Abstract
Coral mucus, tissue, and skeleton harbor compositionally different microbiota, but how these coral compartments shape the microbial evolution remains unexplored. Here, we sampled bacteria inhabiting a prevalent coral species Platygyra acuta and sequenced genomes of 234 isolates comprising two populations in Rhodobacteraceae, an alphaproteobacterial lineage representing a significant but variable proportion (5–50%) of the coral microbiota. The Ruegeria population (20 genomes) contains three clades represented by eight, six, and six isolates predominantly sampled from the skeleton (outgroup), mucus (clade-M), and skeleton (clade-S), respectively. The clade-M possesses functions involved in the utilization of coral osmolytes abundant in the mucus (e.g., methylamines, DMSP, taurine, and L-proline), whereas the clade-S uniquely harbors traits that may promote adaptation to the low-energy and diurnally anoxic skeleton (e.g., sulfur oxidation and swimming motility). These between-clade genetic differences were largely supported by physiological assays. Expanded analyses by including genomes of 24 related isolates (including seven new genomes) from other marine environments suggest that clade-M and clade-S may have diversified in non-coral habitats, but they also consolidated a key role of distinct coral compartments in diversifying many of the above-mentioned traits. The unassigned Rhodobacteraceae population (214 genomes) varies only at a few dozen nucleotide sites across the whole genomes, but the number of between-compartment migration events predicted by the Slatkin–Maddison test supported that dispersal limitation between coral compartments is another key mechanism diversifying microbial populations. Collectively, our results suggest that different coral compartments represent ecologically distinct and microgeographically separate habitats that drive the evolution of the coral microbiota.
Similar content being viewed by others
Introduction
Corals reefs are topographically and ecologically intricate marine habitats that supported diverse and abundant marine life [1]. Scleractinian corals harbor at least four major compartments that microbial populations inhabit (Fig. S1), namely coral mucus, skeleton, tissue, and gastrovascular cavity [2]. The coral mucus is a nutrient and energy-rich mixture of materials [3, 4], including its primary structural macromolecules—glycoproteins, polysaccharides, and lipids [5, 6], as well as abundant organic osmolytes secreted by the coral host and its endosymbiont Symbiodiniaceae [7]. The osmolytes are compatible or counteracting solutes that are accumulated in the cells in response to the fluctuation of osmotic stresses, thermal stress, and hypoxia [8, 9]. Common coral osmolytes include methylamines [e.g., glycine betaine (GBT), choline, choline-O-sulfate (COS), sarcosine and trimethylamine N-oxide (TMAO)], methyl sulfonium [e.g., dimethylsulfoniopropionate (DMSP) and dimethyl sulfoxide (DMSO)], amino acid derivatives (e.g., taurine, L-proline, and ectoine), sugars (e.g., trehalose and arabinose) and polyols (e.g., glycerol) [9]. Of these, GBT, taurine, and DMSP represent the major osmolytes of corals [10]. Corals eliminate or accumulate osmolytes quickly to equilibrate the fluctuated salinity caused by, for example, tides, air exposure, evaporation, and heavy rainfalls [11]. These processes together lead to the enrichment of wasted osmolytes in mucus [12], which serve as organic nutrients to support the resident bacteria [13, 14]. In contrast, the skeleton is a low-energy environment covered by the coral tissue, where up to 99% of the photosynthetically active radiation is either scattered or entrapped by Symbiodiniaceae located within the coral tissue [15]. The porous structure of the skeleton is mainly constructed with inorganic aragonite crystals along with a small fraction (≤0.1% by weight) of the organic matrix [16, 17] which limits skeleton-associated microbes [18]. Although the light penetration is low, there are still endolithic algae that reside inside the skeleton, whose photosynthesis and respiration generate a diel fluctuation of the oxygen and pH levels in the skeleton [15]. Likewise, Symbiodiniaceae in the coral cells causes similar diel changes of the tissue [19]. The gastrovascular cavity is a sac-like external environment of the polyp surrounded by the gastrodermis layer of coral tissue [20]. Compared to seawater, it shows lower levels of oxygen and pH, but a higher concentration of nutrients [20].
The fact that different coral compartments are featured with distinct physicochemical conditions indicates that coral is an ideal system for studying microbial niche adaptation. Previous studies suggest that the community structure of the bacterial associates is largely shaped by the coral anatomy [21, 22]. Among the three compartments, for example, the mucus harbors a higher proportion of compartment-specific bacteria than the tissue [23], and the skeleton accommodates a microbial community of greater diversity compared to the tissue and mucus [15, 24]. While these studies illustrated important differences in terms of the microbial identities and functions across the coral compartments, they provided limited taxonomic resolution in functional differentiation along with coral compartments. Since functional differences can be ascribed to both environmental heterogeneity and phylogenetic diversity, a robust association of metabolic functions with ecological niches may be established when the phylogenetic distance of lineages under comparison is minimized. Leveraging the single nucleotide polymorphism (SNP) distribution at the core genome and gene frequency at the accessory genome, the culture-based population genomics is able to uncover the mechanisms driving population differentiation and niche adaptation [25,26,27,28]. To our knowledge, this approach has not been used to investigate coral-associated microbial populations.
Among the numerous bacterial groups found on corals, Ruegeria, a lineage in the alphaproteobacterial family Rhodobacteraceae, is among the top three genera that are most widely associated with coral species [29]. Members of Ruegeria likely establish mutualistic interactions with the coral hosts. They are constantly associated with both broadcast-spawning and brooding corals in their early developmental stages [30] and can be vertically transferred from parents to larvae in the brooding coral Pocillopora damicornis [31]. Some Ruegeria strains also help the larval settlement in octocoral species [32]. In adult corals, Ruegeria strains can inhibit the growth of pathogenic Vibrio species [33]. Nevertheless, some Ruegeria members may be opportunistic pathogens, which show increased abundance when coral hosts are under stress [34,35,36].
The scleractinian coral species Platygyra acuta is known for its high tolerance to temperature and salinity stresses, as well as its resistance to bleaching in winter [37]. It is not only the dominant species in Hong Kong coastal waters where the samples were collected but also a globally important germplasm resource to understand the survival of marginal reefs [38, 39]. We harvested two Rhodobacteraceae populations from P. acuta: one is a Ruegeria lineage (hereafter, the Ruegeria population) and the other cannot be assigned to any described genus (hereafter, the unassigned Rhodobacteraceae population). Both populations are members of the Roseobacter group, a metabolically versatile lineage that dominates the bacterial communities in coastal ocean environments [40] and encompasses most of the taxonomic diversity reported in Rhodobacteraceae [41]. While the Ruegeria population is represented by only 20 isolates, it harbors a great amount of genetic variation, which is partitioned into two species-level clades colonizing the coral mucus and skeleton respectively. Most of the analyses presented in the current study address this population. On the other hand, the 214 members of the unassigned Rhodobacteraceae population vary at only a few dozen SNP sites across the whole genomes. While this extreme genetic monomorphism makes many population genomics tools less applicable, it nevertheless provides a unique opportunity to test dispersal limitation across coral compartments as a neutral force leading to bacterial population differentiation.
Materials and methods
All the methodological details were described in Supplementary Text 1. Twelve coral samples of P. acuta were collected by SCUBA diving in Hong Kong water at Kiu Tsui Chau (N 22°22′04.4″ E 114°17′42.0″) on 24th April 2017, Wong Wan Chau (N 22°31′31.2″ E 114°19′00.1″) on 12th January 2018 and Ngo Mei Chau (N 22°31′47.2″ E 114°19′02.9″) and Chek Chau (N 22°30′03.3″ E 114°21′22.7″) on 25th February 2018 (Fig. S1A) for bacterial cultivation. Coral rubbles (2–8 cm in diameter) were sampled from three different colonies at each site using a rock chisel, separated in zip-lock bags with their ambient seawater. One sample of ambient seawater was collected at each site. The three coral compartments of each coral rubble were separated following established procedures [6, 42]. Limited by the anatomical feature of the coral (Fig. S1B), the mucus and skeleton samples collected by the current method are both considered clean, whereas the tissue sample may contain residues of mucus and skeleton and cannot be separated from the gastrovascular cavity. Bacteria were subsequently isolated from each compartment and the ambient seawater with a modified marine basal medium (MBM) recipe, and their taxonomic affiliation was classified through 16S rRNA gene analyses.
