Abstract
Endosymbiosis of unicellular eukaryotic algae into previously nonphotosynthetic eukaryotes has established chloroplasts in several eukaryotic lineages. In addition, certain unicellular organisms in several different lineages ingest algae and utilize them as temporal chloroplasts (kleptoplasts) for weeks to months before digesting them. Among these organisms, the dinoflagellate Nusuttodinium aeruginosum ingests the cryptomonad Chroomonas sp. and enlarges the kleptoplast with the aid of the cryptomonad nucleus. To understand how the cryptomonad nucleus is remodeled in the dinoflagellate, here we examined changes in the transcriptome and ploidy of the ingested nucleus. We show that, after ingestion, genes involved in metabolism, translation, and DNA replication are upregulated while those involved in sensory systems and cell motility are downregulated. In the dinoflagellate cell, the cryptomonad nucleus undergoes polyploidization that correlates with an increase in the mRNA levels of upregulated genes. In addition, the ingested nucleus almost loses transcriptional responses to light. Because polyploidization and loss of transcriptional regulation are also known to have occurred during the establishment of endosymbiotic organelles, these changes are probably a common trend in endosymbiotic evolution. Furthermore, we show that the kleptoplast and dinoflagellate are more susceptible to high light than the free-living cryptomonad but that the ingested nucleus reduces this damage.
Similar content being viewed by others
Introduction
Several nonphotosynthetic unicellular eukaryotic lineages are capable of accommodating various lineages of algal endosymbionts or chloroplasts sequestrated from algae (known as kleptoplasts) that are ingested by phagocytosis [1]. Such acquired phototrophy is widespread in aquatic ecosystems although it is generally more frequent in oligotrophic environments [2]. In certain cases, organisms that undergo this process become major primary producers; these include red tide ciliates, which obtain kleptoplasts derived from a cryptophyte prey, and some benthic foraminifera, which host several lineages of algae [1].
The process of acquired phototrophy is also believed to have contributed to multiple independent acquisitions of chloroplasts (and nonphotosynthetic plastids) by previously nonphotosynthetic eukaryotic lineages [3, 4]. More than a billion years ago, the nonphotosynthetic eukaryotic ancestor of Archaeplastida acquired a chloroplast through primary endosymbiosis with a cyanobacterium giving rise to glaucophytes, red algae, and Viridiplantae (green algae and plants). Chloroplasts were then further spread into other eukaryotic lineages through secondary and tertiary endosymbiotic events in which a eukaryotic algae was integrated into a heterotrophic eukaryote. The chloroplasts of red algae were distributed to stramenopiles, dinoflagellates, apicomplexans, chromerids, haptophytes, and cryptophytes, whereas those of green algae were obtained by euglenophytes and chlorarachniophytes. In addition, some species of dinoflagellate further replaced their original chloroplast of red algal origin with those of green algae, cryptophytes, haptophytes, or diatoms [3].
Among the acquired phototrophic organisms, those that are kleptoplastic exhibit different degrees of similarity with those possessing chloroplasts of secondary or tertiary endosymbiotic origin depending on the species as follows [1]. Kleptoplastic organisms have been reported in a wide range of eukaryotic lineages such as ciliates [5], dinoflagellates [6,7,8,9], a katablepharid [10], foraminiferans [11], and metazoans [12, 13]. Some species temporally retain kleptoplasts but immediately digest the nucleus of algal prey after ingestion. In contrast, the nucleus of algal prey is temporally retained with the kleptoplast in other species, including the ciliate Mesodinium rubrum, some dinoflagellates, the katablepharid Hatena arenicola. In addition, among the dinoflagellates Durinskia spp., Durinskia capensis temporally retains the kleptoplast and nucleus of a diatom prey, whereas two other species permanently retain diatom cells in which the diatom nucleus and chloroplasts divide in accordance with the cell division of the dinoflagellate host [14, 15]. The retention of the endosymbiont nucleus and the chloroplast division in accord with the host cell cycle are also observed in cryptophytes and chlorarachniophytes, which possess bona fide chloroplasts with a vestigial nucleus of red and green algal origin, respectively, but with a greatly reduced genome size [16, 17].
