Abstract
Although pyrogenic organic matter (PyOM) generated during wildfires plays a critical role in post-fire ecosystem recovery, the specific mechanisms by which PyOM controls soil microbial community assembly after wildfire perturbation remain largely uncharacterized. Herein we characterized the effect of PyOM on soil bacterial communities at two independent wildfire-perturbed forest sites. We observed that α-diversity of bacterial communities was the highest in wildfire-perturbed soils and that bacterial communities gradually changed along a sequence of unburnt soil → burnt soil → PyOM. The microbial communities reconstructed from unburnt soil and PyOM resembled the real bacterial communities in wildfire-perturbed soils in their α-diversity and community structure. Bacterial specialists in PyOM and soils clustered in phylogenetic coherent lineages with intra-lineage pH-niche conservatism and inter-lineage pH-niche divergence. Our results suggest that PyOM mediates bacterial community assembly in wildfire-perturbed soils by a combination of environmental selection and dispersal of phylogenetic coherent specialists with habitat preference in the heterogeneous microhabitats of burnt soils with distinct PyOM patches.
Similar content being viewed by others
Introduction
Wildfires are ubiquitous across the globe and annually burn an average of 348 Mha of vegetated land worldwide [1]. Given projections for future climate change, wildfires are predicted to become larger and more frequent and intense, causing severe perturbations to terrestrial ecosystems [2]. In particular, wildfires alter all aspects of the soil microbiome, including community structure and diversity [3]. In the days to weeks after fire disturbance, microbial diversity dramatically decreases with a recovery of community structure over time, which is attributed to initial soil sterilization followed by microbial regeneration and/or dispersal in the topsoil [4]. Several years after recovering from fire disturbance and following ecosystem stabilization, microbial diversity may exceed that of unburnt soil, whereas microbial community structure may achieve an alternative stable state [5]. These transformations are usually ascribed to wildfire-induced changes in vegetation and edaphic conditions (e.g., pH, organic carbon, nutrient availability) [6, 7]. Wildfire alterations to soil microbiomes play a vital role in terrestrial ecosystem resilience and recovery [8], highlighting the need to better understand responses of the soil microbiome to wildfire perturbations.
Research interest in pyrogenic organic matter (PyOM) has increased over recent decades in an effort to elucidate the impacts of wildfire perturbation on ecological processes [9]. PyOM is generated during fires, and consists of a broad continuum of carbon-enriched substances ranging from semi-charred biomass to charcoal and soot [10]. It is variously termed charcoal, black carbon or biochar in different contexts as they are all derived from biomass pyrolysis and have similar chemical structure and properties: enriched in carbon and aromatic compounds, and relatively resistant to microbial degradation [11]. Recent studies showed that, much like the long-term effects of wildfires, an application of anthropogenic PyOM (biochar) alters the community structure and diversity of microorganisms [12]. Similar to the soil changes wrought by wildfires, edaphic changes from biochar amendments include increases in nutrient availability and pH [13]. Overall, these studies suggest that long-term wildfire effects may be connected to PyOM through its role in regulating edaphic properties and the resulting soil microbiome, thereby having a strong feedback on terrestrial ecosystems.
It is unknown whether PyOM alters the soil microbiome indirectly through alterations of edaphic conditions or directly by serving as a microhabitat that regulates the colonization of microorganisms. The PyOM appears to be an important microhabitat for microorganisms in fire-perturbed soils as the porous structure of anthropogenic PyOM provides advantageous dwellings for microorganisms [14]. In boreal forests, microbes colonized in PyOM can also facilitate degradation of phenolic compounds retained by PyOM [15]. PyOM-associated microbial communities have a community structure distinctly different from the adjacent soil [16]. Furthermore, PyOM-induced microbial responses, as well as edaphic changes, demonstrate a steep distance-decay relationship at the interface between PyOM and bulk soil [17]. However, current studies on soil microbiome-PyOM interactions usually report the microbial response to overall edaphic changes caused by anthropogenic PyOM addition without specifically addressing habitat effects [17]. Moreover, previous studies either did not report community structure or microbial diversity [16], or lacked a rigorous conclusion due to limitations from inadequate sample size and/or neglected interactions between communities in PyOM and the adjacent soil [18]. Thus, our understanding of the linkages between PyOM microhabitats and fire-perturbed soil remains limited and in need of further study.
This study investigated bacterial communities in PyOM, burnt soils and unburnt soils at two wildfire-perturbed forest sites to examine the relationships between PyOM and soil bacteria, paying particular attention to community assembly processes and phylogenetic coherence. “Coherence,” in this sense, delineates the structured occurrence of habitat specialists under a specific phylogenetic lineage and potentially reflects a lineage’s conserved habitat niche contrasting against diversification. The following hypotheses were tested: (1) PyOM forms a distinct microhabitat for some soil bacteria; (2) the heterogeneous PyOM-soil matrix mediates the bacterial community of burnt soils; and (3) habitat specialists in PyOM and soils have phylogenetic coherence. The relationships between PyOM and the bacterial community were explored through 16 S rRNA gene sequencing of soil samples collected from fire perturbed forests and additionally, through simulations wherein we reconstructed bacterial communities in burnt soils based on the bacterial communities identified in PyOM and unburnt soils to corroborate the bacterial community assembly found after wildfire perturbation. We combined these results to elucidate the role of PyOM in bacterial community assembly in wildfire-perturbed soils and to link observations of bacterial communities in burnt soils with microbial community assembly theories.
