Abstract
Most microorganisms in the biosphere remain uncultured and poorly characterized. Although the surge in genome sequences has enabled insights into the genetic and metabolic properties of uncultured microorganisms, their physiology and ecological roles cannot be determined without direct probing of their activities in natural habitats. Here we employed an experimental framework coupling genome reconstruction and activity assays to characterize the largely uncultured microorganisms responsible for aerobic biodegradation of biphenyl as a proxy for a large class of environmental pollutants, polychlorinated biphenyls. We used 13C-labeled biphenyl in contaminated soils and traced the flow of pollutant-derived carbon into active cells using single-cell analyses and protein–stable isotope probing. The detection of 13C-enriched proteins linked biphenyl biodegradation to the uncultured Alphaproteobacteria clade UBA11222, which we found to host a distinctive biphenyl dioxygenase gene widely retrieved from contaminated environments. The same approach indicated the capacity of Azoarcus species to oxidize biphenyl and suggested similar metabolic abilities for species of Rugosibacter. Biphenyl oxidation would thus represent formerly unrecognized ecological functions of both genera. The quantitative role of these microorganisms in pollutant degradation was resolved using single-cell-based uptake measurements. Our strategy advances our understanding of microbially mediated biodegradation processes and has general application potential for elucidating the ecological roles of uncultured microorganisms in their natural habitats.
Similar content being viewed by others
Introduction
One of the grand challenges of microbial ecology and ecosystem science is to link microbial community structure to microbial functional traits underpinning key ecological processes [1, 2]. To date, the physiological and ecological functions of most microorganisms in natural habitats remain poorly characterized particularly due to their recalcitrance to cultivation [3, 4]. To some extent, the need to cultivate individual taxa has been overcome by recent advances in environmental metagenomic sequencing and computational approaches. This enabled community-level genome reconstruction from a wide range of ecosystems, shedding new insights into phylogenetic identity and metabolic potential of numerous uncultured lineages [5,6,7,8,9]. Consequently, the physiology and ecological roles of uncultured bacteria and archaea have been lately inferred mostly from reconstructed genomes. However, these genome-driven discoveries relied strongly on the predictive understanding of genes and metabolic pathways, which do not always reflect actual functions carried out by microorganisms in their natural habitats. Thus, it became apparent that experimental evidence to support genome-based predictions by direct detection and quantification of function is needed to properly understand the role of uncultured microorganisms in ecologically significant biogeochemical processes [2, 4, 10, 11].
A prominent, microbially mediated environmental process is the biodegradation of widespread, persistent organic pollutants like polychlorinated biphenyls (PCBs), which have broad consequences for human health and ecosystem functioning [12, 13]. The identity of microorganisms involved in such processes in various environments has been extensively investigated by both culture-dependent and culture-independent methods. The formerly provided insights into the biochemical basis of pollutant activation and downstream transformation in pure cultures [14,15,16,17]. The latter targeted active pollutant-metabolizing microorganisms within natural microbial communities, such as those inhabiting PCB-contaminated soils and sediments, through substrate-specific labeling of their DNA by stable isotope probing (SIP). Sequence analysis of marker genes (i.e., 16S rRNA gene) retrieved from labeled fractions of community DNA revealed the phylogenetic identity of metabolically active populations [18,19,20,21,22,23]. These SIP-based gene surveys indicated that pollutant removal in contaminated environments was mostly achieved by uncultured microorganisms; only a small fraction of these are represented by closely related isolates or genomes [24,25,26]. Thus, there is remarkably little reliable information about the full genetic potential and taxon-specific metabolism of the ecologically relevant degraders. Furthermore, DNA-based SIP provided only a qualitative link between phylogenetic identity and biodegradation activity, whereas direct quantification of metabolic rate and cellular abundance has never been obtained for pollutant degraders in real-world habitats like soils.
Here we selected biphenyl as a model compound to document metabolic activities and genomic makeup of biodegradative microbial populations of PCB-impacted soils. Biphenyl has often been used as a proxy to investigate the biodegradation of PCBs since many PCB congeners can be co-metabolized by biphenyl-oxidizing microorganisms via the same oxidation pathways [27, 28]. The capacity of the soil microbial communities to oxidize biphenyl was initially assessed using cultivation approaches. We then coupled stable isotope probing with nanoscale secondary ion mass spectrometry (nanoSIMS) to image and quantify biphenyl assimilation by individual microbial cells. The phylogenetic identity of biphenyl oxidizers was resolved using metaproteomics-SIP based on metagenomic databases constructed for each soil type investigated. Moreover, for the identified microorganisms, we used the metagenome-assembled genomes (MAGs) to reconstruct the complete metabolic pathways involved in biphenyl oxidization.
Materials and methods
Chemicals
The chemicals used include 13C-biphenyl (99 atom% 13C; Cambridge Isotope Laboratories, Inc., USA), unlabeled biphenyl (Sigma-Aldrich), nuclease-free water (ThermoFisher Scientific), dichloromethane (Sigma-Aldrich), sodium cacodylate buffer (Electron Microscopy Sciences, USA), and Nycodenz (Progen).
Soil samples
The four soil samples investigated in this study were collected from Taizhou, Zhejiang Province, China, and were denoted as soil A (river sediment; 28°32’19”N, 121°22’41”E), soil B (paddy soil; 28°32’28”N, 121°22’15”E), soil C (river sediment; 28°37’13”N, 121°24’54”E), and soil D (upland soil; 28°30’32”N, 121°21’49”E), respectively. The sampling sites were historically contaminated with PCBs at concentrations of 0.03 − 5 mg kg−1 (Supplementary Table 1). The samples were stored at 4 °C until analysis or use for incubation experiments. A diagram summarizing our main experimental workflow is presented in Supplementary Fig. 1.
Establishing biphenyl-degrading enrichment cultures
Incubations with biphenyl were set up in 150 ml conical flasks provided with 30 ml aerobic, mineral freshwater medium [29, 30], and 5 g of soil as inoculum. Crystals of biphenyl (approx. 20 mg per flask) were added as the sole carbon and energy source. The flasks were sealed with cotton stoppers to permit the free passage of oxygen. Incubation was done at 28 °C in the dark with gentle horizontal shaking (120 rpm). Controls without the addition of biphenyl were prepared similarly and incubated under the same conditions. Biphenyl-dependent growth was monitored by observing changes of turbidity and by microscopic examinations. Sediment-free enrichment cultures were obtained after five subsequent transfers into fresh culture media with 10% inoculum each. Enrichment cultures were further maintained by inoculating 10% volume of grown cultures to fresh-water medium every three days. Growth experiments with the enrichment cultures were prepared under the same conditions as described above. Growth was monitored by measuring the OD at λ = 600 nm every four hours by subsampling 0.5 ml of culture. Inoculated medium without biphenyl was used as a control.