Generally, kleptoplastic organisms that do not retain their prey’s nucleus are mixotrophic, whereas those that retain the nucleus are functionally closer to phototrophs [1]. In addition, in M. rubrum, the ingested nucleus of the cryptophyte prey is required to enlarge, divide, and sustain the function of kleptoplasts [18]. It has been shown that the ingested nucleus is transcriptionally active in the ciliate cell and its transcriptome changes after ingestion. Such kleptoplast enlargements have not been observed in kleptoplastic organisms that do not retain the nucleus of ingested algae [19]. However, other than in M. rubrum, the function of the ingested nuclei has not yet been studied; thus, it remains unclear whether commonalities exist in the function of ingested nuclei and the evolutionary relationship between kleptoplasty and secondary/tertiary endosymbioses.
Among several kleptoplastic organisms, we have studied kleptoplasty in the dinoflagellates Nusuttodinium spp., which transiently retain kleptoplasts derived from a cryptomonad prey [6, 19,20,21,22,23]. We previously showed that Nusuttodinium poecilochroum digests the cryptomonad nucleus within a few hours of ingestion, while keeping but not enlarging the kleptoplast for about a week. In contrast, Nusuttodinium aeruginosum retains the cryptomonad nucleus, enlarges the kleptoplast more than 20-fold, and keeps it for more than a month after ingestion [19, 20]. In N. aeruginosum, the ingested cryptomonad nucleus is inherited by only one of the two daughter cells during cell division. The cell without the cryptomonad nucleus stops enlarging the kleptoplast and, after three rounds of cell division, starts to digest it [20]. These observations suggest that the retention of the cryptomonad nucleus is required to enlarge the kleptoplast and maintain its function for a longer period. We also found an obligate heterotrophic species feeding on a cryptomonad that is closely related to Nusuttodinium spp. [24]. Taken together, Nusuttodinium spp. are ideal organisms to study the evolution of kleptoplasty.
In order to obtain insights into how the ingested cryptomonad nucleus supports the growth and function of the kleptoplast, we examined the changes in the structure and transcriptome of the nucleus during the course of kleptoplasty in N. aeruginosum. In addition, we compared the effects of high light stress on N. aeruginosum in cells with and without the cryptomonad nucleus. Here we show that similar to M. rubrum, cryptomonad genes involved in metabolism and translation are upregulated after being ingested by N. aeruginosum, which is in accordance with polyploidization of the nucleus. Furthermore, transcriptional responses to illumination are almost lost in the cryptomonad nucleus after ingestion. Moreover, our results suggest that the cryptomonad nucleus reduces high light damage on the kleptoplast and N. aeruginosum. Because polyploidization and loss of transcriptional regulation were also observed in nucleomorph and organelle genomes, these events are common trends in evolution of kleptoplasty and the course of chloroplast establishment by secondary/tertiary endosymbioses.
Materials and methods
Cell cultures
The dinoflagellate N. aeruginosum and the cryptophytes Chroomonas spp. Dc01 and HrL01 were collected in previous studies [19,20,21]. To remove bacteria in respective cultures, the cells were washed several times with sterilized AF-6 medium (an inorganic medium; https://mcc.nies.go.jp/medium/en/af6.pdf) by micropipetting. The cells were incubated at 20 °C under continuous light (10 µmol photons m−2 s−1) and, unless otherwise indicated, in 25-cm2 tissue culture flasks without agitation. N. aeruginosum was fed Chroomonas sp. Dc01 on a regular basis.
RNA preparation for transcriptomic analyses
To analyze the transcriptome of N. aeruginosum in which the nuclei and kleptoplasts derived from Chroomonas sp. Dc01 were being enlarged, free Chroomonas sp. were removed from the coculture using a plankton-net with 5-µm pores (NY5-HC, SEFER) to inhibit N. aeruginosum newly ingesting Chroomonas sp. Then, N. aeruginosum with kleptoplasts (35.2 × 102 ± 1.70 cells/mL; three independent cultures) were inoculated into 175 mL of fresh AF-6 medium in a 75-cm2 cell culture flask. The culture was incubated with illumination for 4 days so that the kleptoplasts were enlarged. After being incubated in the dark for 24 h (hour 0), the culture was illuminated for 12 h and then returned to the dark. For comparison, free-living Chroomonas sp. Dc01 (111.9 × 103 ± 7.23 cells/mL; three independent cultures) were cultured alone under the same conditions.