Materials and methods
Sample collection
Soil samples were collected post-fire in two forests in Zhejiang (30° 2’ 8” N, 119° 39’ 24” E) and Jiangxi (28° 14’ 35” N, 117° 4’ 27” E) provinces, China. The fire at the Zhejiang site occurred in August 2013 with soil sampling in March 2016. Soils were classified as Humults and had a sandy loam texture. The site had a slope of 55°, a mean annual precipitation of 1477 mm and a mean annual temperature of 16.7 °C. The fire at the Jiangxi site occurred in December 2017, and the soil was sampled in October 2018. Soils were classified as Humults and had a loam texture. The site had a slope of 30°, a mean annual precipitation of 1750 mm and a mean annual temperature of 18 °C. No historical wildfires were previously recorded at either site before the current fire events. Prior to the wildfires, both sites were vegetated by subtropical evergreen broad-leafed forests. Both fires burned at a moderate intensity with an estimated average temperature of 250–400 °C, consuming most of the litter layer and leaving an abundance of semi-charred and charred materials (Fig. S1); the wildfire was somewhat more severe at the Jiangxi forest compared to the Zhejiang forest site. Post-fire vegetation at the Zhejiang site was shrubs and low trees (Fig. S1c), while at Jiangxi the post-fire vegetation was primarily grasses or barren soil (Fig. S1d).
Twelve composite soil samples were collected from each site that spanned a gradient from the fire center to the perimeter. Composite soil samples consisting of triplicate cores were taken from the upper 8 cm of the A horizon after removal of any O horizon. We isolated PyOM particles (i.e., charred materials; 0.5–15 mm diameter) by handpicking from each soil sample to characterize their microbiomes and distinct habitat conditions (Fig. S2). To investigate soil microbiomes free of any recent PyOM materials, 12 soil samples were collected from an adjacent unburnt location at each site that had similar soil, topography and pre-burn vegetation. Overall, 24 samples were collected at each site, comprising 12 samples from the unburnt locations (‘unburnt’ samples) and 12 samples from the burnt locations that were further subdivided into 12 “burnt” soil samples and 12 “PyOM” samples. In addition, 9 PyOM samples were taken from the Zhejiang site and washed with Milli-Q water (EMD Millipore, Germany) to investigate whether adhered soil impacted the PyOM microbiome characterization. Each soil sample was sieved through a 2-mm screen and split into three zip-lock bags stored at three different conditions: air-dried, frozen at −20 oC and frozen at −80 oC, for determination of pH and organic carbon (SOC), analysis of dissolvable organic carbon (DOC) and DNA extraction, respectively. Each isolated PyOM sample was split into two zip-lock bags and stored at two conditions: vacuum-dried and −80 oC, for pH analysis and DNA extraction, respectively. Full details are given in Section 1.1 of the Supplemental Information (SI).
Bacterial 16 S rRNA gene sequencing and sequence data processing
Total DNA was extracted from the soil and PyOM samples using a FastDNA™ SPIN Kit for Soil following the manufacturer instructions (MP Biomedicals, USA). The V4–V5 regions (515F–907 R) of the 16 S rRNA gene were amplified with forward (515 F: 5’-GTG CCA GCM GCC GCG G -3’) and reverse primers (907 R: 5’- CCG TCA ATT CMT TTR AGT TT -3’) and sequenced with a MiSeq platform (Illumina Inc., USA). After raw data were quality screened, operational taxonomic units (OTUs) were identified using a 97% similarity criterion based on the UPARSE algorithm [19]. Phylogenies of OTUs were constructed using MUSCLE [20] and FastTree [21] and taxonomies were inferred using EzBioCloud [22]. Full details are provided in SI Section 1.2. OTU sequences from this study were deposited at DDBJ/EMBL/GenBank under the accession numbers KEKP00000000 for Zhejiang and KEKQ00000000 for the Jiangxi site.
Bacterial community analysis
Biodiversity measurements included α-diversity, β-diversity and phylogenetic clustering indices. The α-diversity indices included Shannon index, Faith’s phylogeny diversity [23], Pielou’s J [24] and observed OTU number. β-diversity was characterized using a non-constrained ordination, non-metric multidimensional scaling (NMDS) [25], based on a phylogenetic distance generalized UniFrac (GUniFrac) [26]. Procrustes analysis [27] was performed based on the NMDS results to assess correlations between the community structure of PyOM and adjacent soils. The washing procedure did not discernibly affect the microbial structure of PyOM samples (pseudo-F = 0.65, P = 0.89, PERMANOVA) (Fig. S3); thus, the data of washed and unwashed PyOM samples were pooled. Phylogenetic clustering indices included the α-nearest-taxon-index (αNTI, in each sample) and β-nearest-taxon-index (βNTI, between samples) [28]. Full analytical details are provided in SI Section 1.3. The source code of this and the following analyses are available at https://github.com/Lujun995/Soil-PyOM-microbiome-studies.git.
