Abstract
Fungal communities associated with plants often decrease in similarity as the distance between sampling sites increases (i.e., they demonstrate distance decay). In the southwestern USA, forests occur in highlands separated from one another by warmer, drier biomes with plant and fungal communities that differ from those at higher elevations. These disjunct forests are broadly similar in climate to one another, offering an opportunity to examine drivers of distance decay in plant-associated fungi across multiple ecologically similar yet geographically disparate landscapes. We examined ectomycorrhizal and foliar endophytic fungi associated with a dominant forest tree (Pinus ponderosa) in forests across ca. 550 km of geographic distance from northwestern to southeastern Arizona (USA). Both guilds of fungi showed distance decay, but drivers differed for each: ectomycorrhizal fungi are constrained primarily by dispersal limitation, whereas foliar endophytes are constrained by specific environmental conditions. Most ectomycorrhizal fungi were found in only a single forested area, as were many endophytic fungi. Such regional-scale perspectives are needed for baseline estimates of fungal diversity associated with forest trees at a landscape scale, with attention to the sensitivity of different guilds of fungal symbionts to decreasing areas of suitable habitat, increasing disturbance, and related impacts of climate change.
Similar content being viewed by others
Introduction
The long association between plants and fungi has linked their ecology and evolution for more than 500 million years and has led to the evolution of multiple, plant-associated guilds across the fungal tree of life [1]. Plant-symbiotic fungi—those that live for at least part of their life cycles in affiliation with living plants—occur in association with all plants, in all plant tissues, and in all ecosystems studied to date, echoing their roles in facilitating the colonization of land and the subsequent diversification of plant lineages [1,2,3].
Guilds of symbiotic fungi such as ectomycorrhizal fungi and foliar endophytic fungi (i.e., endophytes that occur in leaves) represent different evolutionary origins and differ in how they colonize their hosts, where they are localized with regard to host tissues, how they reproduce and disperse, and their effects on host phenotypes [2,3,4]. However, both have the capacity to mediate plant health and resilience to stress. For example, certain ectomycorrhizal fungi enhance nutrient acquisition or nutrient use efficiency, increase water uptake, or regulate water use efficiency, generally to the benefit of their hosts [2]. Roles of foliar endophytes, although not as well known, range from beneficial to detrimental among the millions of species globally that occur within healthy photosynthetic tissues [3]. Case studies suggest that endophytes can influence susceptibility, tolerance, or resistance to disease; protect against herbivores; and improve plant growth under diverse stresses [5,6,7,8]. At the ecosystem level, these guilds impact plant-soil feedbacks, soil nutrient availability, carbon cycling, and plant recruitment, shaping ecosystem services provided by forests and other biomes [2, 8,9,10,11,12].
Communities of fungi associated with plants can shift in composition or function when environmental conditions change, with implications for fungal contributions to plant resilience in a rapidly changing world [7, 13]. As such, plant-symbiotic fungi are increasingly considered important when charting the future of terrestrial ecosystems under climate change [14, 15]. Understanding the distributions of plant-symbiotic fungi and their sensitivity to environmental factors is important for linking them to conservation strategies for threatened biomes. By considering both ectomycorrhizal and endophytic fungi that inhabit the same trees, we can assess whether these different guilds merit distinctive conservation approaches.
In the interior west of North America, warming associated with climate change, coupled with shifts in the timing and quantity of precipitation, are anticipated to decrease snow runoff and soil moisture and to increase the frequency and intensity of wildfire [15], with impacts on plant phenology, growth, productivity, demography, and distributions [16]. When tolerable climate conditions are exceeded, plants may be extirpated, adapt, or migrate over generations to remain within their climatic tolerances [15, 16]. Responses of individual plant taxa often are distinct, reflecting niche breadth, dispersal ability, and the availability of compatible symbionts [16,17,18].
Pinus ponderosa Lawson and C. Lawson (sensu lato: Ponderosa pine, Pinaceae) is an economically and ecologically important forest tree of western North America that occurs from southern Canada to northern Mexico. Its range has been shaped by climate shifts over the long term: during the last glacial period (Wisconsin Glaciation, ca. 25,000–15,000 years BP), P. ponderosa occupied refugia in northern Mexico and the southwestern USA [19]. With subsequent warming, its range expanded northward in latitude, with southern distributions contracting and moving upward in elevation [19]. Among the currently recognized varieties of Ponderosa pine, P. ponderosa Lawson and C. Lawson var. brachyptera (Engelm.) Lemmon (southwestern ponderosa pine; hereafter, P. ponderosa) is of particular interest as it occurs in the warmest and driest parts of the range of the species (i.e., forests in Arizona, Nevada, Utah, Colorado, and New Mexico). Ponderosa pine forests of this region are increasingly threatened due to increases in wildfire frequency and intensity, pest outbreaks, decreases in precipitation, and warming [15, 17, 18, 20].
In the southwestern portion of its range (Arizona), P. ponderosa forests cover more than 1.5 million ha [21] and occur in two major landforms: a contiguous forest from north-central Arizona to the border of New Mexico along the Mogollon Rim (see ref. [22]), and forested ‘islands’—disjunct highlands separated by arid and semi-arid biomes such as desert, chaparral, woodlands, and grasslands [21]. In these forested islands, P. ponderosa forms monodominant stands or co-occurs with species such as Douglas fir (Pseudotsuga menziesii) and diverse oaks (Quercus spp.) [23, 24]. The plant and fungal communities in these forests are taxonomically and evolutionarily distinct from those in the lower-elevation biomes that separate them [2, 25, 26].
The forests in which P. ponderosa occurs across the region are broadly similar to one another in climate and in major aspects of their plant communities, offering an opportunity to examine distributions of ectomycorrhizal and foliar endophytic fungi in the same host species across multiple forested areas that are ecologically similar, yet geographically distinct. Previous surveys in forests of the region (Santa Catalina Mountains, Pinaleño Mountains) revealed that P. ponderosa hosts diverse ectomycorrhizal and foliar endophytic fungi at local scales [27, 28]. The dominant fungi in this region typically represent distinct species or genotypes relative to those in other portions of the range of Ponderosa pine (e.g., ref. [28]). However, little is known regarding the regional distributions of ectomycorrhizal or foliar endophytic fungi associated with P. ponderosa or the factors that shape those distributions at a landscape scale.
It is plausible that the long evolutionary associations of fungi with plants in general, and Pinaceae and Pinus more specifically, could select for communities of symbionts associated with roots and leaves that are relatively uniform in composition across the range of P. ponderosa. However, plant-symbiotic fungi typically differ in composition over the range of their host species, reflecting spatial variation in climate, fire history, vegetation, land use, or related factors (e.g., refs. [28,29,30]). In such situations, the decrease in community similarity of symbionts that occurs as a function of geographic distance—that is, distance decay—can be attributed to environmental differences, and conservation approaches should be attentive to maintaining host populations under markedly different environmental conditions to maintain fungal diversity at a regional scale. However, distance decay also can reflect dispersal limitation, such that communities turn over as the distance between samples increases, even if environmental conditions are relatively consistent. In that scenario, conservation approaches might focus on maintaining populations of the host species even if environmental conditions are relatively similar among them: in each, locally distinct, dispersal-limited fungal communities can persist and thus maintain regional biodiversity.
Here, we test the broad hypothesis that ectomycorrhizal fungi and foliar endophytes associated with the same host species demonstrate evidence of distance decay at a landscape scale, but that the drivers of distance decay differ for each when geographic distance, climate differences, and aspects of environmental dissimilarity are considered. We predicted that ectomycorrhizal fungi would demonstrate distance decay as a function of dispersal limitation, but that foliar endophytes would demonstrate distance decay as a function of environmental dissimilarity. Our predictions are based on the observations that ectomycorrhizal fungi typically have a limited ability to disperse, often on the scale of only one or a few meters [31,32,33]. In contrast, foliar endophytes seem to have little dispersal limitation at local to regional scales if no physical barriers are present and compatible hosts are available [26, 30, 34]. Yet endophytes often form locally distinct assemblages with turnover across geographic space [30], consistent with direct or indirect sensitivity to climate or related environmental factors.