Cells were collected at hours 0 (just before illumination), 1, 6 (during the light period), 12 (just before dark), and 13 (1 h after the transfer to dark) by centrifugation at 1500 × g for 5 min. The cell pellet was immediately frozen in liquid nitrogen. Total RNA was extracted using a NucleoSpin RNA XS kit (TaKaRa). To construct a cDNA library of 250–300 bp, 0.7–2.7 μg (N. aeruginosum culture), and 5.2–9.4 μg (free-living Chroomonas sp. Dc01 culture) of total RNA were used. Paired-end sequencing was performed using the Illumina sequencing platform (Novaseq 6000) according to the manufacturer’s instructions (Illumina).
De novo assembly and annotation of Chroomonas sp. RNA-seq data
The RNA-seq raw reads were cleaned up using cutadapt ver. 1.81 [25] by trimming low-quality ends (<QV30) and adapter sequences and by discarding reads shorter than 50 bp. The trimmed reads of the free Chroomonas sp. culture were assembled de novo using Trinity ver. 2.0.6 [26] with the paired-end mode and the option “--min_contig_length 300,” and the output fasta files were clustered together using CD-HIT ver. 4.6.8 [27] with the “-c 0.95” option. When splicing variants of a gene were found, the longest transcript was selected as a representative mRNA sequence. Open reading frames (orfs) were predicted by TransDecoder ver. 5.1.0 (http://transdecoder.github.io). The contigs were used as the reference for the Chroomonas sp. Dc01 transcripts. To obtain gene expression scores, one side of the trimmed paired-end reads was mapped to the reference by Bowtie2 ver. 2.3.4.1 [28]. SAMtools ver. 1.8 [29], BEDtools ver. 2.19.1 [30], and R ver. 3.5.3 [31] were used to calculate the number of reads mapped to the contigs (raw count).
Analyses of transcriptomic changes
To compare the nuclear transcriptome of free-living Chroomonas sp. Dc01 with that of Chroomonas sp. Dc01 ingested by N. aeruginosum, mapped read counts (count per million: CPM) of each contig at five time points were averaged. The genes with low mapped read counts (average CPM < 1) were omitted. The data were then normalized between free-living and ingested Chroomonas sp., and differentially expressed genes (DEGs) were identified by edgeR ver. 3.24.3 [32] in R. Contigs of ingested Chroomonas sp. were defined as DEGs when the false discovery rate (FDR) was <0.01 and the absolute value of logFC (log2-fold-change of ingested state vs. free-living state) was >2. Upregulated and downregulated genes were categorized without redundancy according to the second level terms of the Kyoto Encyclopedia of Genes and Genomes (KEGG) [33]. The KEGG Orthology ID assignment was performed for all orfs in the KAAS pipeline [34].
To compare changes in the transcriptome in light and dark conditions, the count data were analyzed by TCC ver. 1.22.1 [35] in R. The low count contigs (CPM < 1) were omitted. Principal component analysis (PCA) of count data, which were normalized by TCC, was performed using R. DEGs of free-living Chroomonas sp. were confirmed only when FDR was <0.01 and the absolute value of logFC (log2 fold-change of either hour 1, 16, 12, or 13 vs. hour 0) was >2. To compare expression patterns between free-living and ingested Chroomonas sp., the count data of both were normalized together by TCC in R. The logFC (log2 fold-change against the value of free-living Chroomonas sp. at hour 0) was calculated based on the normalized count data + 1. The logFC values of 2708 DEGs were clustered by the k-means method (k = 7).