DNA extraction and community sequencing
DNA was extracted from 0.5 g soil as described previously [31]. The soil samples were suspended in 1.35 ml extraction buffer (100 mM Tris-HCl, 100 mM sodium EDTA, 100 mM sodium phosphate, 1.5 M NaCl, 1% CTAB, pH 8.0) and lysed via three cycles of freezing and thawing at −196 °C and 37 °C, respectively. The resulting lysates were further incubated with 10 mg ml−1 proteinase K at 37 °C for 30 min and with 20% SDS at 65 °C for 1 h, followed by 10 min centrifugation at 12 °C, 15,000 rpm (21,130×g). The resulting supernatant was extracted with an equal volume of chloroform:isoamyl alcohol (24:1, v/v). Nucleic acids were precipitated with isopropanol (0.6 volume, 1 h at room temperature). The yielded DNA was dissolved in 50 μl nuclease-free water and stored at −20 °C before use. For DNA extraction from biphenyl-degrading enrichments, the same procedure was applied to cells harvested from 20 ml grown cultures. The V3-V5 region of bacterial 16S rRNA genes was amplified using primers 341F (5’-CCTACGGGNGGCWGCAG-3’) and 785 R (5’-GACTACHVGGGTATCTAATCC-3’) and sequenced on MiSeq (Illumina) V3 platform (2 × 300 cycles). After removal of adapters and primers, the paired-end reads were joined using BBMap v38.51 (https://sourceforge.net/projects/bbmap/) and processed using the SilvaNGS pipeline [32, 33]. Operational taxonomic units (OTUs) were clustered at 98% sequence identity level and classified taxonomically using the Silva v.132 databases. The OTU table was downloaded from SilvaNGS and rarefied to minimum sequencing depth across samples (36,000 for soils; 10,000 for enrichments). Alpha diversity indices, including the Shannon, Chao1, Simpson, and Simpson’s evenness measure E, were calculated using the Vegan R package (Supplementary Table 2). Beta diversity was assessed using the capscale function in the Phyloseq R package based on Bray-Curtis dissimilarity (Supplementary Fig. 2).
Stable isotope probing in slurry microcosms
To determine the biphenyl-degrading capacity of the original soil samples, slurry microcosms were established in 20-ml sealed serum vials. To enhance biphenyl bioavailability, unlabeled or 13C-labeled biphenyl was dissolved in dichloromethane (DCM) at a concentration of 20 mg/ml. A volume of 0.1 ml of this solution was added per vial (2 mg net biphenyl per vial); the DCM was thereafter completely evaporated, leaving biphenyl crystals homogeneously distributed on the inner wall of vials. Soil slurries were prepared for each soil type by mixing soil samples with freshwater medium to reach ~75% (v/v) pore-water content. Vials were thereafter supplied with 3 ml slurry, sealed with rubber stoppers, and incubated in the dark at 28 °C with continuous shaking (120 rpm). For each soil type, a total of 14 replicate microcosms were prepared, of which 7 contained unlabeled biphenyl and 7 contained 13C-biphenyl. Two replicates were sacrificed for each soil type and substrate combination, at seven time points (0, 2, 4, 8, 24, 48, and 96 h of incubation). The headspace of the microcosm was collected to determine the carbon isotope composition of CO2 [34]. The slurries were either frozen (−20 °C) for molecular biological analysis or fixed for nanoSIMS analysis. For sample fixation, the sediments were incubated with 3% paraformaldehyde (final concentration) for 1 h at room temperature, washed two times with 1× PBS, and stored in 1× PBS:ethanol (1:1, v/v) at −20 °C.
Soil cell extraction and nanoSIMS analysis
To detach cells from soil particles for subsequent nanoSIMS analyses, we used established protocols based on alternating mild chemicals (using tensioactive compounds) and mechanical treatments [35]. In brief, 200 μl of PFA-fixed sediment was mixed with 700 μl TE buffer (10 mM Tris-HCl, 5 mM EDTA, pH 9.0) and 100 μl pyrophosphate (100 mM), followed by incubation at 55 °C for 5 min (water bath of a histological microwave oven operated at 200 W; Microwave Research and Application Inc., Laurel, MD, USA). The samples were then amended with 1 μl Tween 80 and mixed for 15 min at room temperature on a benchtop vortex (level 3, 1060 rpm, Grant-bio). Cells were thereafter separated from soil particles using established Nycodenz density centrifugation protocols [35]. Each sample was transferred to a 50 ml Falcon tube containing 22.5 ml 1× PBS and 2.5 ml pyrophosphate. A volume of 2 ml Nycodenz solution (1.426 g/ml, 80% w/v) was slowly added to the bottom of the Falcon tube using 10 ml pipette tips to avoid mixing of Nycodenz and slurries. High-speed centrifugation (1.5 h, 16,000×g, 4 °C; ROTINA 380R, Hettich) was applied to separate cells from soil particles. Cell-containing water phase (approx. 25 ml) on the top of the Nycodenz layer was carefully collected in clean 50 ml Falcon tubes.
For nanoSIMS analyses, 10 ml of supernatant recovered from the Nycodenz extraction procedure was filtered on gold-palladium-coated polycarbonate filters (0.22 μm pore size). The samples were rinsed twice with sodium cacodylate buffer (0.2 M, pH 7.4; Electron Microscopy Sciences), dehydrated in an ethanol series prepared in the same buffer (30, 50, 70, 80, 90, 96, and 100%), and dried for 20 exchange cycles using critical point drying machine (Critical Point Dryer, LEICA EM CPD300). Filter pieces of 10 mm in diameter were cut using a punching tool and placed on the sample holder of a nanoSIMS-50L instrument (CAMECA, AMETEK). NanoSIMS analyses were performed in negative extraction mode using Cs+ as the primary ion source. Prior to measurements, 100 × 100 μm2 areas were pre-implanted with 16 keV cesium (Cs+) primary ion beam at 200 pA for 10 min. Fields of view of 25 × 25 μm2 were analyzed with a 3 pA primary ion beam, scanning at 512 × 512-pixel resolution with 2 ms per pixel of dwell time. Secondary ion species (12CH−, 16O−, 12C14N−, 13C14N−, 31P−, 32S−, and 31P16O2−) were collected in parallel. A mass resolving power (M/ΔM) between 8000 and 12,000 was achieved as previously described, with 40 μm exit slits. Look@NanoSIMS software (LANS) was used to process the acquired secondary ion images [36]. More than 25 scans of each field of view were accumulated after lateral drift corrections. Region of interest (RoI) was manually defined for each individual cell along its inner margin based on 12C14N− ion images. The raw ion counts of 13C14N− and 12C14N− were used to calculate the fraction of C assimilated by a single cell (KA). Assimilation rates of carbon (Fc) were derived for individual cells as described previously [37]. The total number of analyzed cells and those showing 13C-enrichment in each soil sample are summarized in Supplementary Table 3.