We identified a Ruegeria population (20 isolates) and an unassigned Rhodobacteraceae population (214 isolates) suitable for subsequent population genomic analyses according to two criteria: (i) each contains a number of closely related isolates and (ii) each covers multiple coral compartments. Genomes of all these 234 isolates were sequenced with BGISEQ-500 PE100, assembled, and annotated. The strain HKCCD4315 in the Ruegeria population was additionally sequenced using the PacBio Sequel platform to obtain a complete and closed genome.
The Ruegeria population was initially analyzed to understand its phylogenetic and population genetic structure. The orthologous gene families were identified with OrthoFinder v2.2.1 [43] and the maximum likelihood phylogenomic tree was constructed based on the concatenated single-copy gene alignments at the amino acid level using IQ-TREE v1.6.5 [44]. Two clades (clade-M and clade-S) matched well with the coral mucus and skeleton compartments, respectively, along with an additional clade locating at the outgroup position. The pairwise 16S rRNA gene identity and the whole-genome average nucleotide identity (ANI) between- and within-clades were compared to help determine whether speciation has already occurred between these clades [45]. Phylogenetic separation does not necessarily lead to allelic isolation between the phylogenetic clusters [46, 47]. We, therefore, investigated the population structure of clade-M and clade-S with the coancestry analysis implemented in fineSTRUCTURE v2.0.7 [48] and characterized the potential gene flow barrier by comparing the between-clade to the within-clade relative frequency of recombination to point mutation (ρ/θ) [49] and the relative effect of recombination to mutation (r/m) [49], and by determining the fixation index (Fst) across the core genome shared by clade-M and clade-S [50]. We further tested whether the genetic separation of the two clades occurred at the accessory genome by measuring the similarity of genome content among strains with the Jaccard index and subsequently clustering it with the complete-linkage method [28].
The population differentiation between clade-M and clade-S was further investigated at the functional level. We sought to identify signature genes of potential importance in niche adaptation. In the core genome, novel allelic replacements via homologous recombination with external species at certain loci represent a potentially important and prevalent adaptive mechanism in marine bacterial populations [47, 51]. This type of core genes was also identified here. Briefly, the substitution rate at synonymous sites (dS) across all single-copy core genes and across all paired-strain comparisons between isolates from clade-M and clade-S was clustered using the K-means method, which resulted in a cluster composed of genes each showing unusually large dS values, the genetic signature of an evolutionary history subjected to novel allelic replacement [51]. For the identified core genes, gene trees were built and compared to the genome tree to help determine which clade was subjected to novel allelic replacement. For the accessory genome, we focused on the genes largely specific to clade-M (or clade-S) and tried to resolve two competing hypotheses: whether they resulted from acquisition events at the last common ancestor (LCA) of clade-M (or clade-S) or loss events at the LCA of clade-S (or clade-M). Specifically, the gene gain and loss history of clade-specific genes were inferred based on the gene copy number distribution among extant genomes of the Ruegeria population using BadiRate v1.35 [52]. For genes whose functions were potentially relevant to niche adaptation and which differentiated the clade-M and clade-S at the DNA level, their potential functional outcomes were tested using physiological assays, though there was no direct evidence connecting the genetic and phenotypic differentiation. For substrate utilization assays, 11 compounds each were used as a sole carbon (C) and/or nitrogen (N) source to compare growth rate and yield between clade-M and clade-S. For the motility experiment, swimming, swarming, and twitching motilities were assayed on different concentrations of agar.
To test whether the clade-M and clade-S may have diverged from a single ancestor that initially colonized the coral or these two clades may have already split in marine habitats unrelated to corals and subsequently independently colonized coral compartments, we included in an expanded phylogenomic tree 24 closely related Ruegeria isolates mostly sampled from other marine habitats in Hong Kong. Of these isolates, seven were sequenced in the present study. We additionally estimated the potential time of the split between clade-M and clade-S based on synonymous substitution rate, spontaneous mutation rate, and potential generation time in the wild, and the goal of this analysis is to show its antiquity compared to the coral ages.
For the unassigned Rhodobacteraceae population, the genetic uniformity enabled the Slatkin–Maddison test [53] to measure the degree of compartmentalization on population subdivision. By assuming that the common ancestor of closely related strains occupied a single compartment, a migration event is inferred when a closely related strain is found in a different compartment. The number of migration events can be estimated from real data, and the expected number of migrations can be calculated from randomly generated population structure by permutating the strains across the phylogenomic tree. A high degree of compartmentalization is manifested as a small probability that the former is greater than the latter. We also estimated the time of its origin and compared it to the potential age of the coral animal.
Results and discussion
The Ruegeria population differentiation along with coral compartments
The phylogenomic tree showed that the 20 closely related Ruegeria strains isolated from P. acuta formed three clades (Fig. 1A). Among these, the six clade-S members were all isolated from the skeleton, and the six clade-M members were all collected from mucus except for one from ambient seawater (Fig. 1A). The outgroup clade consists of seven skeleton-associated strains and one tissue-associated strain. Coral hosts of the 20 strains were sampled from three sites in Hong Kong, of which 11, five, and four were from Wong Wan Chau, Kiu Tsui Chau, and Check Chau, respectively (Table S1). As these strains were isolated from the same coral species, interpretation of any strain-level genomic differences ruled out the host species effect. Within each clade, both ANI and 16S rRNA gene identities are greater than the level often used to delineate a bacterial species at 95.0% [54] and 98.7% [45], respectively (Fig. 1B). Between clade-M and clade-S, a maximum of 89.9% ANI (Table 1 and Fig. 1B) claims the two clades as two distinct species, whereas an average of 99.7% identity at the 16S rRNA gene argues against the completion of speciation (Table 1 and Fig. 1B). We, therefore, collected more evidence to support population differentiation between the two clades.
A The rooted maximum-likelihood phylogenomic tree of the 20 strains isolated from the coral species Platygyra acuta (Accession numbers were showed in Table S1). This tree is rooted in the mid-point rooting method. Solid circles at the nodes indicate that the support value (IQ-TREE v1.6.5 ultrafast bootstraps) of the branch is 100%. The LCA of clade-M, the LCA of the five mucus strains within the clade-M, and the LCA of clade-S are shown as a blue triangle, the red star, and the pink triangle, respectively. Strains isolated from different coral compartments are highlighted with different colors. The clade-M and clade-S are also labeled. B The heatmap of the whole-genome average nucleotide identity (ANI) and heatmap of the pairwise identity of 16S rRNA genes of the 20 strains. C fineSTRUCTURE coancestry matrix of the 12 isolates from clade-M and clade-S, with warmer colors representing more ancestry, shared between strains under comparison. Strains assigned to the same fineSTRUCTURE population are connected with a vertical bar on the left of the matrix, and the dendrogram shows the clustering of the fineSTRUCTURE populations. D Heatmap of accessory gene content similarity measured by Jaccard distance, with warmer colors representing the higher similarity between strains. The left dendrogram was generated based on the complete-linkage clustering method (color figure online).
At the core genome level, fineSTRUCTURE assigned the 12 strains from clade-S and clade-M to seven co-ancestry populations, three in clade-S and four in clade-M (Fig. 1C). The proportion of co-ancestry shared within clade-S or clade-M is much greater than that between clade-S and clade-M (Fig. 1C), suggesting a strong barrier to homologous recombination between the two clades. The overall consistent branching pattern between the coancestry populations and the phylogenetic groups indicates that clade-M and clade-S diverged into two species. The decreased gene flow between clades is also supported by decreased ρ/θ ratio and r/m ratio between the two clades compared to those within clades (Supplementary Text 2.1, Table S2). One more evidence for population differentiation is the genome-wide high Fst values (≥0.5), indicating that the speciation between the two clades is approaching completion (Fig. S2, Supplementary Text 2.1). The above metrics are proxies for population differentiation, but they cannot tell the mechanisms leading to differentiation. One evolutionary mechanism demonstrated to drive the differentiation of a pelagic Roseobacter population is novel allelic replacement at the ecologically relevant core genes through homologous recombination with external species [47], which leaves a genetic signature manifested as unusually large dS values at those loci for between-clade comparisons (Fig. S3, Supplementary Text 1.6) [51]. For the Ruegeria population, 167 single-copy genes each show unusually large dS values between clade-S and clade-M (Table S3 and Fig. 2), indicating that novel allele replacement at these loci likely occurred at the LCA of either clade-M (marked with a blue triangle in Fig. 1A) or clade-S (marked with a pink triangle in Fig. 1A). Resolving the exact evolutionary history of allelic replacements requires gene tree analyses (Fig. S4 versus Fig. S5–S10).