Quantification of SYBR green fluorescence and chloroplast fluorescence
Free Chroomonas sp. was removed from the coculture, and N. aeruginosum were starved in the light for 14 days until they had digested most of the kleptoplasts derived from Chroomonas sp. Then, the starved N. aeruginosum were fed Chroomonas sp. (the ratio of N. aeruginosum:Chroomonas sp. was 1:3) for 9 h until the majority of the cells had ingested Chroomonas sp. in the light. After free Chroomonas sp. was removed, N. aeruginosum with kleptoplasts (4.0 × 102 cells/mL) were cultured in 175 mL of AF-6 medium in 75-cm2 cell culture flasks in the light. One milliliter of the culture was fixed with 0.3% glutaraldehyde, stained with 1/10,000-diluted SYBR Green I (LONZA) for 1 h, and observed using fluorescence microscopy. The intensities of kleptoplast red fluorescence and SYBR Green fluorescence of Chroomonas sp. nuclei were quantified with ImageJ [36].
Quantitative PCR (qPCR)
After starvation of N. aeruginosum, it was fed Chroomonas sp. HrL01 in the light instead of Dc01 to avoid amplification of residual Chroomonas sp. Dc01 DNA by qPCR. Free Chroomonas sp. HrL01 were removed, and N. aeruginosum were starved for 5 days in the light until they had digested most of the kleptoplasts derived from Chroomonas sp. HrL01. The starved N. aeruginosum were then fed Chroomonas sp. Dc01 (the ratio of N. aeruginosum:Chroomonas sp. was 1:2) for 9 h until the majority of cells had ingested Chroomonas sp. Free Chroomonas sp. were removed, and the N. aeruginosum with kleptoplasts were inoculated into 240 mL of fresh AF-6 medium in 75-cm2 cell culture flasks (3.33 × 102 ± 0.577 cells/mL; three independent cultures). Subsequently, on day 0, the N. aeruginosum with kleptoplasts were cultured in the light.
Total DNA was extracted with a DNeasy® Plant Mini Kit (QIAGEN). qPCR was performed with 70 pg of total DNA for each sample using a StepOne-Plus Real-Time PCR system (Life Technologies) and Power SYBR Green Master Mix (Life Technologies). The values of the respective genes were normalized with that of N. aeruginosum 18S rDNA. The primer sequences are shown in Supplementary Table 3.
Quantitative reverse transcription-PCR (qRT-PCR)
The N. aeruginosum cells were starved, as described in the “Quantification of SYBR Green fluorescence…” section, and then Chroomonas sp. cells were added to the culture (the ratio of N. aeruginosum:Chroomonas sp. cells was 1:10). After the majority of N. aeruginosum cells had ingested Chroomonas sp. (9 h after the addition of Chroomonas sp.), free Chroomonas sp. cells were removed (day 0), and then N. aeruginosum with kleptoplasts (7.0 × 102 cells/mL) were cultured in 175 mL of AF-6 medium in 75-cm2 cell culture flasks in the light. N. aeruginosum cells were harvested from 50 mL of culture at days 0, 6, and 12, and the cell pellets were stored at −80 °C. To quantify mRNA levels of free Chroomonas sp., 50 mL of culture was harvested just after the addition of Chroomonas sp. to the starved N. aeruginosum, and the cell pellets were stored at −80 °C. Immediately before RNA extraction, 2 mL of Cyanidioschyzon merolae 10D culture (OD750 = 0.406), which was exponentially growing in continuous light [37], was added to each sample as an internal control (i.e., equivalent to the culture volume).
Total RNA was extracted using a NucleoSpin RNA XS kit (TaKaRa). cDNA was synthesized from 500 ng of total RNA using a mixture of a random hexamer and oligo dT primer (1:9 ratio in concentration) with PrimeScript RTase (TaKaRa). qPCR analyses were performed, as described above. The values of the respective genes were normalized with the values of C. merolae DRP3 mRNA and the number of Chroomonas sp. nuclei per culture volume. The primer sequences are shown in Supplementary Table 3.
Cultures at different light intensities
To examine the cellular growth of N. aeruginosum with kleptoplasts, free Chroomonas sp. Dc01 was removed from the coculture, and N. aeruginosum was starved for 12 days in the light until 40% of the cells had lost the nuclei but not the kleptoplasts derived from Chroomonas sp. Then, N. aeruginosum (23.5 × 102 ± 1.55; three independent cultures) and free-living Chroomonas sp. Dc01 (69.3 × 103 ± 2.52 cells/mL; three independent cultures; chlorophyll a concentration, which was determined as described by Jeffrey and Humphrey [38], was equivalent to that of the N. aeruginosum culture) were inoculated respectively into 6 mL of fresh AF-6 medium in a 6-well microplate and cultured in the dark or in the light (10, 50, 100, or 200 µmol photons m−2 s−1) for 5 days.