Metagenomic sequencing and data analysis
DNA extracted from unlabeled soil microcosms after 96 h incubation was sequenced on a MiSeq (Illumina) V3 platform (2×300 cycles), resulting in >2 million paired-end reads for each soil sample (n = 4). The raw reads were quality filtered using Trimmomatic v.0.33 [38] (SLIDINGWINDOW:4:15; MINLEN 36) and assembled into contigs using Spades v3.14.0 [39] with the following the parameters (-meta; -k 21,33,55,77,99,127). Short contigs ≤500 bp were removed. Metagenomic binning was performed using MetaBAT v0.32.4 [40] with -minContig option of 1500. The generated MAGs were further refined using RefineM v0.0.25 [7] based on genomic properties (i.e., tetranucleotide frequency and read coverage) and taxonomic classification. MAGs of interest were further refined by reads mapping, reassembly, and binning. Reads were mapped to the selected MAGs at a minimum alignment identity of 90% using BBMap v.38.51 (minid option). Reassembly and binning followed the same pipeline described above. The quality (completeness and contamination) of each MAG was assessed using CheckM v1.1.2 [41] with the lineage-specific workflow. Taxonomy assignments of MAGs were conducted using GTDB-Tk [42]. The genome tree of bacterial MAGs was inferred from the concatenation of 120 proteins using RAxML v8.2.11 [43] with 100 bootstrap replicates (-m PROTCAMMALG -f a -N 100). For MAGs, protein-coding genes were called using Prodigal [44] (-p single option) and the translated protein sequences were annotated with KEGG [45], Pfam [46], and EggNOG [47] databases. To search for aerobic biphenyl-oxidation pathways in genomes, reference protein sequences from the bph operon of various known biphenyl degraders, including Gram-positive and Gram-negative bacteria (Supplementary Table 4), were queried against MAGs using Blastp [48] (E-value, 1e-20; minimum sequence identity, 30%). The resulting blast hits were manually curated based on sequence length, domain composition, and conserved functional sites. To reconstruct the phylogenetic tree of BphA, protein sequences were aligned using Muscle v3.8.1551 [49], followed by trimming of ambiguous sites in the alignment using trimAl v1.4 [50] with -automated1 option. Maximum likelihood trees were calculated using RAxML with the PROTGAMMALG evolutionary model.
Metaproteomics and protein stable isotope probing
In the protein-SIP experiment, soils incubated with unlabeled and labeled biphenyl (20 atom% 13C) were processed in parallel. Extraction and quantification of proteins from soil microcosms (3 ml) were carried out using NoviPure Soil Protein Kit (QIAGEN) and BCA Protein Assay Kit (QIAGEN), respectively. The extracted proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). For each sample, the corresponding SDS-PAGE gel lane was cut into two pieces. Protein in the gel pieces was reduced with dithionite, carbamidomethylated with iodoacetamide, and digested with trypsin as described by Budhraja et al (2021). Peptides were purified using ZipTip C18-columns (Millipore) and analyzed by an Orbitrap Fusion Tribrid mass spectrometer (Thermo Scientific) equipped with a nanoLC system (Dionex Ultimate 3000RSLC; Thermo Scientific) [8, 51].
The obtained MS/MS spectra were searched against the specific metagenome database (bulk assembly) by XTandem [52] in the OpenMS pipeline [53]. Precursor and fragment ion mass tolerance were set to 5 ppm and 0.02 Da, respectively. Two tryptic miss cleavages were allowed; carbamidomethylation of cysteines was treated as fixed modification and oxidation of methionine as a dynamic modification. Proteins were considered to be identified when at least two unique peptides were recovered with medium confidence (False discovery rate, FDR < 0.05).
Incorporation of 13C was determined for peptides at a high confidence level (FDR < 0.01) using MetaProSIP [54] with a mass window of 10 ppm. Centroid MS1 data, the metagenome database, and feature data annotated with identification (generated by IDMapper in OpenMS) were fed as input for MetaProSIP. A minimum correlation of 0.8 was set as the threshold for reporting 13C-peptides to reduce false positives. Relative isotope abundance (RIA) and labeling ratio (LR) of peptides were retrieved for labeled peptides from the output of MetaProSIP. The isotope pattern of the labeled peptides was further evaluated by comparing the observed mass spectra with its theoretical distributions using Pearson’s chi-squared test (χ2) in R software.
Results
Soil microbial diversity and biphenyl biodegradation potential
Soil samples were collected at four different sites with a long history of contamination with aromatic compounds. Polychlorinated biphenyls (PCBs) were among the most prominent contaminants (Supplementary Table 1). Since microorganisms often use similar biochemical pathways to oxidize both biphenyl and PCBs [16], we hypothesized that the sampled soils contained biphenyl-oxidizing microorganisms. Amplicon sequencing of 16S rRNA genes indicated that all four soils harbored highly diverse microbial communities. A total of 18,188, 29,517, 49,167, and 35,121 operational taxonomic units (OTUs) were observed for soils A, B, C, and D, respectively. Among these, OTUs affiliated with Proteobacteria accounted for more than 20% of all sequences in each soil type. The remaining bacterial sequences were assigned to Acidobacteria (4–33%), Chloroflexi (3–20%), Bacteroidetes (3–9%), and Planctomycetes (4–6%) (Fig. 1a). Archaea (mostly Euryarchaeota and Crenarchaeota) contributed to 8–12% of total sequences in soils A, B, C, and less than 0.5% in soil D. Overall, the dominant phyla detected here matched the microbial diversity typically associated with PCB-contaminated soils [55].
a–b Relative abundance of major microbial lineages in each soil (a) and in the corresponding biphenyl-degrading enrichment cultures (b). c Growth curves of the biphenyl-degrading enrichment cultures when amended with biphenyl (filled circles), vs. control incubations (without biphenyl, open circles). Cultures A and C formed large flocs after 24 h of incubation, which led to high OD scattering. d Evolution of 13CO2 in the headspace of soil microcosms incubated with 13C-biphenyl. The last three times points of soil B and D were replotted as an inset to show the mineralization of 13C-biphenyl. Duplicate assays were done for each soil type. For soil A, only one of the replicates was active.
Cultivation experiments showed that all soils had the potential to oxidize biphenyl, with biphenyl-dependent microbial growth observed within 5 to 10 days of incubation. Sediment-free biphenyl-oxidizing enrichment cultures were obtained from each soil type. These cultures formed typical yellow meta-cleavage intermediates during growth (not shown) [56]. The approximate doubling time calculated from the OD-based growth curves ranged from 1.6 h to 5.7 h. Taxonomic profiling based on 16S rRNA genes revealed that the enrichments were dominated by Pseudomonas, Achromobacter, Rhodococcus, Hydrogenophaga, and Flavobacterium (Fig. 1b). Members of these genera have been frequently shown to degrade biphenyl [12], suggesting a similar function in the obtained cultures.
Short-term labeling experiments with 13C-biphenyl demonstrated that biphenyl-oxidizing microorganisms were present and active in all original soils. The formation of 13CO2 was detected in microcosms of all soil types, albeit at different extent, indicating different biphenyl mineralization rates. The fastest response was measured for soil C, where 13CO2 was detected within 24 h and continued to increase in concentration throughout the experiment, reaching 5.2 atom% labelings within 4 days of incubation (Fig. 1d). In contrast, longer lag phases and lower abundances of 13CO2 were observed for soils A, B, and D.