To test these predictions, we examined ectomycorrhizal fungi and foliar endophytes associated with living, mature P. ponderosa in anciently disjunct forests of Arizona, USA. Our sampling encompassed forest ‘islands’ located across a total of more than 550 km of geographic distance and ca. 4° of latitude, from northwestern to southeastern Arizona. We first measured distance decay in these fungal communities between disjunct forests and, for ectomycorrhizal fungi, within the largest contiguous stand of Ponderosa pine in the southwest (the Mogollon Rim). We then evaluated the relevance of geographic distance and environmental factors as drivers of distance decay and identified factors relevant to diversity, community structure, and composition for each guild. For fungal communities that appeared to be dispersal-limited, we focused on local characteristics such as the degree of isolation of the forest, climate, and plant community composition. For communities limited by environmental conditions, we focused on climate and plant communities. Our work highlights the previously unknown biodiversity of ectomycorrhizal and foliar endophytic fungi in a biodiverse region threatened by climate change, reveals the breadth of symbiotic fungi with which P. ponderosa affiliates in the warmest and driest area of its range, and indicates how different guilds of symbionts associated with the same host differ in ways relevant to forest conservation and management.
Materials and methods
Sampling approach: disjunct forest areas
We collected roots of mature, healthy individuals of P. ponderosa in eight disjunct forest areas of Arizona, USA, between October 2014 and October 2018 (Hualapai Mts., Humphreys Peak, Bradshaw Mts., Mingus Mt., Santa Catalina Mts., Pinaleño Mts., Huachuca Mts., and Chiricahua Mts.) (Fig. 1; Table 1). We concurrently collected mature, healthy leaves of the same trees in six of these areas to survey foliar endophytes (2014–2017) (Table 1). Previous work revealed no evidence of interannual variation in community composition of ectomycorrhizal fungi or foliar endophytes in these forests [27,28,29].
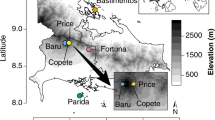
Each point represents a forest area. Each forest area comprised three sites, located 0.5–1.5 km apart. In each site, five or six trees were sampled (see text). Panels show mean annual temperature (MAT; A), mean annual precipitation (MAP; B), and elevation (masl; C). Circles indicate disjunct forest areas where only ectomycorrhizal fungi were sampled (gray circles), or both ectomycorrhizal and foliar endophytes were sampled (black circles). Boxes indicate locations in contiguous forest of the Mogollon Rim where only ectomycorrhizal fungi were sampled (gray boxes), or where both ectomycorrhizal and foliar endophytes were sampled (black box). Areas are numbered as follows, with names corresponding to Table 1: 1, Hualapai Mts.; 2, Mogollon Rim (sites 1.1–1.3); 3, Humphreys Peak; 4, Bradshaw Mts.; 5, Mingus Mt.; 6, Mogollon Rim (sites 2.1–2.3); 7, Mogollon Rim (sites 3.1–3.3); 8, Santa Catalina Mts.; 9, Pinaleño Mts.; 10, Huachuca Mts.; 11, Chiricahua Mts.
In each area, we sampled multiple trees in each of three sites. Sites were located ca. 0.5–1.5 km from each other (Table 1), represented local forest conditions in each area, and had no evidence of recent fire (i.e., within 50 years; [28, 29]) (Table 1, Supplementary Methods S1). Intersite distances were consistent with those used to define sampling replicates in previous landscape-level studies of conifers (e.g., refs. [26, 29, 30]). We obtained climate data from WorldClim based on coordinates [35] and described plant communities by observing the dominant forest canopy in each site (i.e., pine, pine-oak, and pine-Douglas fir forest; Table 1).
In each site, we sampled roots and leaves from mature trees. Initially (2014) we sampled six trees per site. Species accumulation curves [27] showed that sampling five trees per site was sufficient for capturing site-level diversity of ectomycorrhizal fungi and foliar endophytes, such that we sampled five trees per site in 2016–2018 (Table 1). Focal trees in each site were located at least 3 m apart, providing multiple independent samples per site based on sampling protocols defined previously (see refs. [28, 36]). Sampling methods are described below.
Overall, our dataset from disjunct forests represented 24 sites sampled for ectomycorrhizal fungi (123 trees). Of these, 18 sites (93 trees) also were surveyed for foliar endophytes (Table 1). Data were pooled for all trees in a site to generate site-level samples of ectomycorrhizal and endophytic fungi for analyses of distance decay and the factors relevant to fungal community structure at local scales.
Sampling approach: contiguous forest
The Mogollon Rim represents a portion of the largest contiguous forest of P. ponderosa in the southwestern USA (ca. 27,900 km2, [37]). To evaluate distance decay within a contiguous forest, we sampled roots from five trees in each of nine sites at the Mogollon Rim (i.e., Mogollon Rim 1.1–3.3; Table 1, Fig. 1). We focused only on ectomycorrhizal fungi because endophyte communities associated with a single host species typically show little evidence of distance decay at such local scales (Table 1; see refs. [27, 30, 34]). We pooled data by site and compared the nine sites to assess distance decay of ectomycorrhizal communities. We collected leaves from the same trees in three sites (Mogollon Rim 1.1–1.3; Table 1, Fig. 1) for additional insight into the environmental factors shaping endophyte communities, as described below.
Tissue collection
We collected roots as in 28. Briefly, we collected three root cores (5 cm diameter, 15 cm depth) from the canopy dripline of each tree, removing the litter layer prior to sampling. Cores were transported in a cooler at ca. 10 °C and stored at −20 °C within 48 h. When cores were processed for morphotyping (below), roots were intact, turgid, and in good condition, with no signs of damage due to storage or transport, and morphotypes were readily identifiable. Overall, our sampling included 504 root cores collected from trees in 33 sites (24 sites in disjunct forests and nine at the Mogollon Rim).
We collected mature, healthy leaves as in 28. Briefly, we used a Big Shot line launcher (SherrillTree, Greensboro, NC, USA) to catapult a rope saw over branches ca. 3–10 m above ground, and then collected 200 leaves (needles) >2 years old from lateral branches. Leaves were refrigerated (4 °C) within 24 h and processed within 72 h. When processed for endophyte surveys (below), leaves were intact and in good condition. Overall, our sampling included material from >20,000 leaves collected from trees in 21 sites (18 in disjunct forests and three at the Mogollon Rim).
Processing root samples for ectomycorrhizal fungi
Root cores were processed individually as in 28. Briefly, we washed roots, sorted colonized root tips into morphotypes based on characteristics of the fungal mantle, and chose a representative root tip from each morphotype per root core for DNA extraction [38,39,40]. Overall, we sorted 37,332 colonized root tips representing 157 of 168 trees (samples from 11 trees had no colonized roots). Root cores for each tree were processed separately, but data from cores were pooled by tree, and data from trees in each site were pooled for analyses.
We extracted genomic DNA from 1523 representative root tips. See Supplementary Methods S1 and 28 for details of DNA extraction, PCR, PCR cleanup, Sanger sequencing, sequence editing, and confirmation that roots represented P. ponderosa. We sequenced ITSrDNA-partial LSUrDNA successfully for 1481 (97%) of 1523 representative root tips. Before analysis we added 151 sequences from previous sampling at the Santa Catalina Mts. [27]. We clustered ITSrDNA-partial LSUrDNA into operational taxonomic units (OTUs) at 97% sequence similarity with the Mobyle SNAP Workbench [41,42,43]. The final data set for ectomycorrhizal fungi comprised 281 OTUs. All were included in analyses of richness and diversity. For analyses of distance decay and community composition we used OTUs observed ≥4 times (247 OTUs) to avoid biasing our results with rare species or minor sequencing errors.