Results
Establishment of an axenic coculture of the dinoflagellate N. aeruginosum and the cryptomonad Chroomonas sp.
In our previous study, we isolated the kleptoplastic dinoflagellate N. aeruginosum and its prey, Chroomonas sp. Dc01 from a mixture of unicellular organisms in the same freshwater pond (Fig. 1a, b); however, we had not yet established their axenic cultures [19]. Within the scope of our testing to date, N. aeruginosum ingests Chroomonas spp. Dc01 and HrL01 but not other cryptophytes or eukaryotic algae. In the present study, to avoid the possible effects of contaminated bacteria on the activities of the dinoflagellate and cryptomonads, we first removed bacteria from the cultures of N. aeruginosum and Chroomonas spp. by serial micropipetting and transferring the cells to a sterilized inorganic medium under a microscope. The strains were maintained under continuous light conditions (10 µmol photons m−2 s−1) without agitation at 20 °C. N. aeruginosum cells were fed Chroomonas sp. Dc01 (Fig. 1c). For 3 years after the sterilization, we have confirmed that the respective cultures are axenic by phase-contrast and differential interference contrast microscopy with a 100× objective lens and based on no colony formation on Luria Broth agar plate (9 cm in diameter) 3 days after streaking 0.5 mL of cocultured medium and incubation at 20 °C for 7 days.
a The pond in Hokkaido prefecture, Japan from which N. aeruginosum and Chroomonas sp. Dc01 were collected. b Micrograph of a water sample from the pond showing the coexistence of N. aeruginosum and unicellular algae. The cells were collected on September 24, 2010 [19]. Scale bar = 10 μm. c Micrograph showing the axenic coculture of N. aeruginosum and Chroomonas sp. The coculture contains Chroomonas sp. as a source kleptoplast of N. aeruginosum as well as N. aeruginosum cells with or without kleptoplast, and cysts. Scale bar = 50 μm. d Photograph of a plankton-net filter and a schematic drawing showing the separation of N. aeruginosum cells with Chroomonas sp.-derived kleptoplasts from free Chroomonas sp. (those had not been ingested by N. aeruginosum). A 5-µm diameter plankton-net filter was attached to the bottom of the sieve. The coculture of N. aeruginosum and Chroomonas sp. was filtrated to separate free Chroomonas sp. from N. aeruginosum with kleptoplasts. N. aeruginosum cells on the filter were washed several times before further experiments. e Micrographs showing the coculture of N. aeruginosum cells with Chroomonas sp.-derived kleptoplasts and free Chroomonas sp. (left), filtered N. aeruginosum cells that are separated from free Chroomonas sp. (middle), and free Chroomonas sp. in the flow-through fraction. Scale bar = 10 μm.
To compare Chroomonas sp. in the free-living population with those that were ingested by N. aeruginosum, we developed a filtration procedure to fractionate Chroomonas sp. that had and had not been ingested by N. aeruginosum. When the coculture of N. aeruginosum and Chroomonas sp. Dc01 was passed through a filter with 5-µm pores (Fig. 1d), all N. aeruginosum cells remained on the filter, while all free Chroomonas sp. passed through (Fig. 1e).
Genes involved in metabolism, protein synthesis, and DNA replication are upregulated in the cryptomonad nucleus after ingestion by the dinoflagellate
We previously observed that during cell division of N. aeruginosum, the ingested nucleus was inherited by only one of the two daughter cells. The daughter cell with the Chroomonas sp. nucleus continued to enlarge the kleptoplast while that without the nucleus did not. These results suggested that the ingested nucleus is required for kleptoplast enlargement and that the ingested nucleus is probably transcriptionally active [19, 20].