Visualization of soil biphenyl-degraders at a single-cell level
Mineralization of 13C-biphenyl was expected to be accompanied by assimilation of 13C by biphenyl oxidizers. To test this hypothesis, subsamples of the labeling assays collected at 0, 24, 48, and 96 h of incubation were analyzed by nanoSIMS. Individual cells were defined based on secondary ion images of 12C14N− and 13C14N−. The 13C abundance was determined for each cell. After 96 h of incubation, cells showing substantial enrichment in 13C were observed in all four soils (Fig. 2 and Supplementary Figs. 3–7). Most showed 13C enrichment close to the substrate labeling ratio (10 atom%) at the single-cell level. The relatively short incubation time and the high labeling ratio indicated that labeled cells were most likely primary biphenyl oxidizers, although the presence of secondary consumers feeding on excreted metabolites cannot be presently excluded. Biphenyl oxidizers are known to metabolize biphenyl to benzoate or other intermediates, which can be excreted and serve as a growth substrate for other members of the microbial communities [57, 58]. The variations in 13C abundance among individual cells could be caused by metabolic heterogeneity of primary biphenyl oxidizers [59, 60], or by poor substrate availability in the soil matrix. Alternatively, the less 13C-enriched cells may represent secondary consumers. Quantitatively, the cells enriched in 13C accounted for approximately 11%, 3%, 15%, and 8% of the total cell number of soils A, B, C, and D, respectively.
Secondary ion images of 12C14N− (a, d) and 13C14N− (b, e) were used to identify individual cells and to quantify 13C abundance. Arrowhead in 12C14N− ion image (d) point to the cells enriched in 13C in panel e. Raw ion counts of 12C14N− and 13C14N− for individual cells were shown as scatter plots (c, f). Cells having 13C abundance over 2% are presented as red symbols (f), whereas other cells are presented as blue symbols (c, f). Scale bars = 3.5 μm.
Carbon assimilation rate (Fc) was calculated using the cellular 13C14N−/12C14N− ratios and cell volumes calculated for regions of interest of nanoSIMS images [37]. For the calculation, we considered for all individual cells an average carbon density of 93.4 fg × μm−3 (average of several Proteobacteria isolates [61, 62]), since Proteobacteria was the most abundant microbial group in all tested soils. The cells showing enrichment in 13C have assimilation rates up to 0.61 fg cell−1 h−1 for soil A, 0.40 fg cell−1 h−1 for soil B, 2.9 fg cell−1 h−1 for soil C, and 0.96 fg cell−1 h−1 for soil D. To our knowledge, this is the first study reporting carbon assimilation rates for single cells of hydrocarbon-degrading microorganisms directly in soil environments.
Phylogenetic identification of biphenyl degraders using protein-SIP
Since microbial communities of all soils were highly diverse, a FISH-based phylogenetic identification of the 13C-labeled cells via a correlative FISH-SIMS analysis was not possible. Instead, we opted for a high-throughput protein-SIP–metagenomics approach. For this, we used microcosm subsamples collected at 96 h incubation with labeled biphenyl since these showed the highest rates of substrate assimilation (Fig. 2 and Supplementary Fig. 7).
Metagenome sequencing of each sample was conducted to an average depth of 2.8 million reads. For soils A, B, C, and D de novo assembly yielded 274,602, 219,108, 554,918, and 236,566 scaffolds, respectively. Predicted open reading frames were compiled as soil-specific databases for global metaproteomic analyses. Soil proteins extracted from microcosms with labeled and unlabeled biphenyl were analyzed in parallel. Querying mass spectra against the protein databases resulted in the identification of 345, 410, 1070, and 1986 peptides, from 381, 349, 1599, and 1506 proteins for soil A, B, C, and D, respectively. The most frequently detected proteins were affiliated to Proteobacteria, Chloroflexi, Acidobacteria, and Bacteroidetes (Fig. 3a), consistent with taxa abundance revealed by 16S rRNA amplicon sequencing (Fig. 1a).
a Phylogenetic classification of identified proteins in four soil microcosms after 96-h incubation with biphenyl. The number in parenthesis indicates the number of identified proteins in the different soil samples. b Mass spectra of doubly charged peptide VVNAEAGNAR (PAH dioxygenase alpha subunit) in 13C (red lines) and 12C (green lines) microcosms. Black circles represent the best fit of the theoretical isotopic patterns. c Overview of medium-quality to high-quality MAGs obtained in this study. Phylogenetic classification, tree, and soil of origin for each MAG are shown on the left. Phylogeny of MAGs was reconstructed based on a concatenated alignment of 120 bacterial marker genes. Bars indicated the completeness, contamination, number of identified proteins, and number of detected 13C-peptides for each MAG. MAGs encoding 13C-peptides are shaded by pink boxes.
For the identified proteins, we analyzed the 13C incorporation level by comparing mass spectra of identified peptides from 13C-labeled vs. unlabeled microcosms. As an example, in soil A the peptide VVNAEAGNAR which is part of the PAH dioxygenase large subunit, showed a shift of the most abundant isotopomer from m/z of 500.7622 (z = 2) to 504.7756 (z = 2), accompanied by a Gaussian-like shaped isotopic pattern (Fig. 3b). The extent of the mass shift represented a relative isotope abundance (RIA) of 18.4%, close to the 13C abundance of the labeled substrate (20 atom% for protein-SIP). These results demonstrated 13C assimilation into proteins and suggested that the incorporation was achieved via direct substrate utilization, and not indirectly through a food web in which the label would be diluted. A similar 13C incorporation pattern was observed for 5 and 11 peptides in soils A and D (Supplementary Figs. 8–10), whereas no 13C-labeled peptides were detected in soils B and C. Possible reasons include the stringency of the statistical approach, low abundance of biphenyl degraders (i.e., in soil B), high complexity of the microbial community (i.e., soil C) or moderate recovery of soil proteins (soils B and C).
Phylogenetic classification of potential biphenyl degraders was achieved by mapping the 13C-labeled peptides to metagenome-assembled genomes (MAGs). Binning of the four assembled metagenomes resulted in a total of 15 medium to high-quality MAGs (>50% completeness and <10% contamination) comprising taxonomically distinct members of Patescibacteria, Doudnabacteria, Gemmatimonadetes, and Proteobacteria (Fig. 3c and Supplementary Tables 5–6). An average of 40 proteins was identified across all 15 MAGs, demonstrating their viability and activity in soils (Fig. 3c). Of these, the MAGs classified as Alphaproteobacteria (D_bin4) and Azoarcus (D_bin1) encoded 4 and, respectively, 6 uniquely mapped 13C-labeled peptides, indicating active biphenyl assimilation (Fig. 3c). A third MAG classified as uncultured Rugosibacter (A_bin2) encoded three unique 13C-labeled peptides. However, since for this MAG only a total of 5 unique proteins were identified (Fig. 3c), its assignment as active biphenyl oxidizer is tentative.
Biphenyl degradation pathway in SIP-identified biphenyl-degraders
Each of the three MAGs identified by protein-SIP provided further genomic evidence for the utilization of biphenyl (Fig. 4). All three MAGs contained genes encoding homologs of biphenyl dioxygenase (BPDO), the first enzyme of the biphenyl oxidation pathway [16]. To confirm the relatedness of these genes to known BPDO, we constructed a phylogenetic tree using proteins of the BPDO catalytic subunit (BphA). This analysis revealed that the BphAs from D_bin1 and D_bin4 belonged to a toluene/biphenyl dioxygenase (T/B DO) clade closely related to those of biphenyl-degrading Burkholderia xenovorans LB400 and Pseudomonas furukawaii KF707 (Fig. 5a) [63, 64]. In contrast, BphAs encoded by A_bin2 exhibited higher sequence similarity with biochemically characterized BphA from Sphingobium yanoikuyae within the PAH-GN clade (Fig. 5a) [65]. In addition to bphA genes, D_bin4 and A_bin2 contained multiple copies of paralogs that were genetically distant and phylogenetically distinct from known BPDOs (Fig. 5a). These genes could possibly be responsible for the activation of aromatic compounds other than biphenyl (e.g., naphthalene).