Phylogenetic coherence of habitat specialists
Habitat specialists were defined as OTUs preferring PyOM or soil habitats; specialist analysis was conducted after community-level analyses and was aimed to provide detailed information about OTU responses to habitat types. To select specialists, regression analysis was performed between OTU abundance (response variable) and a scale representing the PyOM’s effect (explanatory variable). Scale values were 1, 2, and 3 for unburnt soils, burnt soils and PyOM, representing negligible, medium and high effect of PyOM, respectively. For regression analyses, OTUs with permutation-based false discovery rates below 0.01 [29] were deemed habitat specialists and all others were considered habitat generalists. Specialists that were positively associated with the PyOM scale were considered PyOM specialists, whereas those negatively associated were considered soil specialists. Sensitivity analyses [30] showed that specialists were responsible for 69% and 82% of the differences in ordinations for the Zhejiang and Jiangxi sites, respectively; generalists were responsible for the remaining 31% and 18% (Fig. S4). Furthermore, co-occurrence networks [31] showed potential co-occurrence patterns between specialists (Fig. S5), indicating they were good representatives. Full details are given in SI Section 1.4.
Phylogenetic coherence was used to distinguish heredity-induced niche conservatism from diversification-induced niche plasticity (Fig. 1a). To evaluate phylogenetic coherence of specialists, we visualized a phylogenetic tree and quantified phylogenetic conservatism using Fritz’s D [32]. For convenience, the 1 − D metric was reported [33]; a larger 1 − D value indicates stronger phylogenetic conservatism. We also customized a hierarchical and agglomerative procedure to cluster the coherent lineages of specialists (Fig. 1b). The procedure was based on testing a statistical null hypothesis H0: the occurrence of specialists was random. Full details are presented in SI Section 1.5.
a Conserved niche is a convergent value of one functional trait of genetic variants in some lineages, representing a niche shared by all the genetic variants in the lineages. b Conserved niche is recognized as coherence of habitat-preference in phylogenies. Statistical significance was tested using binominal tests.
To contrast the niches of coherent lineages preferring PyOM versus soil habitats, we reviewed physiological traits of prokaryotic species in these lineages in order to access data on their phenotypic traits (niches) with a focus on convergent traits of growth pH, halotolerance and endospore formation. A coherent lineage was excluded from the review if it was a predicted taxon without a reliable taxonomy or a class whose phenotypic traits were considered too diverse. Full details are given in SI Section 1.6.
Reconstructing the bacterial community of burnt soils
Our second hypothesis implies that the bacterial community in burnt soils originated from the interactions of communities found in PyOM and in unburnt soils. In order to examine this relationship, we reconstructed the bacterial community in burnt soils via simulations that used data from the unburnt soils and PyOM. We projected the co-existence of soil and PyOM specialists by averaging their abundance in PyOM and unburnt soils and generated the generalists by extracting their abundance from either PyOM or unburnt soils. These two components were then combined and rarefied to the sequencing depth of burnt soils. Subsequently, the simulated communities were contrasted to the real communities measured in burnt soils using Shannon, αNTI and GUniFrac distance indices. Full details are presented in SI Section 1.7.
Results
Edaphic properties and α-diversity
The pH among samples followed an order of PyOM ≥ burnt soils > unburnt soils for both sites (Dunn-test, P < 0.05) (Fig. 2a and 2g). However, even though it followed the same pattern, no significant difference was observed between PyOM and burnt soils for the Jiangxi site (Dunn-test, P = 0.41). Burnt soils had decreased DOC concentrations at both sites (Welch’s t test, P = 0.005) (Fig. S6); fire history appeared to increase SOC at Jiangxi (Welch’s t test, P < 0.001), but not at the Zhejiang site (Welch’s t test, P = 0.42) (Fig. S6). Bacterial communities in burnt soils generally had higher α-diversity than unburnt soils and PyOM, although the differences were not always statistically significant (Fig. 2b–2e, 2h–2k). Burnt soils’ Shannon and observed OTU counts were significantly higher than both unburnt soils and PyOM in Zhejiang samples (Dunn-test, P < 0.05). All other α-diversity measures aside Faith’s diversity at the Jiangxi site showed diminished differences at one or both directions (Dunn-test, P ≥ 0.05). Phylogenetic clustering indices indicate that the bacterial communities in PyOM were the most phylogenetically scattered with lower α-NTI values than both burnt and unburnt soils at both sites (pairwise t test, P < 0.01) (Fig. 2f and 2l).
(a–f) The Zhejiang site. (g–l) The Jiangxi site. An αNTI value >2 is considered phylogenetic clustering. The curvilinear polygons indicate estimates of frequency densities. The upper/lower hinges of boxes represent the 75th/25th quantiles, and the lines inside boxes indicate the median value. The whiskers above/below boxes extend to the maximum/minimum values within ≤1.5 inter-quantile-range, whereas data points outside the range are plotted separately. *, ** and *** indicate significance at P ≤ 0.05, ≤ 0.01 and ≤ 0.001, respectively.
The relationship between Shannon indices and pH values followed an inverted-U shape at Zhejiang (quadratic regression, P < 0.001 for the quadratic coefficient) (Fig. 3b), but was relatively flat at Jiangxi (quadratic regression, P = 0.09 for the quadratic coefficient) (Fig. 3d). Neutral environmental pH and low clustering of phylogenetic structure did not necessarily lead to increases in bacterial diversity; PyOM had pH values close to neutral and the least clustered phylogenetic structures (Fig. 2a, 2g, 2f and 2l), but did not have a higher biodiversity than the burnt soil (Fig. 3b and 3d). All biodiversity indices followed an inverted-U relationship with NMDS1 scores (see below; quadratic regression, P < 0.01 for quadratic coefficients) (Fig. 3e–3l).