To examine how the transcriptome changes upon ingestion by N. aeruginosum, we compared the nuclear transcriptome of free (free-living Chroomonas sp.) and ingested Chroomonas sp. (5 days after ingestion by N. aeruginosum) by RNA-seq analysis (Fig. 2a). To this end, cells were precultured in the dark for 24 h, illuminated for 12 h, and again returned to the dark. Mapped read counts (count per million: CPM) of each contig at five time points [hour 0 (just before illumination), 1, 6, 12 (during illumination), and 13 (1 h after the transfer to the dark)] were averaged and then compared (Fig. 2b, c). From a total of 70,969 Chroomonas sp. nuclear contigs, 2505 (3.53%) and 1874 (2.64%) contigs were upregulated (FDR < 0.01, logFC of ingested state vs free-living state > 2) and downregulated (FDR < 0.01, logFC of ingested state vs free-living state < −2), respectively, after ingestion (Fig. 2b; Supplementary Table 1). To further evaluate the functional change of the Chroomonas sp. nucleus following ingestion, the upregulated and downregulated contigs were assigned to the KEGG database [33] (Fig. 2c). Upregulated contigs were predominantly found in “Metabolism” and “Genetic information processing” categories (Fig. 2c). These included contigs annotated as genes encoding components of photosystems, proteins involved in carbon fixation (e.g., carbonic anhydrase and proteins involved in carbohydrate metabolism), proteins involved in translation (e.g., ribosomal proteins), and proteins involved in nuclear DNA replication (MCM proteins and DNA primase) (Fig. 2c; Supplementary Table 1). In addition, these contigs included genes encoding superoxide dismutase, ascorbate peroxidase, and thioredoxin reductase, which are involved in the dissipation of reactive oxygen species (ROS) [39,40,41] (Fig. 2c; Supplementary Table 1). In contrast, “Environmental information processing,” “Cellular Processes,” and “Organismal systems” categories predominantly contained downregulated contigs (Fig. 2c). Contigs annotated as genes encoding actin, tubulin, CDC20 [42], and CDC14 [43], which are involved in M-phase progression, were also downregulated.
a Schematic diagrams showing the culture of free-living Chroomonas sp. Dc01 and that of Chroomonas sp. Dc01 ingested by N. aeruginosum used for comparison of the nuclear transcriptomes of Chroomonas sp. Dc01 in each state. Both cultures were pre-incubated in the dark for 24 h. Then, from hour 0, the cultures were subjected to 12-h-light and subsequently dark. Nuclear transcriptomes of Chroomonas sp. Dc01 were examined at hours 0 (just before illumination), 1, 6 (during the light period), 12 (just before the dark), and 13 (during the dark period). To inhibit N. aeruginosum from ingesting any new Chroomonas sp. Dc01, free-living Chroomonas sp. were removed from the coculture 4 days prior to the dark preincubation. b M-A plot showing 2505 upregulated and 1874 downregulated contigs from Chroomonas sp. ingested by N. aeruginosum cells and free-living Chroomonas sp. cells. The averages of respective mRNA levels at five time points (hours 0, 1, 6, 12, and 13) were compared. Upregulated and downregulated contigs were defined as those that had FDR < 0.01 and logFC > 2 (indicated in red) and FDR < 0.01 and logFC < −2 (indicated in blue), respectively (three independent cultures). The x-axis represents averaged logCPM, and the y-axis represents the base 2 logarithm of the fold change to the free-living condition. c Comparison of the number of upregulated and downregulated contigs whose functions were assigned to respective KEGG functional categories.
The cryptophyte nucleus almost loses its transcriptional response to dark–light shift after ingestion by the dinoflagellate
As described later, both enlargements of the kleptoplast and N. aeruginosum growth depend on light (i.e., photosynthesis by the kleptoplast). In addition, it is well known that illumination largely changes transcriptome in photosynthetic organisms e.g. [44, 45]. To assess how the ingested Chroomonas sp. nucleus responds to dark/light shift, we examined and compared the transcriptomic changes of free and ingested Chroomonas sp. (5 days after ingestion by N. aeruginosum) following illumination after preculture in the dark. Cells were precultured in the dark for 24 h, illuminated for 12 h, and again returned to the dark. The nuclear transcriptome of free and ingested Chroomonas sp. was examined at hours 0 (just before illumination), 1, 6, 12 (during illumination), and 13 (1 h after the transfer to the dark) (Figs. 2a and 3).