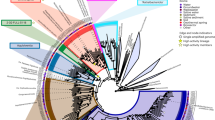
Tree clades containing 13C-labeled MAGs detected in this study are shown in red. Known biphenyl-degrading species are highlighted in green. The genomic tree was built from the concatenation of 120 bacterial marker genes. Filled circles on tree nodes indicated bootstrap values >70%. Scale bar, amino acid substitutions per site. The distribution of bph genes is shown in the right panel. Filled and empty circles represent the presence or absence of bph genes. The gene distribution is not shown for lineages without detectable bph homologs. Abbreviations: bphA: biphenyl dioxygenase subunit alpha; bphE: biphenyl dioxygenase subunit beta; bphF: biphenyl dioxygenase ferredoxin component; bphG: biphenyl dioxygenase system ferredoxin/NAD+ reductase component; bphB: cis-2,3-dihydrobiphenyl-2,3-diol dehydrogenase; bphC: 2,3-dihydroxybiphenyl 1,2-dioxygenase; bphD: 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase; bphI: 4-hydroxy-2-oxovalerate aldolase; bphH: 2-oxopent-4-enoate hydratase; bphJ: acetaldehyde dehydrogenase.
a BphA homologs from metagenomic bins are shown in bold font, and those affiliated to toluene/biphenyl dioxygenase (DO) are highlighted in red color. Filled circles on the tree branches indicated the bootstrap values >50%. The scale bar represents the number of substitutions per site. PAH-GN (GP): PAH dioxygenase from Gram-negative (Gram-positive) bacteria; T/B DO: toluene/biphenyl dioxygenase. b Sequence alignment of BphA from A_bin2, D_bin4, and well-known biphenyl degraders. Sequence regions that influenced the PCB congener preference and regiospecificity of biphenyl dioxygenase, are displayed [79, 80]. B. xenovorans: Burkholderia xenovorans; P. furukawaii: Pseudomonas furukawaii; C. testosteroni: Comamonas testosteroni. The amino acid numbering corresponds to BphA of the B. xenovorans LB400 sequence. c, Modeled substrate binding pocket in the BphA of D_bin4. The predicted BphA structure (yellow sticks) was superimposed on B. xenovorans BphA (blue wireline). The biphenyl molecule is shown as green lines.
Furthermore, all three MAGs contained genes of the biphenyl upper and lower pathways [16]. Specifically, these included genes encoding cis-2,3-dihydrobiphenyl-2,3-diol dehydrogenase (BphB), 2,3-dihydroxybiphenyl 1,2-dioxygenase (BphC), and 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase (BphD) for the upper pathway, and genes coding for 4-hydroxy-2-oxovalerate aldolase (BphI), 2-oxopent-4-enoate hydratase (BphH), and acetaldehyde dehydrogenase (BphJ) for the lower pathway (Figs. 4 and 6; Supplementary Table 7). In D_bin1 and D_bin4, genes for the upper and lower pathways were present in operons showing similar synteny with that of B. xenovorans LB400 [66]. In the Rugosibacter A_bin2, synteny was absent, the same set of genes being located at different genomic regions (Fig. 6).
The biphenyl upper and lower pathways yield eventually benzoate, which can be further metabolized to TCA intermediates via two different pathways. D_bin4 and A_bin2 encoded benzoate 1,2-dixoygenase (BenAB) and/or catechol 2,3-dioxygenase (DmpB) (Supplementary Table 8), suggesting the corresponding microorganisms were able to oxidize benzoate via the catechol meta-cleavage pathway [67]. By contrast, D_bin1 appeared to oxidize benzoate via the benzoyl-CoA pathway, as shown by the presence of genes coding benzoate coenzyme A ligase (BadA) and benzoyl-CoA 2,3-epoxidase (BoxAB) [68]. Finally, all three MAGs encode complete TCA cycle enzymes and multiple aerobic respiration complexes that could enable them to completely oxidize biphenyl to CO2 and conserve energy via oxygen respiration, consistent with a strict or facultative aerobic lifestyle (Supplementary Tables 9–10). These findings further consolidated the identification of the uncultured Alphaproteobacteria (D_bin4) and Azoarcus (D_bin1) as active biphenyl-degrading microorganisms in the tested soils. Although the uncultured Rugosibacter A_bin2 encodes two BphAs highly similar to biochemically characterized enzymes and all genes required for complete biphenyl oxidation, it lacks the syntenic organization of the bph genes typical of biphenyl oxidizers. Its assignment as biphenyl oxidizer remains therefore cautious.
Discussion
Here we explored the largely uncultured soil microbiota mediating organic pollutants degradation via an experimental framework that integrated stable isotope probing with single-cell analysis and isotope-specific molecular biology approaches. Biphenyl-assimilating microorganisms were visualized directly in soils via nanoSIMS chemical imaging. With this approach, we determined both the abundance of active biphenyl oxidizers in different soils and their rates of biphenyl assimilation (Fig. 2). Taxonomic identification of labeled cells based on protein-SIP showed an obvious difference between biphenyl oxidizers active under near in situ conditions and species enriched via repeated sub-cultivation. In contrast to well-known biphenyl degraders yielded by the cultivation-dependent approach, our labeling approach revealed previously unrecognized functions of uncultured yet ecologically relevant species. The clade UBA11222 represents such an uncultivated Alphaproteobacteria lineage whose physiological functions remained so far undocumented, despite their broad distribution in terrestrial and marine environments associated with organic contaminants (Supplementary Fig. 11) [7, 69]. Here we provided both physiological and genomic evidence that members of this clade are able to derive carbon and energy from the oxidation of biphenyl (Figs. 2 and 3). Considering that aromatic hydrocarbon oxidation pathways often have extended substrate spectra and that biphenyl-oxidizing microorganisms can often co-metabolize PCBs, members of this clade may have broader environmental implications in biodegradation processes. In addition, the combination of activity probing and genomic reconstruction indicated greater biodegradation capacities for known hydrocarbon-degrading bacteria. Strains of Azoarcus and Rugosibacter have been frequently retrieved as toluene and pyrene degraders, respectively [70, 71]. Their in situ activities towards biphenyl, as revealed (Azoarcus) or suggested (Rugosibacter) here, thus indicate a broader substrate range and higher metabolic versatility than previously recognized.