Processing leaf samples for foliar endophytic fungi: culture-based approach
We washed leaves from each tree under tap water and cut them into ca. 2 mm segments, which were surface-sterilized following 45. We haphazardly selected two sets of 96 segments from each sample: one for culturing, and one for culture-free analyses. Culturing can reveal taxa not observed readily by culture-free methods (e.g., refs. [9, 31, 45, 46]) but typically results in a smaller data set that may have limited inferential power for landscape-level studies. In turn, culture-free approaches often identify taxa not isolated in culture (e.g., ref. [30]) and may capture more strictly symbiotic fungi relative to the facultative saprotrophs that can dominate culture-based surveys.
We placed each of the 96 leaf segments from each tree onto 2% malt extract agar in an individual 1.7 ml microcentrifuge tube under sterile conditions [46]. We processed 10,368 leaf segments in total for the culture-based survey. After 3 months at room temperature, emergent endophytes were isolated into axenic culture, vouchered in sterile water, and accessioned at the Robert L. Gilbertson Mycological Herbarium (ARIZ; Supplementary Table S1). Endophytes were obtained in culture from 1434 leaf segments (isolation frequency, 13.8%). We sequenced ITSrDNA- partial LSUrDNA successfully for 1307 (91%) of these cultures via the Sanger platform. Details of DNA extraction, PCR, sequencing, and data processing are in Supplementary Methods S1. We added 66 sequences to the data set before OTU clustering, representing previous surveys in the Santa Catalina Mts. [27]. We clustered sequence data into OTUs at 95% sequence similarity with the SNAP Workbench [43, 47], both independent of the culture-free data and with the culture-free data for direct comparisons (below). Cultured endophytes represented 98 OTUs, of which 43 occurred more than once (i.e., were non-singletons).
Processing leaf samples for foliar endophytic fungi: culture-free approach
We extracted total genomic DNA from four sets of 24 leaf segments per tree (i.e., 96 segments per tree) with the PowerPlant Pro Kit (Qiagen, USA) following [48]. We used a dual-barcoded two-step process for library preparation [11, 30, 49, 50]. Briefly, we amplified ITSrDNA in an initial PCR (PCR1) with primers ITS1F and ITS4. We used products as template in a second PCR (PCR2), in which we added Illumina adapters and sample-specific barcodes (IBEST Genomics Resource Core, Moscow, ID, USA) [51, 52]. Details of tissue handling, protocols, DNA quality assessments, negative controls, and mock communities are in Supplementary Methods S1 and Supplementary Table S2.
We normalized and pooled all samples at equimolar concentrations before submitting them to the IBEST Genomics Resources Core for paired-end sequencing with the Reagent Kit v3 (2 × 300 bp) on an Illumina MiSeq (see Supplementary Methods S1 for details). We processed ITS2 of 2,800,421 raw reads with USEARCH [53]. Based on quality assessments and these analyses of mock communities, we trimmed and filtered reads (2017 Illumina: 230 bp; 2019 Illumina: 180 bp; maxEE = 1.0), yielding 844,081 high quality reads overall (see refs. [54, 55]).
Analysis of mock communities (Supplementary Table S2) informed the threshold at which OTU boundaries were defined (95% sequence similarity) and identified potentially spurious OTUs (those with ≤8 occurrences). Evaluation of mock communities also led us to rarify reads with coverage-based rarefaction to minimize uneven coverage of samples without underestimating richness [56, 57]. To remove potential contaminants, we subtracted the number of reads per OTU in negative controls and extraction blanks, removing OTUs in which the negative controls had a higher read number than samples [58]. The final culture-free data set for endophytes comprised 672 OTUs, including 268 OTUs with ≥8 occurrences. Examination of mock communities showed that the same taxa were recovered from both Illumina runs, but that relative abundances differed between runs. We therefore used presence–absence data for community analyses of the culture-free data set. We compared Sanger sequences from cultured endophytes and Illumina sequences from the culture-free endophyte survey as described in Supplementary Methods S1.
Taxonomic identification and phylogenetic inference
Taxonomic placement for sequence data was estimated by querying GenBank [59] with validation for Basidiomycota in UNITE [60] and taxonomic parsing by MEGAN5 [61]. Sanger sequence data representing Ascomycota (20.5% of ectomycorrhizal fungi; 88.9% of cultured endophytes) were placed phylogenetically with the Tree-Based Alignment Selector Toolkit (T-BAS) v2.1 [62] (Supplementary Methods S1).
Measurements of distance decay: disjunct forests
After examining normality and heterogeneity of variance, we used nonparametric Spearman’s rho to assess correlations between pairwise community dissimilarity and pairwise geographic distance between sites. Data for ectomycorrhizal fungi, endophytes isolated in culture, and endophytes observed via the culture-free approach were analyzed separately. We used the Jaccard (presence–absence data) and Morisita–Horn (abundance data) dissimilarity indices for ectomycorrhizal fungi and cultured endophytes, and the Jaccard dissimilarity index for the culture-free survey of endophytes. For ectomycorrhizal fungi and cultured endophytes, we included OTUs with ≥4 occurrences; for culture-free endophyte data, we included OTUs with ≥8 occurrences. We tested a range of cutoff thresholds for each community; our results remained consistent, even when rarely observed OTUs were included (Supplementary Table S3). We included sites in all areas except the Mogollon Rim (i.e., we considered sites in disjunct forest areas only: eight for ectomycorrhizal fungi and six for endophytic fungi). For local measures of distance decay, we compared sites within each isolated forest area. For regional measures, we compared sites in different forest areas, not including within-area comparisons. For overall measures, we considered local and regional comparisons.
Measurements of distance decay: contiguous forest
We used the above approach to examine correlations between pairwise community dissimilarity and pairwise geographic distance between sites in the Mogollon Rim area, focusing only on ectomycorrhizal communities in nine sites. We used the Jaccard (presence–absence data) and Morisita–Horn (abundance data) dissimilarity indices and included OTUs with ≥4 occurrences.
Identifying drivers of distance decay
We first used partial Mantel tests to evaluate whether distance decay reflected geographic distance alone or was consistent with differences in environmental factors (Table 2). We defined environmental factors as the plant community (Table 1) and climate (see Tables 1 and 2) [63]. We examined four climate measures relevant to the region: mean annual precipitation (MAP), mean annual temperature (MAT), mean precipitation during the wettest quarter (MPWQ), and mean precipitation during the driest quarter (MPDQ) (Table 1). To account for covariation and multidimensionality we generated a single environmental eigenvector from a principal component analysis (PCA) of climate and plant community (62.9% of variation explained). We then repeated the PCA without the plant community to isolate the effects of climate (MAP, MAT, MPWQ, and MPDQ as a single eigenvector; 53.6% of variation explained). In both approaches the first principal component was used to create distance matrices based on Euclidean distances, defined as environmental dissimilarity or climate dissimilarity, respectively.
To overcome limitations of Mantel tests, we complemented our analyses with variation partitioning that accounted for spatial autocorrelation via Moran’s eigenvector map method [63,64,65]. This allowed us to focus on how differences in plant communities, which reflect edaphic and local factors, may be relevant to distance decay as compared to climate alone.
Factors relevant to ectomycorrhizal diversity and community structure
To understand factors relevant to the diversity and structure of ectomycorrhizal communities at local scales, we examined environmental factors (plant community and climate, per above) and isolation of the forest area (defined as the distance to the closest Ponderosa pine forest, measured in km). We used MAP to represent climate because MAT, MPWQ, MPDQ, and the eigenvector representing climate in the partial Mantel analyses were correlated with distance to the closest Ponderosa pine forest (Supplementary Table S4). Diversity (Fisher’s alpha) was log-transformed prior to analysis. We used non-metric multidimensional scaling (NMDS) to visualize community structure and permutational analysis of variance (PERMANOVA) to quantify variation.