The RNA-seq data were obtained from the process described in Fig. 2a. a PCA of the transcriptomic changes of the free and ingested Chroomonas sp. following illumination. The raw count data were normalized with TCC using R and then plotted (low count contigs were omitted before normalization). The square and circle correspond to free-living and ingested Chroomonas sp. at the indicated time point, respectively. The color represents the presence (orange) or absence (gray) of light illumination. b Heatmap showing the expression patterns of DEGs (log2-fold-change at any time point from hour 0 > 2; FDR < 0.01; three independent cultures) in the free-living Chroomonas sp. and a comparison with those in the ingested Chroomonas sp. The levels in the free-living Chroomonas sp. at hour 0 were defined as “0.” Yellow and blue indicate upregulation and downregulation of the gene, respectively, compared with the level at hour 0 in the free-living Chroomonas sp. The clustering was performed by the k-means method. c Comparison of the number of contigs whose functions were assigned to respective KEGG functional categories of second level terms in respective clusters. Colors correspond to clusters 1–7 in (b).
PCA showed that a conspicuous difference existed between the free-living and ingested Chroomonas sp. along the PC1 axis (Fig. 3a). Along the PC2 axis, the transcriptomes in free-living states were separated according to the dark (hour 0: just before illumination) and light (hours 1, 6, and 12) conditions. The hour 13 (1 h after the dark shift) transcriptomes moved toward those of hour 0 in both the free-living and ingested states (Fig. 3a). Furthermore, the transcriptomes at any time point were largely different between the free-living and ingested states (Fig. 3a).
In the PCA analysis, there was less difference upon the illumination (PC2 axis) in the ingested state than in the free-living state (Fig. 3a; differences in principal component 2 score between hour 0 and 1 and between hour 0 and 12 in the ingested state were significantly less than those in free-living state; p = 0.0044 and 0.0046, respectively, t-test), which suggests that the transcriptomic response of the Chroomonas sp. nucleus to light was compromised after it was ingested by N. aeruginosum. To further investigate this finding, we identified all DEGs following illumination in the free-living state (FDR < 0.01 and |logFC of either hour 1, 16, 12, or 13 vs. hour 0 | > 2; 2708 contigs, Supplementary Table 2) and then compared their expression patterns with those in the ingested state (Fig. 3b). As suggested by the PCA analysis, changes in levels of mRNAs that were identified as DEGs in the free-living state were abolished or compromised in the ingested state (Fig. 3b). By k-means clustering, the DEGs in the free-living state were classified according to the extent by which the magnitude of the changes was reduced in the ingested state (Fig. 3b). Relative to the free-living state, clusters 1 (208 contigs) and 2 (668 contigs) were highly expressed in the ingested state regardless of dark or light conditions. In these clusters, the majority of the contigs were assigned to the “metabolism” category by KEGG classification. Clusters 3 (812 contigs) and 4 (350 contigs) were slightly upregulated in the ingested state following illumination, and they included a majority of “translation” genes. Relative to the free-living state, clusters 5 (122 contigs) and 6 (375 contigs) were downregulated in the ingested state regardless of dark or light conditions. In the ingested state, Cluster 7 (173 contigs) almost lost the change of the DEGs in the free-living state following illumination (Fig. 3b, c, Supplementary Table 2).
In summary, the analyses of transcriptomes showed that the Chroomonas sp. nucleus almost lost its transcriptional response to illumination following ingestion by N. aeruginosum. Furthermore, contigs annotated as genes involved in metabolism were upregulated in the ingested state relative to the free-living state regardless of light conditions, while contigs annotated as genes involved in translation were slightly upregulated following illumination although the magnitude of the changes was reduced compared with the free-living state.