Genome analysis of taxa that we identified as active hydrocarbon oxidizers provided novel mechanistic insights regarding biodegradation processes in nature. Biphenyl dioxygenase (BPDO), the key enzyme for biodegradation of biphenyl and its analogs (i.e., PCBs), was detected in all three SIP-identified bacteria. The BPDO from the clade UBA11222 apparently belonged to an offset cluster within biphenyl/toluene dioxygenase family [72], with clear separation from those of cultivated biphenyl-utilizing bacteria (Fig. 5a). Their catalytic subunit (BphA) exhibited distinct sequence patterns of amino acids that were known to influence enzyme-substrate specificity and regiospecificity (Fig. 5b). Consistently, structure modeling of BphA from UBA11222 based on the available BphA crystal structure (Burkholderia xenovorans LB400) revealed differences between the amino acid sequences of the substrate-binding pocket (Fig. 5c). Replacement of an aromatic amino acid by an aliphatic residue may allow UBA11222 BPDO to accommodate different sets of PCB substrates. These results extend the genetic diversity of bphA in soil environments and imply a divergent function of BPDO in uncultured biphenyl-degrading microorganisms compared to cultivated counterparts. In two SIP-identified MAGs from soil D, bphA was found at genomic regions enriched in mobile genetic elements (MGEs), including those encoding transposes and integrase (Fig. 6). This concurs with previous findings of close genetic associations between MGEs and catabolic genes in isolated PCB-degrading microorganisms [16]. These observations suggest that horizontal gene transfer (HGT) mediated by MGEs may play a major role in the in situ dissemination of catabolic pathways, potentially allowing rapid adaptation of microbial communities to organic pollutants [73]. In addition, metabolic capacities of complete biphenyl oxidation were apparently conserved across phylogenetically distinct MAGs retrieved from the same soil, suggesting some functional plasticity of the soil microbial communities. This appears distinct from oil-degrading microbial communities of oil-contaminated environments, where the main catabolic pathways are partitioned among individual community members, and complete oxidation of hydrocarbons is achieved by complex coordination between microorganisms carrying out non-redundant reactions [74]. This disparity reflects diverse modes of microbial biodegradation networks in the environment [75].
All the 13C-labeled peptides identified here were mapped to MAGs with a genetic makeup indicating a potential for complete oxidation of biphenyl. However, nanoSIMS analyses showed a small number of cells becoming lightly enriched in 13C during the later incubation times (Fig. 2, Supplementary Figs. 3–7). These could represent secondary consumers feeding on metabolites excreted by the primary biphenyl oxidizers. The most likely reasons secondary consumers were not apparent in the protein-SIP analyses were their relatively low abundance and low 13C enrichment level. Future studies employing time-resolved ecophysiology probing over longer incubation times are needed to resolve metabolic networks involved in the degradation of biphenyl or other contaminants and to reveal the identity of secondary consumers.
Our findings have two kinds of implications. On the one hand, taxonomic identification of resident clades and measurement of their near-in-situ hydrocarbon uptake rates may assist to devise tailored bioremediation strategies. We showed that it was uncultured taxa distinct from those retrieved by enrichment approaches that responded readily to biphenyl additions under near in situ conditions. For example, in slurries of soil D, we identified Azoarcus, while cultivation yielded Pseudomonas and Rhodococcus. Azoarcus and the related Aromatoleum and Thauera genera are best known for their metabolic plasticity and ability to oxidize aromatic hydrocarbons under nitrate-reducing conditions (Supplementary Fig. 12) [76]. Therefore, in environments like soil D, a more effective bioremediation strategy might be to enhance the natural attenuation potential for example by stimulating nitrate reduction, instead of the more common practices of aeration or injection of oxygen [77, 78].
On the other hand, the wealth of metabolic pathways retrieved during the past decade via environmental sequencing is in dire need of direct evidence for their functionality [10, 11]. Extending the approach used here to other substrates and environments may allow testing of predicted ecophysiologies, uncover in situ metabolic networks, or identify metabolically active microorganisms, which, as shown here, could be rather different than those obtained via long-time cultivation. Combined with rapid innovations in cultivation strategies [4], direct in situ or near in situ ecophysiology probing is one of the awaited approaches to expand our knowledge of the metabolic potential and ecological roles of members of the uncultured majority.
Data availability
Sequencing data are archived in the NCBI database under BioProject number PRJNA692092, including the metagenome and 16 S rRNA gene amplicon datasets for soil A (SAMN17313532), B (SAMN17313578), C (SAMN17313580), and D (SAMN17313590). Metagenome-assembled genomes (MAGs) of uncultured biphenyl-degrading bacteria, including Rugosibacter_A_bin2 (SAMN17315487), Azoarcus_D_bin1 (SAMN17315485), and Alphaproteobacteria_D_bin4 (SAMN17315486), have been submitted to NCBI under the same BioProject. The proteomics dataset has been deposited with the ProteomeXchange Consortium identifier PXD023646. All other data are available in the paper or the Supplementary Information.
References
Singer E, Wagner M, Woyke T. Capturing the genetic makeup of the active microbiome in situ. ISME J. 2017;11:1949–63.
Hall EK, Bernhardt ES, Bier RL, Bradford MA, Boot CM, Cotner JB, et al. Understanding how microbiomes influence the systems they inhabit. Nat Microbiol. 2018;3:977–82.
Lloyd KG, Steen AD, Ladau J, Yin J, Crosby L. Phylogenetically novel uncultured microbial cells dominate earth microbiomes. mSystems 2018;3:e00055–18.
Lewis WH, Tahon G, Geesink P, Sousa DZ, Ettema TJG. Innovations to culturing the uncultured microbial majority. Nat Rev Microbiol. 2021;19:225–40.
Hug LA, Baker BJ, Anantharaman K, Brown CT, Probst AJ, Castelle CJ, et al. A new view of the tree of life. Nat Microbiol. 2016;1:16048.
Spang A, Caceres EF, Ettema TJG. Genomic exploration of the diversity, ecology, and evolution of the archaeal domain of life. Science. 2017;357:eaaf3883.
Parks DH, Rinke C, Chuvochina M, Chaumeil P-A, Woodcroft BJ, Evans PN, et al. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat Microbiol. 2017;2:1533–42.
Chen S-C, Musat N, Lechtenfeld OJ, Paschke H, Schmidt M, Said N, et al. Anaerobic oxidation of ethane by archaea from a marine hydrocarbon seep. Nature 2019;568:108–11.
Nayfach S, Roux S, Seshadri R, Udwary D, Varghese N, Schulz F, et al. A genomic catalog of Earth’s microbiomes. Nat Biotechnol. 2021;39:499–509.
Hatzenpichler R, Krukenberg V, Spietz RL, Jay ZJ. Next-generation physiology approaches to study microbiome function at single cell level. Nat Rev Microbiol. 2020;18:241–56.
Baker BJ, De Anda V, Seitz KW, Dombrowski N, Santoro AE, Lloyd KG. Diversity, ecology and evolution of Archaea. Nat Microbiol. 2020;5:887–900.
Abraham WR, Nogales B, Golyshin PN, Pieper DH, Timmis KN. Polychlorinated biphenyl-degrading microbial communities in soils and sediments. Curr Opin Microbiol. 2002;5:246–53.
Galbán-Malagón C, Berrojalbiz N, Ojeda M-J, Dachs J. The oceanic biological pump modulates the atmospheric transport of persistent organic pollutants to the Arctic. Nat Commun 2012;3:862.
Pieper DH. Aerobic degradation of polychlorinated biphenyls. Appl Microbiol Biotechnol. 2005;67:170–91.
Chain PSG, Denef VJ, Konstantinidis KT, Vergez LM, Agulló L, Reyes VL, et al. Burkholderia xenovorans LB400 harbors a multi-replicon, 9.73-Mbp genome shaped for versatility. Proc Natl Acad Sci USA. 2006;103:15280.