The phylogeny and pangenome of the Ruegeria population generated using Circos v0.64 [88]. The gene families are arranged in order of the closed genome of strain HKCCD4315. The circular tracks depict the genomes which are arranged in order of the phylogeny and shown in gray. From the inner to the outer circle: (1)–(6) six genomes of clade-M. The genes gained at the LCA of clade-M are shown in cyan, and those lost at the LCA of clade-S are shown in blue. (7)–(12) Six genomes of clade-S. The genes gained at the LCA of clade-S are shown in magenta, and those lost at the LCA of clade-M are shown in orange. (13)–(20) Eight genomes of an outgroup. The 536 clade-M specific genes and the 365 clade-S specific genes are shown in yellow in all tracks. The genomic islands (GIs) are shown in dark gray in all tracks. Other genes are shown in light gray in all tracks. (21) The genomic region of the chromosome and three plasmids are shown in gray, and the core genes showing unusually large synonymous substitution rate (dS) are shown in green (color figure online).
At the accessory genome level, we showed that the similarity of accessory gene content was higher within clade-M or clade-S than that between the two clades. The congruent branching pattern between the gene content dendrogram (Fig. 1D) and the core gene-based phylogenomic tree (Fig. 1A) suggests that population differentiation has occurred at the accessory genome. Among the 3202 accessory gene families, 194 and 157 families are universally and exclusively present in clade-M and clade-S members, respectively. Using a relaxed definition of “clade-specific gene families” where genes are present in at least two-thirds of the strains in one clade but present in no more than one-third of the strains in the other clade, we found 536 and 365 gene families specific to clade-M and clade-S (Table S4, Table S5 and Fig. 2), respectively. Of the 536 clade-M specific genes, 224 and 64 gene families were inferred to be acquired at the LCA of clade-M and lost at the LCA of clade-S, respectively (Table S4). Since the basal position of clade-M is occupied by a seawater strain (Fig. 1A), we further inferred that 122 additional gene family gains occurred at the LCA (marked with a red star in Fig. 1A) of the remaining five members which are exclusively mucus-associated (Table S4). For the 365 clade-S specific genes, 233 and 97 gene families were inferred to be gained at the LCA of clade-S and lost at the LCA of clade-M, respectively (Table S5). Thus, both gene gains and losses likely played an important role in differentiating the accessory genome. Among the 901 clade-specific gene families, 126 are associated with genomic islands (GIs) (Fig. 2), suggesting that GIs likely facilitated population differentiation.
Metabolic adaptation of the mucus clade to the eutrophic mucus niche
Many of the clade-M-specific genes are likely involved in the utilization of methylamines. Many methylamine compounds are coral osmolytes, such as GBT, COS, glycerophosphocholine (GPC), choline, and sarcosine [55, 56], though TMAO and dimethylglycine (DMG) have not been reported in coral studies. Of these, GBT may be taken up through the ABC transporter ProVWX [57]. The GBT precursors, GPC and choline, may be assimilated through the ABC transporter UgpABCE [58] and the BCCT transporter BetT [59], respectively. Clade-M members specifically contain an extra copy of all three transporters (Table S4 and Fig. 3). The downstream catabolic pathways for GBT, GPC, and choline are largely overlapped (Fig. 3). Once at the cytoplasm, GPC can be converted to choline through glycerophosphodiester phosphodiesterase (Gde1). Choline is either incorporated into phosphatidylcholine (PC) or subjected to demethylative degradation [60, 61]. For the former, the key gene encoding choline kinase (cki1, Table S4) for assimilation is specific to clade-M. For the latter, the choline is oxidized to GBT aerobically before demethylation (Fig. 3), and the key genes including the choline dehydrogenase (betA, Table S4) and betaine aldehyde dehydrogenase (betB, Table S4) are part of the clade-M specific genes (Fig. 3).
The compounds in gray blocks are osmolytes in coral. The solid arrows represent the genes present in the Ruegeria population. The arrows in dashed lines represent the genes missing from the Ruegeria population. The cyan arrows represent the clade-M-specific genes. The green arrows denote the outlier core genes showing unusually large between-clade dS values. Divergent allelic replacement at the LCA of clade-M and clade-S are marked with solid green and red circles, respectively. The open circle in green represents multiple allelic replacements within clade-M. The black circles indicate that the allelic replacement history cannot be resolved based on the available data. The black arrows show the core genes shared by all clade-M and clade-S members. The red arrows indicate the reactions related to nitrogen assimilation. The yellow arrows indicate the reactions related to the oxidation of C1 groups. The blue arrows indicate the reactions related to carbon assimilation. There are no clade-S-specific genes found in these pathways. Only the dominant substrates of the promiscuous transporters are shown in the figure. The conversion from GBT to TMA was previously revealed by a metabolomics study on intestinal microbiota [63], but the underlying gene for this reaction has not been known (color figure online).
The model strain of the Roseobacter group, Ruegeria pomeroyi DSS-3, was shown to grow on GBT as a sole C source and produce intermediate metabolites including DMG, sarcosine and glycine sequentially [61]. This canonical pathway (Fig. 3, the right-hand pathway located downstream of GBT) assimilates a one-carbon (C1) group into methionine and a two-carbon group (glycine) into pyruvate, respectively [61]. It starts from the demethylation of GBT to DMG catalyzed by betaine-homocysteine methyltransferase (bhmt) (Fig. 3) [61]. In the Ruegeria population under study, however, a truncated bhmt gene (HKCCD4315_03106) which lacks the vitamin B12-binding domain is present in the core genome. This truncated bhmt was also identified in the Roseobacter group member Phaeobacter inhihens (previously known as Phaeobacter gallaeciensis), and was considered nonfunctional for this reaction because the bacterium poorly used GBT as a C source [62]. Likewise, none of the six isolates assayed here were able to thrive on choline or GBT without a supplemental C source (Fig. 4A-2, A-3, B-2 and B-4). When provided with the intermediate metabolites downstream of GBT, such as DMG (Fig. 4A-4) and sarcosine (Fig. 4A-5), these isolates thrived, further supporting that the utilization of GBT is blocked at the step catalyzed by BHMT. Specifically, DMG is demethylated to sarcosine through dimethylglycine dehydrogenase (dmgdh), and sarcosine is demethylated to glycine through either sarcosine dehydrogenase (sardh) or sarcosine oxidase (soxABDG). Sarcosine also connects to another coral osmolyte creatine [55]; while creatine is not a downstream metabolite of GBT, it is hydrolyzed to sarcosine and urea through creatinase (cre) and the products follow the sarcosine demethylation pathway and ureolysis (ureABCDEF), respectively (Fig. 3).
A The minimal medium is supplemented with different substrates. (1) Control experiments. In the negative control, bacteria are cultured in the minimal medium without C and N source (open circles). In the positive control, bacteria are cultured in the rich medium, in which the peptone, glucose, and yeast extracts are added as mixed C and N sources (open triangles). Growth experiments of the six strains on 5 mM of choline (2), GBT (3), DMG (4), sarcosine (5), TMA (6), TMAO (7), creatine (8) and urea (9), each used as a sole C and N source (solid triangles). Three replicates are performed for each condition and error bars denote standard deviation. B Different substrates are used as either a sole C source or a sole N source. (1) Control experiments. In the negative control, bacteria are cultured in the minimal medium without C and N source added (open circles). In the positive control, bacteria are cultured using pyruvate as the C source and ammonium as the N source, respectively (open triangles). Growth experiments of the six strains on 5 mM of choline (2), GBT (4), TMA (6), TMAO (8), and urea (10) as the sole C source and each with ammonium (10 mM) as the N source (solid triangles). Growth experiments of six strains on 5 mM of choline (3), glycine betaine (5), TMA (7), TMAO (9), and urea (11) as the sole N source and each with pyruvate (5 mM) as the C source (solid triangles). Three replicates are performed for each condition and error bars denote the standard deviation.