(a, b, e, f, i, j) The Zhejiang site. (c, d, g, h, k, l) The Jiangxi site. Relationships were tested using linear (a and c) or quadratic (b, d, e–l) regressions. A solid curve or line indicates statistical significance at the 0.05 level with the dashed line indicating insignificance. The gray shading represents the 95% confident interval for regressions. Notations and abbreviations: r, Pearson’s r statistics; P2, P values for the quadratic coefficients, which reflect the inverted-U shape.
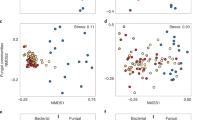
β-diversity of bacterial communities
The bacterial community structure in burnt soils was significantly different from those in unburnt soils and PyOM (PERMANOVA, P < 0.001 for both sites) along the first NMDS axis (Fig. 4a and 4b). The bacterial community structure at Zhejiang was differentiated by the first and third NMDS axes (Fig. 4a), whereas at Jiangxi it was differentiated by the first and second NMDS axes (Fig. 4b). The first NMDS axis was strongly correlated with pH (Pearson correlation, P < 0.001 at both sites) (Fig. 3a and 3c). Bacterial community structure in the burnt soils was strongly related to that in PyOM (Procrustes analysis, P = 0.003 for Zhejiang and P < 0.001 for Jiangxi) (Fig. 4c and 4d).
Unconstraint ordinations (NMDS) among all samples based on generalized UniFrac distances for Zhejiang (a) and Jiangxi (b) sites. Statistical significance was tested using PERMANOVA by assigning 3 (high), 2 (medium) and 1 (no) influence of PyOM, to PyOM, burnt soils and unburnt soils. Curves next to and below the ordinations are estimations of frequency densities for the corresponding axes. Procrustes analysis between PyOM and its adjacent burnt soils at Zhejiang (c) and Jiangxi (d) sites. The adjacency relationship is indicated by arrows.
Relative abundances of phyla differed between sample types at both sites (Fig. 5). Overall, differentiation of bacterial community structure was strongly influenced by the relative abundances of Acidobacteria, Bacteroidetes and Streptophyta at both sites (Kruskal–Wallis test with Bonferroni correction, P < 0.01). The relative abundances of Bacteroidetes and Streptophyta were higher in burnt soils and PyOM, but Acidobacteria abundance followed the order of unburnt > burnt > PyOM (Fig. 5).
(a) The Zhejiang site. (b) The Jiangxi site. Phyla are selected as dominant phyla if their relative abundance is >1% in at least one sample type (unburnt soils, burnt soils or PyOM) at each site. Phyla are labeled with ** if their relative abundance is associated with a specific sample type (Kruskal–Wallis test with Bonferroni correction, P < 0.01).
Simulation-based reconstruction of burnt soil communities
To link the bacterial community found in burnt soils to that in PyOM and unburnt soils, we reconstructed burnt soil communities via simulations based on the identified abundance data from the PyOM and unburnt soils. The simulated communities resembled the measured communities in the burnt soils at both sites when NMDS1 and NMDS3 scores were considered (Kolmogorov-Smirnov test, P ≥ 0.05) (Fig. 6b and 6d). Shannon indices, observed OTU numbers, αNTI of the mock communities were considered equal to (Kolmogorov-Smirnov test, P ≥ 0.05) or moderately different from (Kolmogorov-Smirnov test, 0.01 ≤ P ≤ 0.05) those of measured communities in burnt soils at both sites (Fig. 6). However, disparities were found in NMDS2 scores (Kolmogorov-Smirnov test, P < 0.001) at both sites (Fig. 6b and 6d).
Statistical significance was tested using Kolmogorov-Smirnov tests. Symbols and abbreviations: n.s., * and *** stand for insignificant at P = 0.05, P < 0.05 and P < 0.001, respectively. a, c Biodiversity metrics. The shapes have the same meaning as in Fig. 1. b, d Unconstrained ordinations using NMDS based on generalized UniFrac. The curves next to and below the ordinations are estimations of frequency densities for the corresponding axes.
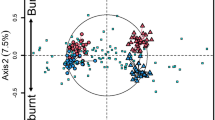
Phylogenetic coherence of specialists in PyOM and soils
Specialists in soil and PyOM had strong phylogenetic signals but the signal strength varied across phyla (Fig. 7). The 1 − D metric (Fritz’s D) for the Zhejiang site was 0.26 for PyOM specialists and 0.51 for soil specialists. That for the Jiangxi site was 0.41 for PyOM specialists and 0.39 for soil specialists (P < 0.01, based on label permutation, Table S1). Notably, 45% of soil specialists and 35% of PyOM specialists were phylogenetically coherent at Zhejiang, while 38% of soil specialists and 22% of PyOM specialists were phylogenetically coherent at Jiangxi (Fig. 7 and Table S2). Coherent lineages found at both sites included some lineages (e.g., EU445199_o and Acidobacteriaceae) in Acidobacteria, EU335336_g (a predicated genus) and Acidibacter in Proteobacteria for soil, and Frankiales, Geodermatophilaceae and Micrococcales in Actinobacteria for PyOM (Table 1). In nearly every coherent lineage, a consistent optimum pH of its bacterial species was found, regardless of year and location of isolation (Tables S2–S5). There was an observable pH-niche difference between coherent lineages preferring soil and PyOM. Coherent lineages of PyOM were chiefly neutrophilic (optimum growing pH 6–8); conversely, lineages of soil were primarily acidophilic (optimum growing pH ≤5.5) (Tables 1 and S2). The relationship between pH niches and habitat preferences was non-random (Fisher-exact-test for count data, P < 0.001). However, sporulation and halotolerance were not associated with habitat preferences of coherent lineages investigated in this study (Fisher’s-exact-test for count data, P ≥ 0.10). Virtually, all the lineages were categorized as non-endospore-forming and non-halotolerant to moderately halotolerant (Tables S2–S5).