We examined variation in taxonomic composition at the class and order levels with multiple regression on distance matrices (MRM) [66]. We created a distance matrix with the Gower metric for plant community data [67] and Euclidean distance matrices for distance to the closest Ponderosa pine forest and MAP. For ordination and taxonomic analyses, we considered OTUs, orders, and classes with ≥4 occurrences.
Factors relevant to endophyte diversity and community structure
We examined factors associated with the diversity and structure of endophyte communities, with a focus on plant community and climate. We defined climate as MAP and MPDQ, which were correlated with endophyte diversity in previous studies [46, 68] and were not correlated with one another. Because we did not detect strong evidence for dispersal limitation in the endophyte data sets at a regional scale, we did not consider isolation in terms of distance to the closest Ponderosa pine forest. Data were analyzed as above. For ordination and taxonomic analyses, we considered taxa with ≥4 occurrences (culture-based) or ≥8 occurrences (culture-free) (Supplementary Methods S1).
Results
We documented over 900 putative species of fungi in association with mature, healthy P. ponderosa across representative forests in Arizona, including diverse Basidiomycota (especially among ectomycorrhizal fungi) and Ascomycota (especially among endophytes). No species was observed both as an ectomycorrhizal fungus and an endophyte, underscoring the distinctiveness of these guilds relative to one another.
Distance decay: ectomycorrhizal communities
We observed distance decay in ectomycorrhizal communities when we considered data from sites in eight disjunct forest areas (Fig. 2, Supplementary Fig. S1). Distance decay was evident at local scales (among sites within areas), regional scales (among sites in different forest areas), and overall (including comparisons among sites within and among forest areas) (Fig. 2).
Data represent pairwise dissimilarity among sites within disjunct forests (bottom row, local scale); among sites in different areas (middle row, regional scale); and taking into account all intersite distances (local and regional, overall). Left column, ectomycorrhizal fungi; center, foliar endophytes, culture-free survey; right column, foliar endophytes, culture-based survey. Dissimilarity values are based on presence/absence data (Jaccard dissimilarity index), with 1 indicating completely distinct communities and 0 representing completely similar communities. Data from the contiguous forest of the Mogollon Rim (boxes in Fig. 1) are in Fig. 3. For site and area details see Table 1.
When we took environmental dissimilarity into account, geographic distance was significantly associated with dissimilarity (abundance data) or approached significance (presence/absence data) (Table 3, Supplementary Tables S5 and S6). When we controlled for variation due to climate, geographic distance remained significant (Table 3, Supplementary Tables S5 and S6). Together these results are consistent with distance decay resulting from dispersal limitation both within and among disjunct forests (Fig. 2). When we considered only sites in a single contiguous forest (Mogollon Rim), evidence for distance decay approached significance based on Jaccard dissimilarity but was less robust based on relative abundance data (Morisita–Horn, Fig. 3).
Ectomycorrhizal diversity, composition, and community structure
Over the entire study, we detected a mean of 4.5 OTUs (±2.9, standard deviation, SD) of ectomycorrhizal fungi per tree. Sites hosted a mean of 17.5 OTUs (±8.9), and areas hosted a mean of 51.0 OTUs (±29.0). Overall, 64.4% of ectomycorrhizal OTUs were only found in one forested area, 15.3% in three or more areas, and only one (OTU9, representing Wilcoxina) in all areas.
Communities across all sites were dominated by Basidiomycota, especially Tomentella (33.0% of root tips), Russula (30.3%), and Inocybe (25.3%). The most common orders were Thelephorales, Russulales, Pezizales, and Agaricales (Fig. 4). Ectomycorrhizal Ascomycota (23.0% of root tips) were dominated by Pezizomycetes (Fig. 5), including Tuber and Wilcoxina (19.7%). Cenococcum geophilum (Dothideomycetes) also was relatively common (3.1%).
Each point in A (diversity) and B (community structure) represents the ectomycorrhizal community associated with a single site. Abundance data (Morisita–Horn) were used for NMDS. In B, green arrow = mean annual precipitation (MAP); blue arrow = distance to the closest Ponderosa pine forest, with the scale in km shown to the right. Symbols correspond to plant communities (see Table 1). In C (taxonomy), Ascomycota are marked with ‡ and distance refers to the nearest Ponderosa pine forest (km), following B.
Inference was conducted in T-BAS [62]. Rings of metadata indicate fungal guild, area of origin, and mean precipitation during the driest quarter (MPDQ). Panels represent Pezizomycotina (A) and the most prevalent classes in that subphylum (B–F).
Diversity of ectomycorrhizal fungi at the site level was correlated positively with MAP and did not vary among plant communities or with distance the closest Ponderosa pine forest (Fig. 4, Table 4). Community structure differed among plant communities and as a function of the interaction of isolation and MAP (Fig. 4, Table 5, Supplementary Fig. S2). The relative abundance of the most common fungal orders differed with distance to the closest forest and MAP, but did not vary markedly among the plant community types surveyed here (Fig. 4). The relative abundance of the most common classes did not differ meaningfully across the study as a whole (data not shown).
Distance decay: foliar endophyte communities
Communities of endophytes observed via the culture-free approach increased in dissimilarity with increasing geographic distance between sites (Fig. 2: significant in the culture-free survey at local scales and overall, and approaching significance at the regional scale). Distance decay reflected environmental dissimilarity (i.e., dissimilarities of plant communities and climate) rather than dispersal limitation (Table 3, Supplementary Tables S5 and S6). Communities observed via culturing showed no evidence of distance decay (Fig. 2), potentially reflecting the relatively low richness observed in culture (see Discussion).
Foliar endophyte diversity, composition, and community structure
We detected a mean of 41.4 (±27.6) and 3.8 (±2.6) OTUs of foliar endophytes per tree via the culture-free and culture-based approaches, respectively. Sites hosted a mean of 81.1 (±47.0) and 11.9 (±4.4) OTUs, and areas hosted a mean of 220.0 (±145.0) and 29.0 (±6.9) OTUs as inferred by culture-free and culture-based approaches. Overall, 38.1% of OTUs in the culture-free survey, and 23.3% of OTUs in the culture-based survey, were unique to a single forested area. A total of 44.2% and 38.4% of OTUs in the culture-free and culture-based data, respectively, occurred in more than three areas. Five OTUs were found across all areas in the culture-based survey (two Leotiomycetes, two Eurotiomycetes, and one member of the Dothideomycetes) and also were found in the culture-free survey. One OTU (Eurotiomycetes) was found across all areas in the culture-free survey but was not observed by culturing.
The majority of endophytes were Ascomycota, representing Dothideomycetes, Eurotiomycetes, Leotiomycetes, Pezizomycetes, and Sordariomycetes (Figs. 5, 6). Two OTUs represented Basidiomycota, both Atractiellomycetes (Pucciniomycotina): OTU23 (1.4% of isolates in culture) and OTU3 (9.6% of isolates and listed as OTU904 in the culture-free data). The relative abundance of fungal classes was not associated robustly with MDPQ, MAP, or plant community in the culture-free data set but varied as a function of these variables in the culture-based data (Fig. 6).
Left column, culture-free survey; right column, culture-based survey. A richness; B diversity; C, D community structure; E, F taxonomy. Each point in A–D represents the endophyte community associated with a single site. In C, D green arrow = mean annual precipitation (MAP); black arrow = mean precipitation during the driest quarter (MPDQ). Plant communities are listed in Table 1. In E, F Ascomycota are marked with ‡. Atra., Atractiellomycetes, Doth., Dothideomycetes, Euro., Eurotiomycetes, Leot., Leotiomycetes, Pezi., Pezizomycetes, Sord., Sordariomycetes. E No significant relationship of relative abundance with MPDQ, MAP, or plant community in the culture-free data set. F Significant relationship in the culture-based data set, reflecting MPDQ, MAP, and plant community; MRM, F = 15.45, p = 0.0010, R2 = 0.16).
