41396-021-01024-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-021-01024-7
This article is cited by
-
Coral thermal stress and bleaching enrich and restructure reef microbial communities via altered organic matter exudation
Communications Biology (2024)
-
Ecology of Endozoicomonadaceae in three coral genera across the Pacific Ocean
Nature Communications (2023)
-
Coral holobiont cues prime Endozoicomonas for a symbiotic lifestyle
The ISME Journal (2022)