On the other hand, all assayed isolates can use choline (Fig. 4B-3) and GBT (Fig. 4B-5) as sole N sources, suggesting that they may acquire N from choline and GBT through an alternative pathway. One possibility is that GBT is initially transformed to trimethylamine (TMA) which can be used as a sole N source (Fig. 4B-7). This reaction was experimentally demonstrated in a metabolomics study on intestinal microbiota [63], but the genetic basis has not been known (Fig. 3). TMA can be sequentially demethylated to dimethylamine (DMA) and monomethylamine (MMA) through dimethylamine/trimethylamine dehydrogenase (dmd-tmd) [64]. The C1 groups yielded by demethylation may be an energy source to the Ruegeria population, as supported by their presence of the H4F-linked C1 oxidation pathway consisting of 5,10-methylene-H4F dehydrogenase/methenyl-H4F cyclohydrolase (folD), formyl-H4F synthetase (fhs), and formate dehydrogenase (fdh) [61, 65]. However, these C1 groups cannot be used as a C source since these bacteria lack methyltransferase/corrinoid-binding protein (cmuA) as the key gene for C1 assimilation [66]. Following demethylation, MMA is further oxidized to release ammonium via a pathway comprising the gamma-glutamylmethylamide synthetase (gmaS), N-methylglutamate synthase (mgsABC), and N-methylglutamate dehydrogenase (mgdABCD) [65]. TMAO is another methylamine osmolyte that is chemically related to choline. It can be reduced to TMA through either anaerobic dimethyl sulfoxide/trimethylamine oxide reductase (dmsABC) [67] or aerobic methionine sulfoxide reductase (yedYZ) [68], and then enters the TMA demethylation pathway (Fig. 3).
While the above-mentioned genes involved in methylamine-related osmolytes are common to all members of the Ruegeria population, many of them have at least one additional copy specific to the clade-M members. The latter includes ugpABCE for GPC uptake, betABT and cki1 for choline acquisition and catabolism, proVWX for GBT uptake, cre for converting creatine to sarcosine, sardh and soxABDG for sarcosine demethylation, and gmaS and mgsABC for MMA oxidation (Fig. 3). The majority of these genes were likely acquired at the LCA of clade-M (12 out of 23) or lost at LCA of clade-S (four out of 23), while the remaining genes were acquired at the LCA of the five clade-M mucus members (seven out of 23) (Table S4). Among the core gene copies, the ugpABE, betA, dmgdh, and dmsABC were subjected to novel allelic replacements, though different component genes in each gene cluster went through the different evolutionary histories of homologous recombination (Fig. 3 and Table S6). In line with these between-clade differences at the genetic level, the clade-M members exhibited a significantly higher ability than the clade-S members to utilize the abundant osmolytes in corals, such as using choline (p < 0.05, one-way repeated-measures ANOVA; the same test used below unless stated otherwise; Fig. 4B-3) and GBT (p < 0.05; Fig. 4B-5) as a sole N source, and using sarcosine and creatine as a sole N and C source (p < 0.05 for each; Figs. 4A-5, A-8). While the osmolytes less common in corals (e.g., DMG, TMA, and TMAO) were also used by the Ruegeria population, no significant differences between clade-M and clade-S were observed (p > 0.05 for each; Figs. 4A-4, B-7, and B-9). Detailed results of the physiological assays can be found in Supplementary Text 2.2.
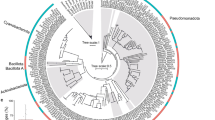
A The gene arrangement of the fla1 clusters in members of exp-clade-M and exp-clade-S. The pink arrows represent the clade-S-specific genes, and the gray arrows denote core genes shared by exp-clade-M and exp-clade-S, respectively. The pseudogenes are marked with red crosses on the arrows. B The motility assays of three clade-M strains and three clade-S strains. Swimming and swarming motilities are tested on 0.3% (w/v) soft agar and 0.6% (w/v) agar for eight days, respectively; twitching motility is tested on 1.0% (w/v) agar in the humidified box for 10 days. C An expanded genome phylogeny of the Ruegeria population based on 44 strains including those collected from various ecological niches. The exp-clade-M, exp-clade-S, and exp-outgroup expand from the clade-M, clade-S, and outgroup in Fig. 1A. Solid circles at the nodes indicate that the ultrafast bootstrap support value of the branch is 100%. The scale bar indicates the number of substitutions per site. The root of the tree is determined using a larger phylogenomic tree (Fig. S14) composed of 454 Ruegeria and related strains [86]. The strains containing the complete fla1 cluster are marked with black boxes. The strains with partial fla1 clusters are marked with white boxes, and different types of partial fla1 clusters in exp-clade-M and exp-clade-S are distinguished by the numbers in the white boxes (color figure online).
In addition to methylamines, L-proline, DMSP, taurine, and L-fucose are also important coral osmolytes [10, 56, 69, 70]. Using similar bioinformatic analyses and physiological assays, we showed that the catabolic genes of these non-methylamine coral osmolytes also differentiated the two clades (Supplementary Text 2.3) and that clade-M members grew better than clade-S members on these substrates (Fig. S11; Supplementary Text 2.2). Furthermore, the clade-M members uniquely possess metabolic potential in utilizing other mucus-related substrates (e.g., aromatics, Supplementary Text 2.4) and adapting to the densely-populated habitat (e.g., quorum sensing), which may also help the clade-M inhabiting the mucus compartment (Supplementary Text 2.5).
Metabolic adaptation of the skeleton clade to anoxic and low-energy skeleton niche
The clade-S members evolved clade-specific strategies for energy conservation in the energy-limited skeleton. Members of the Roseobacter group commonly conserve energy through the oxidation of inorganic reduced sulfur and carbon monoxide through the sox and cox gene clusters, respectively [40]. We identified a complete sox gene cluster in all members of the clade-S but in only one strain (HKCCD4884) of the clade-M (Table S5), and inferred that this sox gene cluster was lost at the LCA of clade-M. In addition, two cox gene clusters encoding the form I (HKCCD4315_03676-03684) and form II (HKCCD4315_03987-03970) carbon monoxide dehydrogenase (codh) are shared by both clades. Among these, the form I cox gene cluster (Table S6), the indispensable component for carbon monoxide oxidation [71], showed unusually large between-clade dS values, and phylogenetic analysis showed that the LCA of clade-S was subjected to divergent allele replacements at the cox gene cluster (Fig. S4 versus Fig. S8).
A second major trait unique to clade-S is motility. In the Roseobacter group, the swimming motility in liquids is widely observed, but swarming motility on solid surfaces has not been reported [72, 73]. Both swimming and swarming may be conferred by any of the three homologous flagellar gene clusters (FGCs), termed fla1, fla2, and fla3 [72, 73]. Another type of motility on the solid surface is called twitching motility, which results in a dendritic-shaped phenotype and was previously observed in seven Roseobacter species, but the genetic basis has not been clear [72]. All clade-S members except for HKCCD7318 carry the complete set of fla1 consisting of 36 genes (Fig. 5A). In contrast, two DNA segments encompassing 17 continuous genes and part of the flgI gene (Table S5) are missing from the clade-M members (Fig. 5A), and they were inferred to be lost at the LCA of clade-M. These lost genes are involved in the assembly of multiple components of the flagellum, including components of the type III secretion system (flhA, flhB, fliQ, and fliR), P- and L-rings (flgA and flgH), motor proteins (motB), basal body proteins (flgB, flgC, flgG, and fliI), hook (flgE, flgK, flgL, and fliE) and flagellum-specific ATP synthase (fliI). The strain HKCCD7318, an exception of the clade-S members, also carries a truncated fla1 cluster but encompasses five more genes encoding components of the type III secretion system and basal body compared to the clade-M members (Table S5 and Fig. 5A). Our assays showed that the clade-S members with a complete set of fla1 displayed larger swimming circles than the clade-M members and the HKCCD7318 which carried a truncated fla1 cluster (Fig. 5B). No swarming or twitching motility was observed in all tested isolates (Fig. 5B). Previous studies on Rhodobacter sphaeroides showed that the expression of the fla1 cluster is positively regulated under anaerobic conditions which could be part of the aerotaxic response to oxygen or alternative electron acceptors in response to the anoxic condition [74]. Since the coral skeleton is periodically anoxic, the maintained motility may facilitate scavenging oxygen and alternative electron acceptors in the skeleton. Related to potential adaptation to periodic anoxia in the skeleton, clade-S members further possess several pathways of anaerobic respiration (Supplementary Text 2.6).