Richness of endophytes was correlated negatively with MPDQ in the culture-free survey (Fig. 6, Table 6, Supplementary Fig. S3; see Discussion). Endophyte community structure varied with plant community and precipitation (Table 7, Supplementary Fig. S3).
Discussion
Forests at a global scale have been shaped by climate over the long term, and today are shifting in composition and ecosystem services as the climate changes rapidly [14, 15]. In the southwestern USA, forests dominated by P. ponderosa are threatened by increasing disturbance, warming temperatures, and shifts in the seasonality and quantity of precipitation [69]. It is thought that P. ponderosa may retract to higher elevations in ranges in which this species does not yet populate the highest locations [70], with little capacity to cross the semi-arid lands that surround these isolated forests [71]. Against this backdrop of current climate change and historical fragmentation via range retraction, we examined ectomycorrhizal and foliar endophytic fungi from the same individual trees in some of the warmest and driest regions in which Ponderosa pine occurs.
We anticipated that ectomycorrhizal fungi, with limited dispersal, would show strong evidence of distance decay consistent with geographic distance among sites. In contrast, we anticipated that endophytic fungi would be widespread across the region but structured strongly at local scales by environmental factors. These predictions were upheld by our surveys, which illustrated that the factors governing the distributions, composition, and taxonomy of ectomycorrhizal and endophytic fungi can differ markedly, even when these fungi inhabit the same host species at a regional scale, and even the same individual trees at a local scale.
Previous work suggests that the inferences presented here should not be sensitive to interannual variation over the course of our study, nor to minor differences in the timing of sampling within the growing season (see refs. [27,28,29]). Although the methods differed somewhat for examining ectomycorrhizal and endophytic fungi, we sampled each guild to statistical completion, used methods consistent with the literature for each, and analyzed the data in parallel with multiple quality-control steps. Thus, we interpret our results as suggesting differences between the guilds that are consistent with differences in their capacity to disperse, coupled with the additional environmental exposure that endophytes experience as they land as propagules on leaves and must survive prior to infection (see also ref. [27]).
Overall, strong geographic signature in ectomycorrhizal symbioses, and strong environmental signature in endophyte symbioses, suggests that regional-scale perspectives that span a large number of disjunct sampling areas (ectomycorrhizae) and diverse environmental conditions (endophytes) are needed for conservation and management plans to include plant-symbiotic fungi. Such regional perspectives can help establish baseline estimates of fungal diversity associated with forest trees at a landscape scale and frame hypotheses regarding the sensitivity of such fungal communities to disturbance and related impacts of climate change.
Perspectives on ectomycorrhizal fungi
Dissimilarity of ectomycorrhizal fungal communities increased with increasing geographic distance between samples, consistent with their typically short dispersal distances. Distance decay of ectomycorrhizal fungi associated with disjunct forests was observed at the scale of ≤2 km, ca. 500 km, and at the overall scale of the study (≤2 to ca. 500 km) (Fig. 2, Supplementary Fig. S1). We observed weaker but detectable evidence of distance decay among samples from the Mogollon Rim (Fig. 3). Taken together our data suggest that the separation of forested ‘islands’ by lowland regions likely constrains present communities within isolated forests, which in turn have spatially structured ectomycorrhizal communities at local scales.
When we examined communities in detail to understand factors associated with their structure, we found that ectomycorrhizal fungal diversity is relatively consistent among the forests studied here (comparison of diversity among forest areas: F = 2.10, df = 8, 24, P = 0.0762). This may reflect our focus on mature forests across the region without recent fire or other major disturbances (see ref. [30]). Across all ranges, communities of ectomycorrhizal fungi were dominated by Tomentella, Russula, and Inocybe, which contain many host generalists and are typical of later stage, undisturbed forests [2, 72, 73]. Our data suggest that for conservation or management approaches that include ectomycorrhizal fungi, inclusion of many isolated forests across the range of P. ponderosa would be important.
Perspective on sampling structure and spatial scales
In the present study we used sites within forest areas as replicates, each comprising multiple trees for which multiple root cores were collected. If sites within forest areas were not sufficiently independent from one another, then this approach could result in a falsely inflated sample size due to pseudoreplication. We examined this issue carefully, considering three sources of information. First, in our study design, we followed the robust precedent in the mycological literature for spatial replication at the scale of ca. 1 km (or less) in forests (e.g., refs. [28, 31, 38, 74,75,76,77,78]). Second, we evaluated environmental conditions for sites within each area, anticipating that non-independence would be concerning if environmental conditions were consistently more similar among sites within areas vs. among sites in different areas. To test this, we first used a PCA to determine PC1 for all climate data shown in Table 1. Then, we used PC1 (which described 43.1% of the variation in climate) and plant community (Table 1) to cluster all sites according to similarity. The analysis included 24 sites representing eight disjunct forests and three areas (nine sites) on the Mogollon Rim, for a total of 11 areas and 33 sites. We found no evidence that sites consistently clustered within forest areas: only two of 11 areas (18.2%; Mogollon 1 and Mingus Mt.) had sites that clustered together (Supplementary Fig. S4). Finally, at the conclusion of our study we analyzed the data with a bootstrap analysis, considering the equivalent of one site per forested area for analysis of distance decay at a regional scale. The results presented in Fig. 2 remained consistent for ectomycorrhizal fungi, endophytes observed via the culture-free approach, and endophytes observed via the culture-based approach. Notably Spearman’s rho increased from 0.23 (Fig. 2) to 0.42 in the bootstrapped data for ectomycorrhizal fungi, but decreased for endophytes observed by the culture-free data set, from 0.1 to 0.01. This underscores our observation of strong local signatures of ectomycorrhizal fungal communities in each area, and highlights the widespread distributions of endophytes that, when sampled thoroughly (as in the culture-free survey), appear to be sensitive to environmental conditions within and among disjunct forests.
Perspectives on endophytes
Unlike ectomycorrhizal fungi, endophytes showed evidence of distance decay that could not be connected directly with dispersal limitation, and instead was consistent with environmental dissimilarity. This distance decay was detectable with the culture-free data set at local scales and overall, with weaker but suggestive evidence at regional scales (Fig. 2). The culture-based data set was generally less informative but suggested with limited support a trend for distance decay at regional scales (Fig. 2, Supplementary Fig. S1).
These horizontally transmitted symbionts typically have airborne spores that can traverse large distances on air currents [27, 29]. However, data from diverse biomes show that certain endophytes are restricted in their distributions by climate and associated factors. Climate filters (see ref. [79]) may include direct impacts on fungi themselves via abiotic factors (e.g., ultraviolet radiation, heat, low humidity, or infrequent rainfall; (see ref. [4]), or indirect impacts driven by the responses of plants to climate stress (e.g., slower growth rates, changes in metabolite production, shifts in stomatal regulation that can influence fungal entry; (see ref. [80]). This observation is generally in line with the observation that locally distinctive endophyte assemblages associate with particular hosts under particular environmental conditions ([30]; see also ref. [27]). These endophytes are rarely host specific in the strict sense but have the capacity to establish distinctive functional roles for hosts given particular environmental stressors (see ref. [3]).
When we examined communities in detail, we found that endophyte species richness, community composition, and taxonomy varied with climate and plant community composition. This is consistent with work in other biomes and suggests that general rules for the structure of endophyte communities at a global scale (see ref. [30]) apply in these disparate forests: the community of endophytes in a given host may differ according to the plants in the surrounding plant community [80], and communities vary with climate directly and indirectly through the mechanisms described above.