(a) The Zhejiang site. (b) the Jiangxi site. Markers in the outer ring of the aura indicate habitat preference of OTUs with blank areas indicating a neutral response. The coherent markers in the inner ring indicate coherent lineages (Table S2) and their habitat preference. The statistic inside the aura is Fritz’s 1 – D, which is noted in the 6 largest phyla of phylogenetic conservationism (P < 0.01, based on label permutation). A higher 1 – D value indicates stronger phylogenetic conservatism.
Discussion
PyOM-mediated bacterial community assembly after wildfire perturbation
Bacterial community structure in burnt soils differed from those in unburnt soils and PyOM, but could not be fully explained by changes in edaphic conditions. Given that meta-communities inhabit burnt soils comprising both soil and PyOM matrices, we propose an explanation incorporating spatial co-existence [34, 35] and community assembly theories [36] to reveal the effects of PyOM on bacterial community assembly in burnt soils.
Habitat heterogeneity between PyOM and soil was a strong selection factor for microbiome community assembly. At the community level, the bacterial community progressively changed along the unburnt soil → burnt soil → PyOM gradient (Fig. 4a and 4b). These results are consistent with recent observations that PyOM alters bacterial communities, although changes in microbial abundance demonstrated no clear trends [9]. In our present study, community differences were influenced by changes in the relative abundances of Acidobacteria, Bacteroidetes and Streptophyta at both sites (Fig. 5). These changes are in accordance with those induced by variations in environmental pH [37]. pH is a likely driver of the selection process induced by PyOM-soil habitat heterogeneity, given that NMDS1 scores were strongly correlated with this environmental factor (Fig. 3a and 3c). Significant (NTI > 2) phylogenetic clustering indices αNTI and βNTI (Figs. 2f, 2l and S7) provide further support for pH being a strong selection factor [38]. In contrast to these common tendencies, site-specific responses to PyOM and wildfire perturbation (Figs. 2b-2f, 2h-2l, 3b, 3d and 5) may reflect the contribution of site biogeographical history (e.g. vegetation, topography or indigenous microbiomes) to bacterial community assemblage [12].
The close association between bacterial communities in PyOM and the surrounding burnt soils suggests that dispersal accounts for the relatedness between PyOM and its surrounding fire-perturbed soil (Fig. 4c and 4d). Dispersal homogenizes the local communities in regions between heterogeneous habitat patches, resulting in a decrease of β-diversity and an increase of community similarity [39]. Therefore, a necessary indicator of dispersal requires that wildfire-perturbed soil communities appeared to be a homogenization of PyOM and unburnt soil communities; this argument is in line with our homogenized reconstructed communities resembling the measured communities in NMDS1 and NMDS3 (Fig. 6b and 6d). Microorganisms appear to primarily distribute passively, such as via water flow, aerosols [40], dust [41] or fauna transport/mixing [42]. However, it is noteworthy that PyOM can influence surrounding edaphic conditions [16], thus modifying the soil niche via the dispersal of ions and organic compounds, in addition to dispersal of microorganisms.
Wildfire induced higher α-diversity for bacterial communities
Bacterial α-diversities displayed an inverted U relationship with the PyOM gradient (Fig. 3e–3l) and tended to be higher in burnt soils than the unburnt soils or PyOM (Fig. 2). However, such a result should be interpreted with caution due to the narrow differences in α-diversity and the inconsistent result for Faith’s diversity at Jiangxi. The increased biodiversity in burnt soils is frequently attributed to an increase in pH induced by PyOM [43]. This follows as soils with neutral pH generally have higher bacterial biodiversity than acidic soils [44]. Similarly, higher bacterial biodiversity is frequently reported in soils amended with anthropogenic PyOM (i.e., biochar), especially in acidic soils [43].
Bearing the connection between pH and increased biodiversity in mind, our observations in this study suggest how more microbes may co-exist. The burnt soils have a more heterogeneous environment providing both soil and PyOM microhabitats (Figs. S1e and S1f). Accordingly, microbial communities inhabiting more heterogeneous environments with diverse microhabitats will develop into a meta-community with higher overall biodiversity. Hence, bacterial diversity in burnt soils is expected to be characterized as the co-existence of generalists, PyOM specialists and soil specialists. Specialists preferring PyOM or soil microhabitats may spatially co-exist in burnt soils via their pH-niche differences, which is consistent with the environmental pH tendency (unburnt soils < burnt soils ≤ PyOM) (Fig. 2, Table 1 and S2). Our simulations projected such a co-existence by averaging abundance data of specialists in PyOM and unburnt soils and remarkably reproduced the biodiversity patterns found in the microbial communities of burnt soils (Fig. 6a and 6c). These inferences are congruous with previous studies investigating ecotones, the transition zone between two or more ecosystems with distinct traits, such as rhizospheres among rhizocompartments of bulk soil, plant roots and root nodules [45] and seawater-hydrothermal fluid mixing zones at hydrothermal vents [46]. A typical community in an ecotone would comprise specialists from both habitats and some specialists of the ecotone itself [46].