One more interesting trait that differentiates clade-S from clade-M is the potential for urea utilization. A gene cluster encoding the urea transporter (urtABCDE, Table S5) is specific to the clade-S, and four (urtABCD) out of the five subunits were likely lost at the LCA of clade-M (Table S5). Besides, the genes (ureABCDEF, Table S6) encoding the urease for urea hydrolysis are part of the core genes showing unusually large between-clade dS values. Phylogenetic analyses suggest that either clade-S or clade-M acquired alleles from a different genetic origin, depending on the component gene (Fig. S4 versus Fig. S10). These results suggest that the clade-M and clade-S have differentiated on the urea decomposition, and the clade-S may show a higher potential to take advantage of this function as it kept the transporter for urea uptake. However, the growth assay showed that both clade-M and clade-S members grew poorly on urea under the tested conditions and did not exhibit differential growths (Figs. 4A-9, B-10, B-11). We argue that generating an N source may not be the primary purpose of the Ruegeria population to use urea. This is because urea is less likely a preferred N source due to the high energy cost in using it [75], especially when the coral skeleton is enriched with dissolved inorganic nitrogen with its concentration exceeding that in ambient seawater by a factor of 10 [76]. On the other hand, ureolysis leads to the rise of pH and carbonate concentration [77], which facilitates calcification by increasing pH and supplying carbonate in environments such as soil, estuarine, coastal seawater, animal rumen, and gut [78,79,80,81]. In the coral skeleton, the calcification process can also be enhanced by ureolysis [82, 83]. We, therefore, hypothesize that the conservation of genetic traits in urea mineralization by skeleton-associated members might be beneficial for the calcification of the coral host. Future experiments are needed to validate this hypothesis.
The evolutionary origin of the mucus and skeleton clades and the role of coral compartments in driving their divergent evolution
The metabolic differences between clade-M and clade-S provide evidence that the diversification of the two clades is likely driven by heterogeneous conditions associated with different coral compartments. However, the splitting time of the two clades was estimated to be 7.52 million years ago (Supplementary Text 1.11), indicating that the split of clade-M and clade-S far predated the birth of their host coral which likely emerged several decades to a century ago [84]. The mismatch of the two timescales thus leaves the origin of clade-M and clade-S an unanswered question: did they evolve de novo on coral, or had they already occurred in other environments before their colonization on the different compartments of the coral host?
In the former case, different compartments are the direct force driving speciation. The bacteria have a chance to undergo a long evolutionary time on corals even beyond the longevity of coral because bacteria could be transmitted vertically between coral generations (Fig. S12) [85]. In this model, the LCA shared by clade-M and clade-S colonized the ancient coral host, then the two clades diverged in situ through the acquisition or loss of metabolic traits and thrived in their preferred coral compartments along with the growth cycle of the coral host. The predominant bacterial clade in one compartment might still be able to survive in another as they are spatially close, but with a relatively lower abundance due to compartment-specific selection for its own inhabitants. Next, members of both clades have a chance to be transmitted together with the coral gametes and the planula and thrive in their optimum compartments after the coral recruit is settled and starts to secret mucus and develop the skeleton.
If the second hypothesis is true (Fig. S12), distinct coral compartments may not be the initial force driving the speciation between clade-M and clade-S. Instead, the two clades might have already diverged in other environments and repeatedly colonized different compartments of the coral host throughout their evolutionary processes. The environmental heterogeneity of different coral compartments might impose distinct selective forces on the localized bacteria and enrich the best-adapted members, and this, in turn, may accelerate the diversification of the two clades.
To help resolve these competing hypotheses, we included 24 additional closely related Ruegeria isolates. Of these, 22 inhabit a variety of ecological niches in Hong Kong including brown algal ecosystem niches (e.g., seawater, sediment, and algal tissue), mangrove rhizosphere, coastal seawater, intertidal sediments, and another batch of the P. acuta coral sample (Table S1). According to the updated phylogenomic tree (Fig. 5C), the newly added strains expand all three major clades of the Ruegeria population (Fig. 5C). Members in the expanded clade-M (hereafter “exp-clade-M”) are equally partitioned into two subclades, in which subclade-M1 is dominated by the mucus members (equivalent to the clade-M in Fig. 1A) and subclade-M2 consists of members largely from non-coral niches (Fig. 5C). This pattern prevented us from inferring the ancestral habitat of the LCA of exp-clade-M. Members in the expanded clade-S (hereafter “exp-clade-S”) are unequally grouped into two subclades, with the singleton subclade-S2 derived from seawater and the subclade-S1 composed of phylogenetically mixed strains inhabiting the coral skeleton and other non-coral niches (Fig. 5C). Thus, we cannot conclusively determine the habitat of the LCA of the exp-clade-S. In the case of the expanded outgroup clade, the coral-associated strains are embedded in the early-branching lineages that were not derived from corals (Fig. 5C), suggesting that the ancestral habitat of the LCA of this expand outgroup clade was not related to coral. When these three clades were viewed collectively, together with the deeply-branching singleton clade represented by a seawater strain Ruegeria sp. 6PALISEP08 located at the middle of the tree (Fig. 5C), the available isolates’ habitat information and phylogenetic structure are more consistent with the hypothesis that the original skeleton clade, mucus clade, and outgroup clade each independently evolved from ancestors colonizing non-coral habitats.
Since the separation between the original clade-M and clade-S was likely completed in habitats other than the coral compartments, some between-clade differences of metabolic potential could have existed before ancestral lineages were independently transited to coral mucus and skeleton. We, therefore, sought evidence that allows singling out the metabolic differences imposed by different coral compartments from those by non-coral environments. In brief, we screened the gene families that show clade-M (or clade-S) specific distribution but are not prevalent in exp-clade-M (or exp-clade-S), because the evolution of these genes is more likely driven by coral compartments. We then inferred the evolutionary history of the eligible gene families along the expanded phylogeny (Fig. 5C).
This new analysis revealed that over half of the osmolyte utilization genes specific to clade-M discussed above were acquired at the LCA of the subclade-M1 in the expanded phylogeny (Table S7). These genes include ugpABCE for GPC uptake, betAB for choline oxidation, gmaS and mgsABC for MMA oxidation, cre for creatine degradation, tauABC for taurine uptake, and dddP for DMSP lysis. This is evidence that the coral mucus environment represents an important selective force shaping the genome of mucus-associated bacteria. This expanded analysis, however, rendered the evolution of the remaining osmolyte degradation genes specific to the original clade-M uncoupled from the coral mucus habitat, since these genes are prevalent among the exp-clade-M members and were largely acquired at the LCA of exp-clade-M (Table S7). Similarly, the expanded analysis uncorrelated the evolution of urea transporter genes and sulfur oxidation genes to the coral skeleton habitat, since these genes are commonly found in the exp-clade-S members and were either acquired at the LCA of exp-clade-S or lost at the LCA of exp-clade-M (Table S8). In the case of the type I flagellar gene cluster (fla1) composed of 18 genes, it is difficult to infer its evolutionary history based on the gene presence and absence pattern. This is because while a few members in exp-clade-S including most skeleton-associated members possess a complete set of the genes, the basal branch of this clade and all members of exp-clade-M each contain a subset of the genes (Fig. 5). In the strains carrying an incomplete fla1 cluster (Fig. 5C), the genes adjacent to the missing part of the fla1 were mostly pseudogenized (Fig. 5A). Because pseudogenization is unequivocal evidence of ongoing gene loss [86], the missing part of fla1 is more likely a result of gene loss (Table S8). Therefore, the conservation of a complete fla1 cluster in most skeleton-associated members is evidence that coral skeleton acts as a selective force to maintain fla1 in clade-S.