Relative to previous studies it was surprising that endophyte richness did not vary with MAP. This may reflect the relatively small range of MAP among our sites compared to other studies (e.g., refs. [46, 81]; see also ref. [30]). The negative association of richness with MPDQ also was surprising, as we anticipated that endophyte richness would scale positively with more mesic conditions. We predict that endophytes in biomes of the southwestern USA are relatively robust with regard to drought but that their life cycles are most active in periods of high humidity, as in other seasonal forests (see ref. [4]). Overall, our data suggest that for conservation or management approaches that include endophytic fungi, inclusion of many forests with different environmental conditions would be important.
Culture-based and culture-free approaches to study endophyte communities
The culture-free approach to survey endophytes yielded ca. tenfold more species per individual tree than the culture-based approach, even after stringent quality control and even when subsets of the same plant tissue were used for both methods. This mirrors previous studies of other pines in the region (e.g., P. leiophylla, [82]). As in previous work, we found that the two approaches were complementary for estimating total richness of endophytes (see refs. [30, 44, 82,83,84,85,86]): only a subset of the OTUs obtained by each method was found by the other approach.
Strikingly, five OTUs represented nearly three quarters of all isolates in our culture-based survey (Fig. 7). Other culture-based studies of endophytes associated with Pinus spp. found similar patterns of dominance [44, 83, 87, 88]. These dominant OTUs may have intraspecific variation or cryptic species that could not be observed with our data. They were not dominant in the culture-free data, suggesting that these may be particularly amenable to culturing (see ref. [44]). A benefit of the culture-based approach is that infra-OTU variation and/or functional trait differences among members of the same OTU can be explored readily (e.g., ref. [86]).
Our culture-free and culture-based approaches recorded the same major lineages of endophytes but suggested that they occur in different proportions (Fig. 6). This could reflect PCR bias in the culture-free approach, but we did not observe such bias in sequence data from our mock community (Supplementary Fig. S5). The disparity could reflect differences in the culturability of particular endophyte taxa on a standard medium (see ref. [44]), though the medium used here was chosen because it consistently supports diverse endophytes in culture (see refs. [25, 45]). Finally, the phytochemicals in Pinus leaves (e.g., ref. [89]) could play a role, selecting for a specialists that may not grow well on culture media.
Notably, the culture-free and culture-based datasets yielded different ecological insights with regard to distance decay and associated processes (Figs. 2 and 6). This result differs from previous studies in which these two approaches were congruent, albeit with less robustness in the typically smaller culture-based data sets [30, 44, 82,83,84,85]. It is plausible that the fungi most likely to be isolated in culture are substrate generalists that are ecologically and geographically widespread. In future work we propose testing substrate generalism and genome composition of the most commonly isolated endophytes obtained in culture; modifying our culturing approach to capture endophytes observed only via culture-free methods; and then testing the prediction that the latter would have narrower substrate use, ecological tolerances, and/or host ranges.
Conclusions
Symbiotic fungi associated with forest trees are important components of forest health and resilience. Extant ectomycorrhizal fungal communities exhibit dispersal limitation in the context of anciently disjunct forests that today are separated by arid and semi-arid biomes. Climate-driven retraction of the range of Ponderosa pine, or loss of large stands due to climate-related disturbances such as wildfire in isolated ranges, may lead to local extirpation of the ectomycorrhizal fungi that are vital for forest health and re-establishment. In turn, foliar endophytes do not show signatures of dispersal limitation per se, but appear to be limited in their establishment by environmental factors. Here, major shifts in environmental conditions may limit the capacity of endophytes to establish the symbioses that, in turn, influence tree health, resistance to pests, and productivity.
Data availability
The datasets generated during and/or analyzed during the current study are available in the Bowman-Arnold_DistanceDecaySymbioticFungi repository on GitHub, https://doi.org/10.5281/zenodo.470848810.5281/zenodo.4708488. Illumina data are archived at the GenBank Short Read Archive (SAMN18836769 to SAMN18836812). The BioProject is PRJNA723903: Foliar endophyte communities in Ponderosa pine in Arizona (USA). GenBank accession numbers for endophyte cultures and ectomycorrhizal fungi are listed in Supplementary Table S1.
References
Lutzoni F, Nowak MD, Alfaro ME, Reeb V, Miadlikowska J, Krug M, et al. Contemporaneous radiations of fungi and plants linked to symbiosis. Nat Commun. 2018;9:1–11.
Smith SE and Read D. Mycorrhizal symbiosis, 3rd ed. New York, New York, USA; Academic Press: 2008.
Rodriguez RJ, White JF, Arnold AE, Redman RS. Fungal endophytes: diversity and functional roles. N. Phytol. 2009;182:314–30.
Arnold AE, Herre EA. Canopy cover and leaf age affect colonization by tropical fungal endophytes: ecological pattern and process in Theobroma cacao (Malvaceae). Mycologia. 2003;95:388–98.
Bailey JK, Deckert R, Schweitzer JA, Rehill BJ, Lindroth RL, Gehring C, et al. Host plant genetics affect hidden ecological players: links among Populus, condensed tannins, and fungal endophyte infection. Can J Bot. 2005;83:356–61.
Arnold AE, Mejia LC, Kyllo D, Rojas EI, Maynard Z, Robbins N, et al. Fungal endophytes limit pathogen damage in a tropical tree. Proc Natl Acad Sci USA. 2003;100:15649–54.
Giauque H, Connor EW, Hawkes CV. Endophyte traits relevant to stress tolerance, resource use, and habitat of origin predict effects on host plants. N. Phytol. 2018;221:2239–49.
Aschehoug E, Callaway R, Newcombe G, Tharayil N, Chen S. Fungal endophyte increases the allelopathic effects of an invasive forb. Oecologia. 2012;93:285–91.
U’Ren JM, Arnold AE. Diversity, taxonomic composition, and functional aspects of fungal communities in living, senesced, and fallen leaves at five sites across North America. PeerJ. 2016;4:e2768.
Bennett JA, Maherali H, Reinhart KO, Lekberg Y, Hart MM, Klironomos J. Plant-soil feedbacks and mycorrhizal type influence temperate forest population dynamics. Science. 2017;355:181–4.
Sarmiento C, Zalamea PC, Dalling JW, Davis AS, Stump SM, U’Ren JM, et al. Soilborne fungi have host affinity and host-specific effects on seed germination and survival in a lowland tropical forest. Proc Natl Acad Sci USA. 2017;114:11458–63.
Song Z, Kennedy PG, Liew FJ, Schilling JS. Fungal endophytes as priority colonizers initiating wood decomposition. Funct Ecol. 2017;31:407–18.
Patterson A, Flores-Rentería L, Whipple A, Whitham T, Gehring C. Common garden experiments disentangle plant genetic and environmental contributions to ectomycorrhizal fungal community structure. N. Phytol. 2018;221:493–502.
Bonan GB. Forests and climate change: Forcings, feedbacks, and the climate benefit of forests. Science. 2008;320:1444–9.
USGCRP. Climate Science Special Report: Fourth National Climate Assessment, Volume I. Wuebbles DJ, Fahey DW, Hibbard KA, Dokken DJ, Stewart BC, Maycock TK, editors. Washington, DC, USA: U.S. Global Change Research Program; 2017. p. 470.
van Mantgem PJ, Stephenson NL, Byrne JC, Daniels LD, Franklin JF, Fulé PZ, et al. Widespread increase of tree mortality rates in the western United States. Science. 2009;323:521–4.
Ganey JL, Vojta SC. Tree mortality in drought-stressed mixed-conifer and Ponderosa pine forests, Arizona, USA. Ecol Manag. 2011;261:162–8.
Mathys A, Coops NC, Waring RH. Soil water availability effects on the distribution of 20 tree species in western North America. Ecol Manag. 2014;313:144–52.
Roberts DR, Hamann A. Glacial refugia and modern genetic diversity of 22 western North American tree species. Philos Trans R Soc Lond B Biol Sci. 2015;282:20142903.