Phylogenetic coherence in shaping habitat preference of specialists in PyOM and soils
Coherence, synonymous with conservatism [47] and congruence [48], is typically portrayed as an association between phylogenetic relatedness and functional traits of species. Several previous studies emphasize using a universal overall indicator for coherence of the phylogenetic tree [47]. This study also found that phylogenetic coherence may reflect the conserved niche of specific lineages (Fig. 7a and 7b), in contrast to diversification and phenotype plasticity. Indicators for phylogenetic coherence of specialists in both PyOM and soils were statistically significant at both sites (Table S1). Notably, many of the specialists fall within coherent lineages (Table 1 & Fig. 7) in which pH niches are greatly conserved (Tables S2–S5). Therefore, in these lineages, even if diversification plays a role in forming bacterial pH niche and habitat preference, it is expected to play a minor role compared with coherence.
Alternatively, selection may be dominated by the diversification process, yielding incoherent phylogenetic patterns. Although overall coherence indicators were significant in this study (Table S1), the strength was not uniform (Fig. 7) when some lineages, such as Edaphobacter at Jiangxi and Conexibacter at Zhejiang (Fig. S8), were considered. Since Edaphobacter is consistently reported as acidophilic [49], members of this clade are expected to proliferate in acidic forest soils, becoming soil specialists. As expected, OTUs associated as Edaphobacter were coherent soil specialists at the Zhejiang site. In comparison, some OTUs of Edaphobacter preferred either a PyOM or a soil habitat at the Jiangxi site (Fig. S8a). A corresponding phenomenon was also found for Conexibacter; some were presumed to be PyOM specialists because Conexibacter is neutrophilic [50], but some were found as soil specialists at Zhejiang (Fig. S8b). Such phylogenetic incoherence is likely caused by diversification (or speciation) [36], which is a universal mechanism for bacteria to adapt to their environment [51, 52].
Overall, both coherence and diversification shape niches of bacteria. Therefore, the contribution of coherence is relative to that of diversification under various scenarios where the ancestors’ niche might be more or less conserved in the descendants. In addition to phylogenetic lineage characteristics, for example, more complex traits, such as pH range expansion and salinity preference, appear to be more conserved than less complex traits, such as viral resistance [47], which often evolves independently via diversification [53]. Altogether, extensive recent observations of strong phylogenetic coherence in microbial studies [54] call for further examination and application of phylogenetic methods in microbial ecology, since phylogenetic coherence indicates microbial niche conservatism [55].
There are some limitations and caveats associated with our approach. First, the dynamics of meta-communities in soil are still largely unknown because we understand little about the dispersal of bacteria. While some studies have explored this topic [40], there is a lack of dispersal rates and mechanisms for different microorganisms in complex soil systems. Thus, future investigations are critically needed to address bacterial dispersal, especially following major soil/ecosystem perturbations. Second, diversification is a dynamic process that may generate coherent lineages during the long course of evolution, forming new species [56]. However, when clustering coherent lineages, we regarded diversification as static and independent of the phylogenetic tree. Thirdly, there were differences in other attributes besides PyOM between the burnt and unburnt soils at each site, such as differences in vegetation, litter, microclimate (i.e., soil water and temperature) and soil nutrient availability. These additional factors potentially shaped the soil microbiome [57] and might contribute to the differences associated with NMDS2 and NMDS3 (Fig. 4b and 4d) at the community level. Although these differences were mathematically orthogonal to NMDS1 (Fig. S4), they might substantially influence soil specialists. Furthermore, even though we collected phenotypical traits to the best of our knowledge, our communal understanding of the microbial universe is limited and some coherent lineages in Tables S2–S5 are without reliable taxonomy and/or phenotypical traits. These missing values may cause a bias in Table 1 and our analyses. Finally, the microbiome in burnt soils likely displays a temporal pattern during ecosystem succession following a fire event [4]. However, a previous study indicated that such a microbial succession would not last for more than 1 year [58].
Overall, this study bridges the gaps between observations of soil physical-chemical alterations by wildfire, microbial community perturbations from wildfire and ecological community assembly theories. A determinant role of PyOM in soil bacterial community assembly after wildfire perturbation was clearly demonstrated. PyOM microhabitats were distinctly different from those of the adjacent wildfire-perturbed soil and soil-PyOM heterogeneity was a strong selection factor for microbial assembly. Ecological dispersal likely contributed to the relatedness between PyOM and adjacent soils. The role of PyOM in bacterial community assembly in burnt soils was supported by reconstructing bacterial communities in burnt soils based on the specialists identified in PyOM and unburnt soil. A strong contrast in pH niche was found in coherent lineages preferring soil or PyOM. Habitat preferences of bacterial taxa were associated with both their conserved niche and diversification, resulting in a relative coherence in different lineages. Our findings highlight that PyOM contributes a strong control on bacterial community assembly processes in burnt soils after wildfire perturbation through the selection and dispersal of specialists with divergent pH-niche preference in heterogeneous microhabitats.
References
Giglio L, Randerson JT, Werf GR. Analysis of daily, monthly, and annual burned area using the fourth-generation global fire emissions database (GFED4). J Geophys Res Biogeosci. 2013;118:317–28.
Abatzoglou JT, Williams AP. Impact of anthropogenic climate change on wildfire across western US forests. Proc Natl Acad Sci USA. 2016;113:11770–5.
Alcañiz M, Outeiro L, Francos M, Úbeda X. Effects of prescribed fires on soil properties: a review. Sci Total Environ. 2018;613:944–57.