Furukawa K, Suenaga H, Goto M. Biphenyl dioxygenases: functional versatilities and directed evolution. J Bacteriol. 2004;186:5189–96.
McLeod MP, Warren RL, Hsiao WWL, Araki N, Myhre M, Fernandes C, et al. The complete genome of Rhodococcus sp. RHA1 provides insights into a catabolic powerhouse. Proc Natl Acad Sci USA. 2006;103:15582.
Lee TK, Lee J, Sul WJ, Iwai S, Chai BC, Tiedje JM, et al. Novel biphenyl-oxidizing bacteria and dioxygenase genes from a Korean tidal mudflat. Appl Environ Microbiol. 2011;77:3888–91.
Sul WJ, Park J, Quensen JF, Rodrigues JLM, Seliger L, Tsoi TV, et al. DNA-stable isotope probing integrated with metagenomics for retrieval of biphenyl dioxygenase genes from polychlorinated biphenyl-contaminated river sediment. Appl Environ Microbiol. 2009;75:5501–6.
Uhlik O, Jecna K, Mackova M, Vlcek C, Hroudova M, Demnerova K, et al. Biphenyl-metabolizing bacteria in the rhizosphere of horseradish and bulk soil contaminated by polychlorinated biphenyls as revealed by stable isotope probing. Appl Environ Microbiol. 2009;75:6471.
Jiang LF, Luo CL, Zhang DY, Song MK, Sun YT, Zhang G. Biphenyl-Metabolizing microbial community and a functional operon revealed in e-waste-contaminated soil. Environ Sci Technol. 2018;52:8558–67.
Tillmann S, Strompl C, Timmis KN, Abraham WR. Stable isotope probing reveals the dominant role of Burkholderia species in aerobic degradation of PCBs. FEMS Microbiol Ecol. 2005;52:207–17.
Leigh MB, Pellizari VH, Uhlik O, Sutka R, Rodrigues J, Ostrom NE, et al. Biphenyl-utilizing bacteria and their functional genes in a pine root zone contaminated with polychlorinated biphenyls (PCBs). ISME J. 2007;1:134–48.
Chen S-C, Duan G-L, Ding K, Huang F-Y, Zhu Y-G. DNA stable-isotope probing identifies uncultivated members of Pseudonocardia associated with biodegradation of pyrene in agricultural soil. FEMS Microbiol Ecol. 2018;94:fiy026.
Neufeld JD, Dumont MG, Vohra J, Murrell JC. Methodological considerations for the use of stable isotope probing in microbial ecology. Micro Ecol. 2007;53:435–42.
Neufeld JD, Vohra J, Dumont MG, Lueders T, Manefield M, Friedrich MW, et al. DNA stable-isotope probing. Nat Protoc. 2007;2:860–6.
Mohn WW, Westerberg K, Cullen WR, Reimer KJ. Aerobic biodegradation of biphenyl and polychlorinated biphenyls by Arctic soil microorganisms. Appl Environ Microbiol. 1997;63:3378–84.
Wagner-Dobler I, Bennasar A, Vancanneyt M, Strompl C, Brummer I, Eichner C, et al. Microcosm enrichment of biphenyl-degrading microbial communities from soils and sediments. Appl Environ Microbiol. 1998;64:3014–22.
Allen MB. Studies with cyanidium caldarium, an anomalously pigmented chlorophyte. Arch Mikrobiol. 1959;32:270–7.
Rabus R, Widdel F. Anaerobic degradation of ethylbenzene and other aromatic hydrocarbons by new denitrifying bacteria. Arch Microbiol. 1995;163:96–103.
Zhou J, Bruns MA, Tiedje JM. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316.
Pruesse E, Peplies J, Glöckner FO. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 2012;28:1823–9.
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 2013;41:D590–D6.
Ouyang WY, Su JQ, Richnow HH, Adrian L. Identification of dominant sulfamethoxazole-degraders in pig farm-impacted soil by DNA and protein stable isotope probing. Environ Int. 2019;126:118–26.
Tischer K, Zeder M, Klug R, Pernthaler J, Schattenhofer M, Harms H, et al. Fluorescence in situ hybridization (CARD-FISH) of microorganisms in hydrocarbon contaminated aquifer sediment samples. Syst Appl Microbiol. 2012;35:526–32.
Polerecky L, Adam B, Milucka J, Musat N, Vagner T, Kuypers MMM. Look@NanoSIMS–a tool for the analysis of nanoSIMS data in environmental microbiology. Environ Microbiol. 2012;14:1009–23.
Stryhanyuk H, Calabrese F, Kümmel S, Musat F, Richnow HH, Musat N. Calculation of single cell assimilation rates from SIP-NanoSIMS-derived isotope ratios: a comprehensive approach. Front Microbiol. 2018;9:2342.
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014;30:2114–20.
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77.
Kang DD, Li F, Kirton E, Thomas A, Egan R, An H, et al. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 2019;7:e7359–e.
Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–55.
Chaumeil P-A, Mussig AJ, Hugenholtz P, Parks DH. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 2020;36:1925–7.
Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014;30:1312–3.
Hyatt D, Chen G-L, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinforma. 2010;11:119.
Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30.
Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42:D222–D30.
Huerta-Cepas J, Szklarczyk D, Heller D, Hernández-Plaza A, Forslund SK, Cook H, et al. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019;47:D309–D14. (D1)
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC Bioinforma. 2009;10:421.
Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7.
Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–3.
Budhraja R, Karande S, Ding C, Ullrich MK, Wagner S, Reemtsma T, et al. Characterization of membrane-bound metalloproteins in the anaerobic ammonium-oxidizing bacterium “Candidatus Kuenenia stuttgartiensis” strain CSTR1. Talanta. 2021;223:121711.
Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20:1466–7.
Röst HL, Sachsenberg T, Aiche S, Bielow C, Weisser H, Aicheler F, et al. OpenMS: a flexible open-source software platform for mass spectrometry data analysis. Nat Methods. 2016;13:741–8.
Sachsenberg T, Herbst F-A, Taubert M, Kermer R, Jehmlich N, von Bergen M, et al. MetaProSIP: automated inference of stable isotope incorporation rates in proteins for functional metaproteomics. J Proteome Res. 2015;14:619–27.
Liu J, He XX, Lin XR, Chen WC, Zhou QX, Shu WS, et al. Ecological effects of combined pollution associated with e-waste recycling on the composition and diversity of soil microbial communities. Environ Sci Technol. 2015;49:6438–47.
Kumamaru T, Suenaga H, Mitsuoka M, Watanabe T, Furukawa K. Enhanced degradation of polychlorinated biphenyls by directed evolution of biphenyl dioxygenase. Nat Biotechnol. 1998;16:663–6.
Garrido-Sanz D, Manzano J, Martín M, Redondo-Nieto M, Rivilla R. Metagenomic analysis of a biphenyl-degrading soil bacterial consortium reveals the metabolic roles of specific populations. Front Microbiol. 2018;9:232.
Kikuchi Y, Nagata Y, Ohtsubo Y, Koana T, Takagi M. Pseudomonas fluorescens KKL101, a benzoic acid degrader in a mixed culture that degrades biphenyl and polychlorinated biphenyls. Biosci Biotechnol Biochem. 1995;59:2303–4.