Peltier DMP, Ogle K. Legacies of more frequent drought in Ponderosa pine across the western United States. Glob Change Biol. 2019;25:3803–16.
McClaran MP, Brady WW. Arizona’s diverse vegetation and contributions to plant ecology. Rangelands. 1994;16:208–18.
Moir WH, Geils B, Benoit MA, Scurlock D. Ecology of southwestern Ponderosa pine forests. In: Block WM, Finch DM, tech. cords. Songbird ecology in southwestern Ponderosa pine forests: a literature review. Tucson AZ. Fort Collins CO: USDA Forest Service General Technical Report RM GTR-292, Rocky Mountain Forest and Range Experiment Station; 1997. pp. 3–17.
Felger RS, Johnson MB. Trees of the northern Sierra Madre Occidental and sky islands of southwestern North America. In: DeBano FL, Ffolliott PF, Ortega-Rubio A, Gottfried GJ, Hamre RH, editors. Biodiversity and management of the Madrean Archipelago: The sky islands of southwestern United States and northwestern Mexico. Fort Collins, Colorado, USA: U.S. Department of Agriculture, U.S. Forest Service, Rocky Mountain Forest and Range Experiment Station; 1995. pp 71–83.
Willyard A, Gernandt DS, Potter K, Hipkins V, Marquardt P, Mahalovich MF, et al. Pinus Ponderosa: a checkered past obscured four species. Am J Bot. 2017;104:161–81.
Massimo NC, Devan MMN, Arendt KR, Wilch MH, Riddle JM, Furr SH, et al. Fungal endophytes in above-ground tissues of desert plants: infrequence in culture, but highly diverse and distinctive symbionts. Micro Ecol. 2015;70:1–76.
Huang YL, Bowman EA, Massimo NC, Garber NP, U’Ren JM, Sandberg DC, et al. Using collections data to infer biogeographic, environmental, and host structure in communities of endophytic fungi. Mycologia. 2018;110:47–62.
Bowman EA, Arnold AE. Distributions of ectomycorrhizal and foliar endophytic fungal communities associated with Pinus ponderosa along a spatially constrained elevation gradient. Am J Bot. 2018;105:687–99.
Bowman EA, Hayden DR, Arnold AE. Fire and local factors shape ectomycorrhizal fungal communities associated with Pinus ponderosa in mountains of the Madrean Sky Island Archipelago. Fungal Ecol. 2020;49:101013.
Huang Y, Nandi Devan MM, U’Ren JM, Furr SH, Arnold AE. Pervasive effects of wildfire on foliar endophyte communities in montane forest trees. Micro Ecol. 2016;71:452–68.
U’Ren JM, Lutzoni F, Miadlikowska J, Zimmerman NB, Carbone I, May G, et al. Host availability drives distributions of fungal endophytes in the imperiled boreal forest. Nat Ecol Evol. 2019;3:1–8.
Peay KG, Bruns TD, Kennedy PG, Bergemann SE, Garbelotto M. A strong species-area relationship for eukaryotic soil microbes: island size matters for ectomycorrhizal fungi. Ecol Lett. 2007;10:470–80.
Peay KG, Schubert MG, Nguyen NH, Bruns TD. Measuring ectomycorrhizal fungal dispersal: macroecological patterns driven by microscopic propagules. Mol Ecol. 2012;21:4122–36.
Galante TE, Horton TR, Swaney DP. 95% of basidiospores fall within 1 m of the cap: a field-and modeling-based study. Mycologia. 2011;103:1175–83.
Oono R, Rasmussen A, Lefèvre E. Distance decay relationships in foliar fungal endophytes are driven by rare taxa. Environ Microbiol. 2017;19:2794–805.
Fick SE, Hijmans RJ. Worldclim 2: New 1-km spatial resolution climate surfaces for global land areas. Int J Climatol. 2017;37:4302–15.
Lilleskov E, Bruns TD, Horton TR, Taylor DL, Grogan P. Detection of forest stand-level spatial structure in ectomycorrhizal fungal communities. FEMS Microbiol Ecol. 2004;49:319–32.
Shinneman DJ, Means RE, Potter KM, Hipkins VD. Exploring climate niches of Ponderosa pine (Pinus ponderosa Douglas ex Lawson) haplotypes in the western united states: implications for evolutionary history and conservation. PLoS One. 2016;11:e0151811.
Agerer R. Characterization of ectomycorrhiza. Methods Microbiol. 1991;23:25–73.
Agerer R. Anatomical characteristics of identified ectomycorrhizas: an attempt towards a natural classification. In: Varma A, Hock B, editor. Mycorrhiza. Berlin, Heidelberg, Germany: Springer; 1995. p 685–734.
Agerer R. Exploration types of ectomycorrhizae. Mycorrhiza. 2001;11:107–14.
Izzo A, Agbowo J, Bruns TD. Detection of plot-level changes in ectomycorrhizal communities across years in an old-growth mixed-conifer forest. N. Phytol. 2005;166:619–29.
Smith ME, Douhan GW, Rizzo DM. Intra-specific and intra-sporocarp ITS variation of ectomycorrhizal fungi as assessed by rDNA sequencing of sporocarps and pooled ectomycorrhizal roots from a Quercus woodland. Mycorrhiza. 2007;18:15–22.
Monacell JT, Carbone I. Mobyle SNAP Workbench: A web-based analysis portal for population genetics and evolutionary genomics. Bioinformatics. 2014;30:1488–90.
Arnold AE, Henk DA, Eells RL, Lutzoni F, Vilgalys R. Diversity and phylogenetic affinities of foliar fungal endophytes in loblolly pine inferred by culturing and environmental PCR. Mycologia. 2007;99:185–206.
Oita S, Carey J, Kline I, Ibáñez A, Yang N, Hom EFY, et al. Methodological approaches frame insights into endophyte richness and community composition. Microb Ecol. 2021; https://doi.org/10.1007/s00248-020-01654-y.
U’Ren JM, Lutzoni F, Miadlikowska J, Laetsch AD, Arnold AE. Host and geographic structure of endophytic and endolichenic fungi at a continental scale. Am J Bot. 2012;99:898–914.
U’Ren JM, Dalling JW, Gallery RE, Maddison DR, Davis EC, Gibson CM, et al. Diversity and evolutionary origins of fungi associated with seeds of a neotropical pioneer tree: a case study for analysing fungal environmental samples. Mycol Res. 2009;113:432–49.
U’Ren JM, Arnold AE. DNA Extraction Protocol for Plant and Lichen Tissues Stored in CTAB. 2017a; https://doi.org/10.17504/protocols.io.fs8bnhw.
U’Ren JM, Arnold AE. Illumina MiSeq Dual-barcoded Two-step PCR Amplicon Sequencing Protocol. 2017b; https://doi.org/10.17504/protocols.io.fs9bnh6.
Daru BH, Bowman EA, Pfister DH, Arnold AE. A novel proof of concept for capturing the diversity of endophytic fungi preserved in herbarium specimens. Philos Trans R Soc Lond B Biol Sci. 2018;374:1–10.
Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes - application to the identification of mycorrhizae and rusts. Mol Ecol. 1993;2:113–8.
White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. New York, USA: Academic Press; 1990. pp. 315–22.
Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–1.
Andrew S. FastQC: a quality control tool for high throughput sequence data. 2010. http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
Ewels P, Magnusson M, Lundin S, Käller M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32:3047–8.
Chao A, Jost L. Coverage-based rarefaction and extrapolation: standardizing samples by completeness rather than size. Ecology. 2012;93:2533–47.
Okazaki Y, Fujinaga S, Tanaka A, Kohzu A, Oyagi H, Nakano S. Ubiquity and quantitative significance of bacterioplankton lineages inhabiting the oxygenated hypolimnion of deep freshwater lakes. ISME J. 2017;11:2279–93.