Ferrenberg S, O’Neill SP, Knelman JE, Todd B, Duggan S, Bradley D, et al. Changes in assembly processes in soil bacterial communities following a wildfire disturbance. ISME J. 2013;7:1102–11.
Taş N, Prestat E, McFarland JW, Wickland KP, Knight R, Berhe AA, et al. Impact of fire on active layer and permafrost microbial communities and metagenomes in an upland Alaskan boreal forest. ISME J. 2014;8:1904–19.
Mikita-Barbato RA, Kelly JJ, Tate RL. Wildfire effects on the properties and microbial community structure of organic horizon soils in the New Jersey Pinelands. Soil Biol Biochem. 2015;86:67–76.
Rodríguez J, González-Pérez JA, Turmero A, Hernández M, Ball AS, González-Vila F, et al. Wildfire effects on the microbial activity and diversity in a Mediterranean forest soil. Catena. 2017;158:82–88.
Pérez-Valera E, Verdú M, Navarro-Cano JA, Goberna M. Soil microbiome drives the recovery of ecosystem functions after fire. Soil Biol Biochem. 2020;149:107948.
Li Y, Hu S, Chen J, Müller K, Li Y, Fu W, et al. Effects of biochar application in forest ecosystems on soil properties and greenhouse gas emissions: a review. J Soils Sediment. 2018;18:546–63.
Santín C, Doerr SH, Kane ES, Masiello CA, Ohlson M, de la Rosa JM, et al. Towards a global assessment of pyrogenic carbon from vegetation fires. Glob Chang Biol. 2016;22:76–91.
Lehmann JD, Joseph S. Biochar for environmental management: An introduction. In: Lehmann JD, Joseph S, editors. Biochar for environmental management. Earthscan, London: Sterling, VA; 2009. pp. 1–12.
Woolet J, Whitman T. Pyrogenic organic matter effects on soil bacterial community composition. Soil Biol Biochem. 2020;141:107678.
Biederman LA, Harpole WS. Biochar and its effects on plant productivity and nutrient cycling: a meta-analysis. Glob Change Biol Bioenerg. 2013;5:202–14.
Quilliam RS, Glanville HC, Wade SC, Jones DL. Life in the ‘charosphere’ – Does biochar in agricultural soil provide a significant habitat for microorganisms? Soil Biol Biochem. 2013;65:287–93.
Zackrisson O, Nilsson M, Wardle D. Key ecological function of charcoal from wildfire in the Boreal forest. Oikos. 1996;77:10–19.
Yu M, Meng J, Yu L, Su W, Afzal M, Li Y, et al. Changes in nitrogen related functional genes along soil pH, C and nutrient gradients in the charosphere. Sci Total Environ. 2019;650:626–32.
Whitman T, Whitman E, Woolet J, Flannigan MD, Thompson DK, Parisien M-A. Soil bacterial and fungal response to wildfires in the Canadian boreal forest across a burn severity gradient. Soil Biol Biochem. 2019;138:107571.
Dai Z, Barberán A, Li Y, Brookes PC, Xu J. Bacterial community composition associated with pyrogenic organic matter (biochar) varies with pyrolysis temperature and colonization environment. mSphere. 2017;2:e00085–17. mSphere.00085-17
Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–8.
Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acid Res. 2004;32:1792–7.
Price MN, Dehal PS, Arkin AP. FastTree 2 – Approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5:e9490.
Yoon S-H, Ha S-M, Kwon S, Lim J, Kim Y, Seo H, et al. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol. 2017;67:1613–7.
Faith DP. Conservation evaluation and phylogenetic diversity. Biol Conserv. 1992;61:1–10.
Pielou EC. An introduction to mathematical ecology. New York, USA: Wiley Interscience; 1969.
Anderson MJ. A new method for non‐parametric multivariate analysis of variance. Austral Ecol. 2001;26:32–46.
Chen J, Bittinger K, Charlson ES, Hoffmann C, Lewis J, Wu GD, et al. Associating microbiome composition with environmental covariates using generalized UniFrac distances. Bioinformatics. 2012;28:2106–13.
Peres-Neto PR, Jackson DA. How well do multivariate data sets match? The advantages of a Procrustean superimposition approach over the Mantel test. Oecologia. 2001;129:169–78.
Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD, et al. Picante: R tools for integrating phylogenies and ecology. Bioinformatics. 2010;26:1463–4.
Xie Y, Pan W, Khodursky AB. A note on using permutation-based false discovery rate estimates to compare different analysis methods for microarray data. Bioinformatics. 2005;21:4280–8.
Iman RL, Hora SC. A robust measure of uncertainty importance for use in fault tree system analysis. Risk Anal. 1990;10:401–6.
Delgado-Baquerizo M, Oliverio AM, Brewer TE, Benavent-González A, Eldridge DJ, Bardgett RD, et al. A global atlas of the dominant bacteria found in soil. Science. 2018;359:320–5.
Fritz SA, Purvis A. Selectivity in mammalian extinction risk and threat types: a new measure of phylogenetic signal strength in binary traits. Conserv Biol. 2010;24:1042–51.
Barberán A, Caceres Velazquez H, Jones S, Fierer N. Hiding in plain sight: mining bacterial species records for phenotypic trait information. mSphere. 2017;2:00237–17. mSphere
Leibold MA, Holyoak M, Mouquet N, Amarasekare P, Chase JM, Hoopes MF, et al. The metacommunity concept: a framework for multi-scale community ecology: the metacommunity concept. Ecol Lett. 2004;7:601–13.