Different coral compartments act as a microscale geographic barrier
For the above Ruegeria population, we provided evidence that different compartments of the coral host act as distinct selective pressures that diversify the genomes of their localized bacteria. On the other hand, the available results also suggest that this habitat-driven selection may explain only a portion of the genetic differences found between clade-M and clade-S. We next asked whether different coral compartments may act as physical barriers of gene flow, leading to the neutral diversification between the microscale geographically separated populations. Given the considerable amount of genetic diversity harbored in the Ruegeria population, however, it is not amenable to use this population to test the neutral mechanism. We, therefore, turned to a genetically monomorphic Rhodobacteraceae population, which is composed of 214 isolates sampled from four different coral individuals of the same coral species P. acuta (Table S1), each collected from a different location in Hong Kong (Fig. S1A). Among the 214 strains, 129, 60, 22, and three were cultured from coral mucus, skeleton, tissue, and the ambient seawater, respectively (Table S1). Regarding the sampling location, 109 isolates were from Wong Wan Chau (WWC), 87 from Ngo Mei Chau (NMC), 17 from Check Chau, and one from Kiu Tsui Chau (Table S1). Of these, only the WWC and NMC subpopulations covered multiple compartments. This new population represents a distinct lineage from all known roseobacters (Fig. S4), showing an identity of 97.7% at the 16S rRNA gene to its closest relative, Rhodobacteraceae bacterium MA-7-27 (GenBank Assembly Accession Number: GCA_003688285.1). Members of this population share identical 16S rRNA genes, show the pairwise ANI at ~99.99% (Table 1) and differ by only 43 SNPs across around 4.39 Mbp core genome sequences shared by the 214 strains according to kSNP3 [87] (Table 1). Phylogenomic construction based on these SNPs showed that the isolates are not clustered according to the coral compartments or the sampling locations (Fig. S13). The extremely high genetic identity among isolates indicates a very recent origin of this population, only 130 and 94 years ago estimated for the WWC and the NMC subpopulations, respectively (Supplementary text 1.11), which likely spanned only one or a few generations of the coral animals in Hong Kong waters [84]. These results suggest that members of each subpopulation, despite currently distributed among multiple coral compartments (Fig. 6A, B), likely evolved from an ancestral bacterium that colonized one of the compartments. These subpopulations thus provide a unique opportunity to test whether dispersal limitation between coral compartments acts as a neutral force driving population differentiation. The Slatkin–Maddison test (see “Methods”) showed that in both subpopulations fewer migrations were inferred than expected by chance (29 migrations in the WWC subpopulation, p < 0.01, Fig. 6C; 12 migrations in the NMC subpopulation, p < 0.001, Fig. 6D), indicating that members from distinct compartments of the same coral individual are compartmentalized and that differentiation of this unclassified Rhodobacteraceae population likely started from limited migration between coral compartments.
A, B Phylogenomic trees constructed by IQ-TREE v1.6.5 using the core SNPs identified with kSNP3 for the population associated with the coral individual collected at Wong Wan Chau and the other associated with the coral individual at Ngo Mei Chau. Strains isolated from different coral compartments are highlighted with different colors. Solid circles at the nodes indicate that the ultrafast bootstrap support value of the branch is ≥80%. The scale bar indicates the number of SNPs per variable site. C, D The frequency distribution of the number of migrations required to produce each of the 100,000 permuted trees in the two subpopulations. The blue arrow denotes the inferred number of migrations based on the real data (color figure online).
Concluding remarks
Stony corals contain three major compartments: mucus, tissue, and skeleton. They represent neighboring but physicochemically distinct habitats for microbes and thus are excellent models to investigate key mechanisms of microbial evolution such as natural selection and dispersal limitation. Of the two Rhodobacteraceae populations examined here, the Ruegeria population, while represented by only 20 isolates, contains a substantial amount of genetic variation and has already diverged into two distinct species primarily colonizing the coral mucus and skeleton respectively. While the mucus clade (clade-M) acquired novel genes to make use of methylamines and other coral osmolytes, the skeleton clade (clade-S) exclusively maintained a few metabolic traits of vital importance to survive in skeletons, such as potential oxidation of inorganic sulfur through sox, swimming motility, and potential urea utilization to facilitate skeleton calcification. Although some traits may have diversified before their colonization of the coral host, the differentiation in motility and in the utilization of abundant coral osmolytes is evidence that skeleton and mucus environments impose distinct selective pressures that shape the divergent evolution of the Ruegeria population. In contrast, the unassigned Rhodobacteraceae population sampled much more isolates (n = 214) than the Ruegeria population, but it carries only a few dozen SNP sites across the whole genomes. This extreme genetic monomorphism enabled the use of a statistical phylogenetic approach to conclude that the coral compartments act as a physical barrier of bacterial gene flow, suggesting that neutral divergence between populations from different compartments is an important force of bacterial evolution in coral habitats. While this neutral mechanism is revealed through the analysis of the unassigned Rhodobacteraceae population, it may be a prevalent mechanism and may in part account for some of the genetic differences found between clade-M and clade-S that does not result from historical events (e.g., some differences traced back to the time when the two clades diverged in non-coral environments) or that cannot be ascribed to adaptation to distinct physicochemical conditions in mucus and skeleton.
While we have been trying to emphasize the benefit of using coral as a natural laboratory to study mechanisms of bacterial evolution, the knowledge gained through our analyses may in turn help improve our understanding of the coral host. For example, clade-M members exclusively contain the protocatechuate pathway (Table S4) for degradation of aromatic compounds, a common type of pollutants in Hong Kong as one of the busiest seaports in the world, suggesting that the clade-M members may help remove some potentially toxic aromatics trapped in the coral mucus (Supplementary Text 2.4). Clade-S members possess the genetic potential to acquire and decompose urea, which might confer benefits to the coral host by promoting calcification of the skeleton when the environment acidifies. These predictions, if confirmed, would be the building blocks to further understand the whole nature of the bacteria-coral association and would help to improve our strategies in coral conservation by more carefully considering the role of bacteria in maintaining the health of coral holobiont.
Data availability
The assembled genomic sequences and raw reads of the coral-associated Ruegeria population, the seven Ruegeria isolates from non-coral marine habitats, and the coral-associated unassigned Rhodobacteraceae population are made publicly available at NCBI under GenBank assembly accession number PRJNA596594, PRJNA682389, and PRJNA596592, respectively.
References
Bourne DG, Morrow KM, Webster NS. Insights into the coral microbiome: underpinning the health and resilience of reef ecosystems. Annu Rev Microbiol. 2016;70:317–40.
Ainsworth TD, Thurber RV, Gates RD. The future of coral reefs: a microbial perspective. Trends Ecol Evol. 2010;25:233–40.
Huettel M, Wild C, Gonelli S. Mucus trap in coral reefs: formation and temporal evolution of particle aggregates caused by coral mucus. Mar Ecol Prog Ser. 2006;307:69–84.
Coffroth M. Mucous sheet formation on poritid corals: an evaluation of coral mucus as a nutrient source on reefs. Mar Biol. 1990;105:39–49.
Brown BE, Bythell JC. Perspectives on mucus secretion in reef corals. Mar Ecol Prog Ser. 2005;296:291–309.
Sweet M, Croquer A, Bythell J. Bacterial assemblages differ between compartments within the coral holobiont. Coral Reefs. 2011;30:39–52.
Yancey PH. Organic osmolytes as compatible, metabolic and counteracting cytoprotectants in high osmolarity and other stresses. J Exp Biol. 2005;208:2819–30.
Burg MB, Ferraris JD. Intracellular organic osmolytes: function and regulation. J Biol Chem. 2008;283:7309–13.