Musat N, Halm H, Winterholler B, Hoppe P, Peduzzi S, Hillion F, et al. A single-cell view on the ecophysiology of anaerobic phototrophic bacteria. Proc Natl Acad Sci USA. 2008;105:17861.
Calabrese F, Voloshynovska I, Musat F, Thullner M, Schlömann M, Richnow HH, et al. Quantitation and comparison of phenotypic heterogeneity among single cells of monoclonal microbial populations. Front Microbiol. 2019;10:2814.
Robertson BR, Button DK, Koch AL. Determination of the biomasses of small bacteria at low concentrations in a mixture of species with forward light scatter measurements by flow cytometry. Appl Environ Microbiol. 1998;64:3900–9.
Troussellier M, Bouvy M, Courties C, Dupuy C. Variation of carbon content among bacterial species under starvation condition. Aquat Micro Ecol. 1997;13:113–9.
Furukawa K, Miyazaki T. Cloning of a gene cluster encoding biphenyl and chlorobiphenyl degradation in Pseudomonas pseudoalcaligenes. J Bacteriol. 1986;166:392–8.
Seeger M, Timmis KN, Hofer B. Conversion of chlorobiphenyls into phenylhexadienoates and benzoates by the enzymes of the upper pathway for polychlorobiphenyl degradation encoded by the bph locus of Pseudomonas sp. strain LB400. Appl Environ Microbiol. 1995;61:2654–8.
Chadhain SM, Moritz EM, Kim E, Zylstra GJ. Identification, cloning, and characterization of a multicomponent biphenyl dioxygenase from Sphingobium yanoikuyae B1. J Ind Microbiol Biotechnol. 2007;34:605–13.
Hofer B, Backhaus S, Timmis KN. The biphenyl/polychlorinated biphenyl-degradation locus (bph) of Pseudomonas sp. LB400 encodes four additional metabolic enzymes. Gene 1994;144:9–16.
Harwood CS, Parales RE. The beta-ketoadipate pathway and the biology of self-identity. Annu Rev Microbiol. 1996;50:553–90.
Rather LJ, Knapp B, Haehnel W, Fuchs G. Coenzyme A-dependent aerobic metabolism of benzoate via epoxide formation. J Biol Chem. 2010;285:20615–24.
Stegen JC, Fredrickson JK, Wilkins MJ, Konopka AE, Nelson WC, Arntzen EV, et al. Groundwater-surface water mixing shifts ecological assembly processes and stimulates organic carbon turnover. Nat Commun. 2016;7:1–12.
Corteselli EM, Aitken MD, Singleton DR. Rugosibacter aromaticivorans gen. nov., sp. nov., a bacterium within the family Rhodocyclaceae, isolated from contaminated soil, capable of degrading aromatic compounds. Int J Syst Evol Microbiol 2017;67:311–8.
Fernandez H, Prandoni N, Fernandez-Pascual M, Fajardo S, Morcillo C, Diaz E, et al. Azoarcus sp. CIB, an anaerobic biodegrader of aromatic compounds shows an endophytic lifestyle. PLoS ONE. 2014;9:e110771.
Iwai S, Johnson TA, Chai BL, Hashsham SA, Tiedje JM. Comparison of the specificities and efficacies of primers for aromatic dioxygenase gene analysis of environmental samples. Appl Environ Microbiol. 2011;77:3551–7.
Top EM, Springael D. The role of mobile genetic elements in bacterial adaptation to xenobiotic organic compounds. Curr Opin Biotechnol. 2003;14:262–9.
Dombrowski N, Donaho JA, Gutierrez T, Seitz KW, Teske AP, Baker BJ. Reconstructing metabolic pathways of hydrocarbon-degrading bacteria from the Deepwater Horizon oil spill. Nat Microbiol. 2016;1:1–7.
de Lorenzo V. Systems biology approaches to bioremediation. Curr Opin Biotechnol. 2008;19:579–89.
Rabus R, Wöhlbrand L, Thies D, Meyer M, Reinhold-Hurek B, Kämpfer P. Aromatoleum gen. nov., a novel genus accommodating the phylogenetic lineage including Azoarcus evansii and related species, and proposal of Aromatoleum aromaticum sp. nov., Aromatoleum petrolei sp. nov., Aromatoleum bremense sp. nov., Aromatoleum toluolicum sp. nov. and Aromatoleum diolicum sp. nov. Int J Syst Evol Microbiol. 2019;69:982–97.
Vogt C, Richnow HH. Bioremediation via in situ microbial degradation of organic pollutants. Adv Biochem Engin/Biotechnol. 2014;142:123–46.
Cunningham JA, Rahme H, Hopkins GD, Lebron C, Reinhard M. Enhanced in situ bioremediation of BTEX-contaminated groundwater by combined injection of nitrate and sulfate. Environ Sci Technol. 2001;35:1663–70.
Mondello FJ, Turcich MP, Lobos JH, Erickson BD. Identification and modification of biphenyl dioxygenase sequences that determine the specificity of polychlorinated biphenyl degradation. Appl Environ Microbiol. 1997;63:3096–103.
Gomez-Gil L, Kumar P, Barriault D, Bolin JT, Sylvestre M, Eltis LD. Characterization of biphenyl dioxygenase of Pandoraea pnomenusa B-356 as a potent polychlorinated biphenyl-degrading enzyme. J Bacteriol. 2007;189:5705–15.
Acknowledgements
We acknowledge B. Scheer for proteomics analyses and Dr. L. Polerecky (Utrecht University) for support with the LANS software. Funding for this project was provided by Helmholtz Association and by the National Natural Science Foundation of China (41991332). Further financial support was provided by the Helmholtz Association (grant ERC-RA-0020 to FM). We acknowledge the Centre for Chemical Microscopy (ProVIS) at the Helmholtz Centre for Environmental Research for the use of protein mass spectrometry and nanoSIMS. ProVIS is supported by European Regional Development Funds (EFRE–Europe funds Saxony).
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
S-CC, G-LD, H-HR, Y-GZ, and FM designed the research. S-CC, G-LD, and FM retrieved the original soil sample and contributed to the establishment of the enrichment cultures. S-CC and FM performed labeling experiments. S-CC, FC, H.S., F.M., and N.M. designed and performed nanoSIMS analyses. S-CC, RB, H-HR, and LA. performed metaproteomics analyses. S-CC, G-LD, Y-GZ, and FM performed phylogenetic and metagenomics analyses. S-CC, Y-GZ, LA, and FM wrote the manuscript with contributions from all co-authors.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chen, SC., Budhraja, R., Adrian, L. et al. Novel clades of soil biphenyl degraders revealed by integrating isotope probing, multi-omics, and single-cell analyses. ISME J 15, 3508–3521 (2021). https://doi.org/10.1038/s41396-021-01022-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-021-01022-9
This article is cited by
-
Characterization and environmental applications of soil biofilms: a review
Environmental Chemistry Letters (2024)
-
Disentangling effects of multiple agricultural stressors on benthic and hyporheic nitrate uptake
Biogeochemistry (2024)
-
DNA stable isotope probing on soil treated by plant biostimulation and flooding revealed the bacterial communities involved in PCB degradation
Scientific Reports (2022)