Ngyuen NH, Smith D, Peay K, Kennedy P. Parsing ecological signal from noise in next generation amplicon sequencing. N. Phytol. 2015;205:1389–93.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10.
Kõljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AFS, Bahram M, et al. Towards a unified paradigm for sequence-based identification of fungi. Mol Ecol. 2013;22:5271–7.
Huson DH, Mitra S, Ruscheweyh H-J, Weber N, Schuster SC. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 2011;21:1552–60.
Carbone I, White JB, Miadlikowska J, Arnold AE, Miller MA, Magain N, et al. T-BAS version 2.1: Tree-Based Alignment Selector toolkit for evolutionary placement of DNA sequences and viewing alignments and specimen metadata on curated and custom trees. Microbiol Resour Announc. 2019;8:e00328–19.
Legendre P, Legendre L. Numerical Ecology, 3rd ed. Amsterdam, the Netherlands: Elsevier; 2012.
Dray S, Legendre P, Peres-Neto PR. Spatial modelling: a comprehensive framework for principal coordinate analysis of neighbour matrices (PCNM). Ecol Model. 2006;196:483–93.
Legendre P, Borcard D, Roberts DW. Variation partitioning involving orthogonal spatial eigenfunction submodels. Ecology. 2012;93:1234–40.
Lichstein J. Multiple regression on distance matrices: A multivariate spatial analysis tool. Plant Ecol. 2007;188:117–31.
Gower JC. A general coefficient of similarity and some of its properties. Biometrics. 1971;27:857–74.
Zimmerman N, Vitousek P. Fungal endophyte communities reflect environmental structuring across a Hawaiian landscape. Proc Natl Acad Sci USA. 2012;109:13022–7.
Garfin G, Jardine A, Merideth R, Black M, LeRoy S. Assessment of climate change in the southwest United States: a report prepared for the National Climate Assessment. Washington, DC, USA: Island Press; 2013.
Rehfeldt GE, Jaquish BC, López-Upton J, Sáenz-Romero C, St. Clair JB, Leites LP, et al. Comparative genetic responses to climate for the varieties of Pinus ponderosa and Pseudotsuga menziesii: Realized climate niches. Ecol Manag. 2014;324:126–37.
Vander Wall SB. On the relative contributions of wind versus animals to seed dispersal of four Sierra Nevada pines. Ecology. 2008;89:1837–49.
Timling I, Dahlberg A, Walker DA, Gardes M, Charcosset JY, Welker JM, et al. Distribution and drivers of ectomycorrhizal fungal communities across the North American Artic. Ecosphere. 2012;3:3258–72.
Bruns TD, Bidartondo MI, Taylor DL. Host specificity in ectomycorrhizal communities: what do the exceptions tell us? Integr Comp Biol. 2002;42:352–9.
Izzo A, Agbowo J, Bruns TD. Detection of plot-level changes in ectomycorrhizal communities across years in an old-growth mixed-conifer forest. N. Phytol. 2005;2:619–30.
Talbot JM, Bruns TD, Smith DP, Branco S, Glassman SI, Erlandson S, et al. Independent roles of ectomycorrhizal and saprotrophic communities in soil organic matter decomposition. Soil Biol Biochem. 2013;57:282–91.
Matsuoka S, Mori AS, Kawaguchi E, Hobara S, Osono T. Disentangling the relative importance of host tree community, abiotic environment, and spatial factors on ectomycorrhizal fungal assemblages along an elevation gradient. FEMS Microbiol Ecol. 2016;92:fiw044.
Varenius K, Lindahl BD, Dahlberg A. Retention of seed trees fails to lifeboat ectomycorrhizal fungal diversity in harvested Scots pine forests. FEMS Microbiol Ecol. 2017;93:fix105.
Harrison JG, Griffin EA. The diversity and distribution of endophytes across biomes, plant phylogeny and host tissues: how far have we come and where do we go from here? Environ Microbiol. 2020;22:2107–23.
Oita S, Ibánez A, Lutzoni F, Miadlikowska J, Geml J, Lewis LA, et al. Climate and seasonality drive the richness and composition of tropical fungal endophytes at a landscape scale. Commun Biol. 2021;4:313.
Saunders M, Glenn AE, Kohn LM. Exploring the evolutionary ecology of fungal endophytes in agricultural systems: using functional traits to reveal mechanisms in community processes. Evol Appl. 2010;3:525–37.
Lau MK, Arnold AE, Johnson NC. Factors influencing communities of foliar fungal endophytes in riparian woody plants. Fungal Ecol. 2013;6:365–78.
U’Ren JM, Riddle JM, Monacell JT, Carbone I, Miadlikowska J, Arnold AE. Tissue storage and primer selection influence pyrosequencing-based inferences of diversity and community composition of endolichenic and endophytic fungi. Mol Ecol Resour. 2014;14:1032–48.
Oono R, Lefèvre E, Simha A, Lutzoni F. A comparison of the community diversity of foliar fungal endophytes between seedlings and adult loblolly pines (Pinus taeda). Fungal Biol. 2015;119:1–12.
Raizen NL Fungal endophyte diversity in foliage of native and cultivated Rhododendron species determined by culturing, ITS sequencing, and pyrosequencing. Master’s Thesis. Corvallis, USA: Oregon State University; 2013.
Higgins KL, Coley PD, Kursar TA, Arnold AE. Culturing and direct PCR suggest prevalent host generalism among diverse fungal endophytes of tropical forest grasses. Mycologia. 2011;103:247–60.
Harrington AH, Del Olmo-Ruiz M, U’Ren JM, Garcia K, Pignatta D, Wespe N, et al. Coniochaeta endophytica sp. nov., a foliar endophyte associated with healthy photosynthetic tissue of Platycladus orientalis (Cupressaceae). Plant Fungal Syst. 2019;64:65–79.
Ganley RJ, Newcombe G. Fungal endophytes in seeds and needles of Pinus monticola. Mycol Res. 2006;110:318–27.
Gray AE. A molecular characterization of the fungal endophytes within the needles of ponderosa pine (Pinus ponderosa). M.S. thesis. Cheney, WA: Eastern Washington University; 2016.
Hinejima M, Hobson KR, Otsuka T, Wood DL, KuBo I. Antimicrobial terpenes from oleoresin of ponderosa pine tree Pinus ponderosa: a defense mechanism against microbial invasion. J Chem Ecol. 1992;18:1809–18.
Acknowledgements
We thank the National Science Foundation (Graduate Research Fellowship DGE-1143953, EAB), the United States Department of Agriculture (ARZT-1361340-H25-242, AEA), the School of Plant Sciences at the University of Arizona (EAB), and the Mycological Society of America (EAB) for financial support. We thank A. Leo, K. Garcia, A. Ndobegang, A. Harrington, J. Carey, D. Kissell, E. Pat, S. Fischer, H. Siewiora, A. Scull, L. Bowman, A. Peñuñuri, and D. Cordova for assistance with sampling and in the lab, M.M. Lee for laboratory support, and J. U’Ren, I. Carbone, D. New, and M. Fagnan for technical support. We thank K. Dlugosch, C. Gehring, M. Orbach, and S. Smith for guidance and comments on this paper, part of the PhD dissertation by EAB in Plant Pathology at the University of Arizona. We are grateful to Dr. Britt Koskella and anonymous reviewers for insightful feedback that greatly improved this manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Bowman, E.A., Arnold, A.E. Drivers and implications of distance decay differ for ectomycorrhizal and foliar endophytic fungi across an anciently fragmented landscape. ISME J 15, 3437–3454 (2021). https://doi.org/10.1038/s41396-021-01006-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-021-01006-9
This article is cited by
-
Plant and fungal species interactions differ between aboveground and belowground habitats in mountain forests of eastern China
Science China Life Sciences (2023)
-
Climate, host and geography shape insect and fungal communities of trees
Scientific Reports (2023)