Singh LR, Dar TA, editors. Cellular osmolytes: from chaperoning protein folding to clinical perspectives. 1st ed. Singapore: Springer Nature Singapore Pte Ltd.; 2017.
Yancey PH, Heppenstall M, Ly S, Andrell RM, Gates RD, Carter VL, et al. Betaines and dimethylsulfoniopropionate as major osmolytes in cnidaria with endosymbiotic dinoflagellates. Physiol Biochem Zool. 2010;83:167–73.
Mayfield AB, Gates RD. Osmoregulation in anthozoan—dinoflagellate symbiosis. Compar Biochem Physiol A. 2007;147:1–10.
Rublee PA, Lasker HR, Gottfried M, Roman MR. Production and bacterial colonization of mucus from the soft coral Briarium asbestinum. Bull Mar Sci. 1980;30:888–93.
Wild C, Woyt H, Huettel M. Influence of coral mucus on nutrient fluxes in carbonate sands. Mar Ecol Prog Ser. 2005;287:87–98.
Coles SL, Strathmann R. Observations on coral mucus “flocs” and their potential trophic significance. Limnol Oceanogr. 1973;18:673–8.
Pernice M, Raina J-B, Rädecker N, Cárdenas A, Pogoreutz C, Voolstra CR. Down to the bone: the role of overlooked endolithic microbiomes in reef coral health. ISME J. 2020;14:325–34.
Falini G, Fermani S, Goffredo S. Coral biomineralization: a focus on intra-skeletal organic matrix and calcification. Semin Cell Dev Biol. 2015;46:17–26.
Constantz B, Weiner S. Acidic macromolecules associated with the mineral phase of scleractinian coral skeletons. J Exp Zool. 1988;248:253–8.
Muscatine L, Goiran C, Land L, Jaubert J, Cuif JP, Allemand D. Stable isotopes (delta C-13 and delta N-15) of organic matrix from coral skeleton. Proc Natl Acad Sci USA. 2005;102:1525–30.
Sorek M, Díaz-Almeyda EM, Medina M, Levy O. Circadian clocks in symbiotic corals: the duet between Symbiodinium algae and their coral host. Mar Genom. 2014;14:47–57.
Agostini S, Suzuki Y, Higuchi T, Casareto B, Yoshinaga K, Nakano Y, et al. Biological and chemical characteristics of the coral gastric cavity. Coral Reefs. 2012;31:147–56.
Rosenberg E, Koren O, Reshef L, Efrony R, Zilber-Rosenberg I. The role of microorganisms in coral health, disease and evolution. Nat Rev Microbiol. 2007;5:355–62.
Ritchie KB. Bacterial symbionts of corals and Symbiodinium. In: Rosenberg E, Gophna U editors. Beneficial microorganisms in multicellular life forms. 1st ed. Berlin Heidelberg: Springer-Verlag Berlin Heidelberg; 2012. pp 139–50.
Apprill A, Weber LG, Santoro AE. Distinguishing between microbial habitats unravels ecological complexity in coral microbiomes. mSystems. 2016;1:e00143–00116.
Pollock FJ, McMinds R, Smith S, Bourne DG, Willis BL, Medina M, et al. Coral-associated bacteria demonstrate phylosymbiosis and cophylogeny. Nat Commun. 2018;9:1–13.
Shapiro BJ, Friedman J, Cordero OX, Preheim SP, Timberlake SC, Szabó G, et al. Population genomics of early events in the ecological differentiation of bacteria. Science. 2012;336:48–51.
Youngblut ND, Wirth JS, Henriksen JR, Smith M, Simon H, Metcalf WW, et al. Genomic and phenotypic differentiation among Methanosarcina mazei populations from Columbia River sediment. ISME J. 2015;9:2191–205.
Wielgoss S, Didelot X, Chaudhuri RR, Liu X, Weedall GD, Velicer GJ, et al. A barrier to homologous recombination between sympatric strains of the cooperative soil bacterium Myxococcus xanthus. ISME J. 2016;10:2468–77.
Chase AB, Arevalo P, Brodie EL, Polz MF, Karaoz U, Martiny JB. Maintenance of sympatric and allopatric populations in free-living terrestrial bacteria. Mbio. 2019;10:e02361–02319.
Huggett MJ, Apprill A. Coral microbiome database: integration of sequences reveals high diversity and relatedness of coral-associated microbes. Environ Microbiol Rep. 2019;11:372–85.
Apprill A, Marlow HQ, Martindale MQ, Rappe MS. The onset of microbial associations in the coral Pocillopora meandrina. ISME J. 2009;3:685–99.
Epstein HE, Torda G, Munday PL, van Oppen MJH. Parental and early life stage environments drive establishment of bacterial and dinoflagellate communities in a common coral. ISME J. 2019;13:1635–8.
Freire I, Gutner-Hoch E, Muras A, Benayahu Y, Otero A. The effect of bacteria on planula-larvae settlement and metamorphosis in the octocoral Rhytisma fulvum fulvum. PLoS ONE. 2019;14:e0223214.
Miura N, Motone K, Takagi T, Aburaya S, Watanabe S, Aoki W, et al. Ruegeria sp. strains isolated from the reef-building coral Galaxea fascicularis inhibit growth of the temperature-dependent pathogen Vibrio coralliilyticus. Mar Biotechnol. 2019;21:1–8.
Apprill A, Hughen K, Mincer T. Major similarities in the bacterial communities associated with lesioned and healthy Fungiidae corals. Environ Microbiol. 2013;15:2063–72.
Sekar R, Kaczmarsky LT, Richardson LL. Microbial community composition of black band disease on the coral host Siderastrea siderea from three regions of the wider Caribbean. Mar Ecol Prog Ser. 2008;362:85–98.
Casey JM, Connolly SR, Ainsworth TD. Coral transplantation triggers shift in microbiome and promotion of coral disease associated potential pathogens. Sci Rep. 2015;5:11903–11903.
Tsang RHL, Ang PO. Resistance to temperature stress and Drupella corallivory may promote the dominance of Platygyra acuta in the marginal coral communities in Hong Kong. Mar Environ Res. 2019;144:20–27.
Tam TW, Ang PO Jr. Repeated physical disturbances and the stability of sub‐tropical coral communities in Hong Kong, China. Aquat Conserv. 2008;18:1005–24.
Ang Jr PO, Choi LS, Choi MM, Cornish A, Fung HL, Lee MW et al. Hong Kong. In: Centre JWR editors. Status of coral reefs of the East Asian Seas region: 2004. Tokyo: Ministry of the Environment; 2005. pp 121–52.
Luo H, Moran MA. Evolutionary ecology of the marine Roseobacter clade. Microbiol Mol Biol Rev. 2014;78:573–87.
Pujalte MJ, Lucena T, Ruvira MA, Arahal DR, Macián MC. The family Rhodobacteraceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F editors. The Prokaryotes: alphaproteobacteria and Betaproteobacteria. 4th ed. Berlin Heidelberg: Springer-Verlag Berlin Heidelberg; 2014. pp 439–512.
Johannes RE, Wiebe WJ. Method for determination of coral tissue biomass and composition. Limnol Oceanogr. 1970;15:822–4.
Emms DM, Kelly S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019;20:1–14.
Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–74.
Achtman M, Wagner M. Microbial diversity and the genetic nature of microbial species. Nat Rev Microbiol. 2008;6:431–40.
Feil EJ, Spratt BG. Recombination and the population structures of bacterial pathogens. Annu Rev Microbiol. 2001;55:561–90.
Wang X, Zhang Y, Ren M, Xia T, Chu X, Liu C, et al. Cryptic speciation of a pelagic Roseobacter population varying at a few thousand nucleotide sites. ISME J. 2020;14:3106–19.
Lawson DJ, Hellenthal G, Myers S, Falush D. Inference of population structure using dense haplotype data. PLoS Genet. 2012;8:e1002453.
Didelot X, Wilson DJ. ClonalFrameML: efficient inference of recombination in whole bacterial genomes. PLoS Comput Biol. 2015;11:e1004041.
Excoffier L, Lischer HE. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–7.