Kneitel JM, Chase JM. Trade-offs in community ecology: linking spatial scales and species coexistence. Ecol Lett. 2004;7:69–80.
Zhou J, Ning D. Stochastic community assembly: does it matter in microbial ecology? Microbiol Mol Biol Rev. 2017;81:e00002–17. e00002-17
Lauber CL, Hamady M, Knight R, Fierer N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol. 2009;75:5111–20.
Tripathi BM, Stegen JC, Kim M, Dong K, Adams JM, Lee YK. Soil pH mediates the balance between stochastic and deterministic assembly of bacteria. ISME J. 2018;12:1072–83.
Grainger TN, Gilbert B. Dispersal and diversity in experimental metacommunities: linking theory and practice. Oikos. 2016;125:1213–23.
Michaud JM, Thompson LR, Kaul D, Espinoza JL, Richter RA, Xu ZZ, et al. Taxon-specific aerosolization of bacteria and viruses in an experimental ocean-atmosphere mesocosm. Nat Commun. 2018;9:2017.
Choudoir MJ, Barberán A, Menninger HL, Dunn RR, Fierer N. Variation in range size and dispersal capabilities of microbial taxa. Ecology. 2018;99:322–34.
Vannette RL, Fukami T. Dispersal enhances beta diversity in nectar microbes. Ecol Lett. 2017;20:901–10.
Xu HJ, Wang XH, Li H, Yao HY, Su JQ, Zhu YG. Biochar impacts soil microbial community composition and nitrogen cycling in an acidic soil planted with rape. Environ Sci Technol. 2014;48:9391–9.
Thompson LR, Sanders JG, McDonald D, Amir A, Ladau J, Locey KJ, et al. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature. 2017;551:457–63.
Han Q, Ma Q, Chen Y, Tian B, Xu L, Bai Y, et al. Variation in rhizosphere microbial communities and its association with the symbiotic efficiency of rhizobia in soybean. ISME J. 2020;14:1915–28.
Meier DV, Pjevac P, Bach W, Hourdez S, Girguis PR, Vidoudez C, et al. Niche partitioning of diverse sulfur-oxidizing bacteria at hydrothermal vents. ISME J. 2017;11:1545–58.
Martiny AC, Treseder K, Pusch G. Phylogenetic conservatism of functional traits in microorganisms. ISME J. 2013;7:830–8.
Oton EV, Quince C, Nicol GW, Prosser J, Gubry-Rangin C. Phylogenetic congruence and ecological coherence in terrestrial Thaumarchaeota. ISME J. 2016;10:85–96.
Xia F, Cai Y, Chen D, Qiu L. Edaphobacter acidisoli sp. nov., an acidobacterium isolated from forest soil. Int J Syst Evol Microbiol. 2017;67:4260–5.
Lee SD. Conexibacter stalactiti sp. nov., isolated from stalactites in a lava cave and emended description of the genus Conexibacter. Int J Syst Evol Microbiol. 2017;67:3214–8.
Rainey PB, Travisano M. Adaptive radiation in a heterogeneous environment. Nature. 1998;394:69–72.
Puentes-Téllez PE, Hansen MA, Sørensen SJ, van Elsas JD. Adaptation and heterogeneity of Escherichia coli MC1000 growing in complex environments. Appl Environ Microbiol. 2013;79:1008–17.
Welch RA, Burland V, Plunkett G, Redford P, Roesch P, Rasko D, et al. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc Natl Acad Sci USA. 2002;99:17020–4.
Holt CC, van der Giezen M, Daniels CL, Stentiford GD, Bass D. Spatial and temporal axes impact ecology of the gut microbiome in juvenile European lobster (Homarus gammarus). ISME J. 2020;14:531–43.
Losos JB. Phylogenetic niche conservatism, phylogenetic signal and the relationship between phylogenetic relatedness and ecological similarity among species. Ecol Lett. 2008;11:995–1003.
Morlon H. Phylogenetic approaches for studying diversification. Ecol Lett. 2014;17:508–25.
Fierer N. Embracing the unknown: disentangling the complexities of the soil microbiome. Nat Rev Microbiol. 2017;15:579–90.
Vazquez FJ, Acea MJ, Carballas T. Soil microbial populations after wildfire. FEMS Microbiol Ecol. 1993;13:93–103.
Acknowledgements
This research was jointly supported by the National Natural Science Foundation of China (41520104001, 41721001), 111 Project (B17039) and Fundamental Research Funds for Central Universities in China.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Zhang, L., Ma, B., Tang, C. et al. Habitat heterogeneity induced by pyrogenic organic matter in wildfire-perturbed soils mediates bacterial community assembly processes. ISME J 15, 1943–1955 (2021). https://doi.org/10.1038/s41396-021-00896-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-021-00896-z
This article is cited by
-
Contrasting response of rice rhizosphere microbiomes to in situ cadmium-contaminated soil remediation
Soil Ecology Letters (2024)
-
Wildfire-dependent changes in soil microbiome diversity and function
Nature Microbiology (2022)
-
Ecological and genomic responses of soil microbiomes to high-severity wildfire: linking community assembly to functional potential
The ISME Journal (2022)
-
Association of biochar properties with changes in soil bacterial, fungal and fauna communities and nutrient cycling processes
Biochar (2021)