Sun Y, Luo H. Homologous recombination in core genomes facilitates marine bacterial adaptation. Appl Environ Microbiol. 2018;84:e02545–02517.
Librado P, Vieira FG, Rozas J. BadiRate: estimating family turnover rates by likelihood-based methods. Bioinformatics. 2011;28:279–81.
Slatkin M, Maddison WP. A cladistic measure of gene flow inferred from the phylogenies of alleles. Genetics 1989;123:603–13.
Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun. 2018;9:1–8.
Lohr KE, Khattri RB, Guingab-Cagmat J, Camp EF, Merritt ME, Garrett TJ, et al. Metabolomic profiles differ among unique genotypes of a threatened Caribbean coral. Sci Rep. 2019;9:1–11.
Hill R, Li C, Jones A, Gunn J, Frade P. Abundant betaines in reef-building corals and ecological indicators of a photoprotective role. Coral Reefs. 2010;29:869–80.
Gowrishankar J. Nucleotide sequence of the osmoregulatory proU operon of Escherichia coli. J Bacteriol. 1989;171:1923–31.
Chandravanshi M, Gogoi P, Kanaujia SP. Computational characterization of TTHA0379: A potential glycerophosphocholine binding protein of Ugp ATP-binding cassette transporter. Gene. 2016;592:260–8.
Ziegler C, Bremer E, Krämer R. The BCCT family of carriers: from physiology to crystal structure. Mol Microbiol. 2010;78:13–34.
Geiger O, López-Lara IM, Sohlenkamp C. Phosphatidylcholine biosynthesis and function in bacteria. Biochim Biophys Acta Mol Cell Biol Lipids. 2013;1831:503–13.
Lidbury I, Kimberley G, Scanlan DJ, Murrell JC, Chen Y. Comparative genomics and mutagenesis analyses of choline metabolism in the marine Roseobacter clade. Environ Microbiol. 2015;17:5048–62.
Thole S, Kalhoefer D, Voget S, Berger M, Engelhardt T, Liesegang H, et al. Phaeobacter gallaeciensis genomes from globally opposite locations reveal high similarity of adaptation to surface life. ISME J. 2012;6:2229–44.
Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, DuGar B, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63.
Jones M, Talfournier F, Bobrov A, Grossmann JG, Vekshin N, Sutcliffe MJ, et al. Electron transfer and conformational change in complexes of trimethylamine dehydrogenase and electron transferring flavoprotein. J Biol Chem. 2002;277:8457–65.
Chen Y. Comparative genomics of methylated amine utilization by marine Roseobacter clade bacteria and development of functional gene markers (tmm, gmaS). Environ Microbiol. 2012;14:2308–22.
Schäfer H, McDonald IR, Nightingale PD, Murrell JC. Evidence for the presence of a CmuA methyltransferase pathway in novel marine methyl halide‐oxidizing bacteria. Environ Microbiol. 2005;7:839–52.
McNicholas PM, Chiang RC, Gunsalus RP. Anaerobic regulation of the Escherichia coli dmsABC operon requires the molybdate‐responsive regulator ModE. Mol Microbiol. 1998;27:197–208.
Loschi L, Brokx SJ, Hills TL, Zhang G, Bertero MG, Lovering AL, et al. Structural and biochemical identification of a novel bacterial oxidoreductase. J Biol Chem. 2004;279:50391–50400.
Hillyer KE, Dias DA, Lutz A, Wilkinson SP, Roessner U, Davy SK. Metabolite profiling of symbiont and host during thermal stress and bleaching in the coral Acropora aspera. Coral Reefs. 2017;36:105–18.
Rösgen J. Molecular basis of osmolyte effects on protein and metabolites. Methods Enzymol. 2007;428:459–86.
Cunliffe M. Correlating carbon monoxide oxidation with cox genes in the abundant marine Roseobacter clade. ISME J. 2011;5:685–91.
Bartling P, Vollmers J, Petersen J. The first world swimming championships of Roseobacters—phylogenomic insights into an exceptional motility phenotype. Syst Appl Microbiol. 2018;41:544–54.
Michael V, Frank O, Bartling P, Scheuner C, Goker M, Brinkmann H, et al. Biofilm plasmids with a rhamnose operon are widely distributed determinants of the ‘swim-or-stick’ lifestyle in roseobacters. ISME J. 2016;10:2498–513.
Armitage JP. Behavioural responses of bacteria to light and oxygen. Arch Microbiol. 1997;168:249–61.
Jorgensen NOG. Uptake of urea by estuarine bacteria. Aquat Micro Ecol. 2006;42:227–42.
Pernice M, Raina J-B, Rädecker N, Cárdenas A, Pogoreutz C, Voolstra CR. Down to the bone: the role of overlooked endolithic microbiomes in reef coral health. ISME J. 2019: 1–10.
Krajewska B, Ureases I. Functional, catalytic and kinetic properties: a review. J Mol Catal B Enzym. 2009;59:9–21.
Cheng L, Cord-Ruwisch R. In situ soil cementation with ureolytic bacteria by surface percolation. Ecol Eng. 2012;42:64–72.
Cho BC, Park MG, Shim JH, Azam F. Significance of bacteria in urea dynamics in coastal surface waters. Mar Ecol Prog Ser. 1996;142:19–26.
Jin D, Zhao SG, Zheng N, Beckers Y, Wang JQ. Urea metabolism and regulation by rumen bacterial urease in ruminants—a review. Ann Anim Sci. 2018;18:303–18.
Collier JL, Baker KM, Bell SL. Diversity of urea-degrading microorganisms in open-ocean and estuarine planktonic communities. Environ Microbiol. 2009;11:3118–31.
Biscéré T, Ferrier-Pagès C, Grover R, Gilbert A, Rottier C, Wright A, et al. Enhancement of coral calcification via the interplay of nickel and urease. Aquat Toxicol. 2018;200:247–56.
Crossland C, Barnes D. The role of metabolic nitrogen in coral calcification. Mar Biol. 1974;28:325–32.
Goodkin NF, Switzer AD, Mccorry D, Devantier L, True J, Hughen KA, et al. Coral communities of Hong Kong: long-lived corals in a marginal reef environment. Mar Ecol Prog Ser. 2011;426:185–96.
Bernasconi R, Stat M, Koenders A, Paparini A, Bunce M, Huggett MJ. Establishment of coral-bacteria symbioses reveal changes in the core bacterial community with host ontogeny. Front Mirobiol. 2019;10:1529.
Chu X, Li S, Wang S, Luo D, Luo H. Gene loss through pseudogenization contributes to the ecological diversification of a generalist Roseobacter lineage. ISME J. 2020;15:489–502.
Gardner SN, Slezak T, Hall BG. kSNP3. 0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics. 2015;31:2877–8.
Krzywinski M, Schein JE, Birol I, Connors JM, Gascoyne RD, Horsman D, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–45.
Acknowledgements
We thank Ryan Ho-Leung Tsang for the coral sample collection, Tsz-Yan Ng for guiding coral sample processing, Xiao Chu and Shuangfei Zhang for their help on bacteria cultivation, Hao Zhang and Minglei Ren for their help in data analysis, and Xinqin Lin for her advice in experimental design. We thank Xiao Chu, Minglei Ren, and Zhichao Zhou for providing isolates from the brown algae ecosystem, sediments, and mangrove ecosystem. This work was supported by the Shenzhen Science and Technology Committee (JCYJ20180508161811899), the National Natural Science Foundation of China (41776129), the Hong Kong Environment and Conservation Fund (15/2016), the Hong Kong Research Grants Council General Research Fund (14163917), and the Hong Kong Research Grants Council Area of Excellence Scheme (AoE/M-403/16).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Luo, D., Wang, X., Feng, X. et al. Population differentiation of Rhodobacteraceae along with coral compartments. ISME J 15, 3286–3302 (2021). https://doi.org/10.1038/s41396-021-01009-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-021-01009-6
This article is cited by
-
The coral microbiome in sickness, in health and in a changing world
Nature Reviews Microbiology (2024)
-
Probiotics reshape the coral microbiome in situ without detectable off-target effects in the surrounding environment
Communications Biology (2024)