Increase in the ploidy of the cryptomonad genome and mRNAs after ingestion by the dinoflagellate
The transcriptome analyses showed that genes involved in nuclear DNA replication (S phase) were upregulated while certain M-phase genes were downregulated in Chroomonas sp. after ingestion by N. aeruginosum. Thus, we hypothesized that the Chroomonas sp. nucleus continued to replicate in N. aeruginosum. To test this prediction, we examined the change in the size of the nucleus and the ploidy of nuclear and organellar genomes of Chroomonas sp. after ingestion by N. aeruginosum. In addition, we examined the relationship between the change in the nucleus and kleptoplast after ingestion after removing free Chroomonas sp. (Fig. 4).
a Schematic diagrams showing the coculture of N. aeruginosum and Chroomonas sp. Dc01 used to investigate the change in the ploidy level of Chroomonas sp. Dc01 ingested by N. aeruginosum in (b–e). Chroomonas sp. Dc01 was removed from the coculture for N. aeruginosum cells to digest most of the kleptoplasts derived from Chroomonas sp. (~95 and 60% of N. aeruginosum cells had completely digested the nucleus and kleptoplast derived from Chroomonas sp., respectively). Then, the starved N. aeruginosum cells were again fed Chroomonas sp. until approximately 75% of the cells had ingested Chroomonas sp. After this, free-living Chroomonas sp. was removed from the coculture (day 0) and N. aeruginosum cells with kleptoplasts were cultured for 14 days under continuous light (10 µmol photons m−2 s−1). b Micrographs showing the change in the nucleus and kleptoplast derived from Chroomonas sp. in N. aeruginosum cells. Cells were stained with SYBR Green. Images of differential interference contrast (DIC), SYBR Green staining, and kleptoplast red fluorescence (Chl) are shown. The arrowhead indicates a nucleus derived from Chroomonas sp. The cell in the image from day 3 ingested two nuclei. Scale bar = 10 μm. c Change in N. aeruginosum cell density. d Change in the ratio of N. aeruginosum cells retaining the nucleus derived from Chroomonas sp. and kleptoplast fluorescence intensity per N. aeruginosum cell. For kleptoplast fluorescence, the average intensity at day 0 was defined as “1.0.” The kleptoplast fluorescence intensity was quantified both in N. aeruginosum cells with and without the nucleus derived from Chroomonas sp. The error bars represent standard deviation (n = 30 cells for each time point). e Change in SYBR Green fluorescence intensity per Chroomonas sp. nucleus ingested by N. aeruginosum. The average at day 0 was defined as “1.0.” The error bars represent standard deviation (n = 30 nuclei). f Schematic diagrams showing the coculture of N. aeruginosum and Chroomonas sp. used to investigate the change in the ploidy level of Chroomonas sp. Dc01 ingested by N. aeruginosum in (g, h). To avoid qPCR amplification of residual DNA from Chroomonas sp. Dc01 after starvation of N. aeruginosum, Chroomonas sp. HrL01 instead of Dc01 was fed to N. aeruginosum before starvation and then Dc01 was added to the culture. Others are the same as in (a). g Micrographs showing that described in (b) except that they show the cells from (f) instead of (a). h qPCR showing changes in ploidy of nuclear (rpl8 gene), mitochondrial (cox1 gene), nucleomorph (gidA gene), and kleptoplast (rpoB gene) genomes of Chroomonas sp. Dc01 ingested by N. aeruginosum. The values were normalized with values of N. aeruginosum 18S rDNA. The average at day 0 was defined as “1.0” for respective genomes. The error bars represent standard deviation (three independent cultures).
The size and fluorescence intensity of kleptoplasts per N. aeruginosum cell peaked at 5–6 days and then decreased gradually (Fig. 4b, c; note that some N. aeruginosum ingested two or more Chroomonas sp. cells during feeding in Fig. 4a). After removal of free Chroomonas sp. (day 0), the percentage of N. aeruginosum with Chroomonas sp. nuclei increased until day 5 because of the death of cells that had not ingested Chroomonas sp. From then on, the percentage decreased as a result of unequal inheritance of the Chroomonas sp. nuclei by N. aeruginosum daughter cells during cell division [20], which coincided with the reduction of kleptoplast size per N. aeruginosum cell on average (Fig. 4a–d).
The size and fluorescent intensity of SYBR Green staining of the ingested nucleus continued to increase by up to ten times the original size before reaching a plateau at day 12 (Fig. 4b, e). qPCR analysis also showed that the copy number of Chroomonas sp. Dc01 nuclear rpl8 gene (per N. aeruginosum nuclear rDNA gene) increased until day 6 and then decreased (Fig. 4f–h). A discrepancy between the change in nuclear size and SYBR Green intensity (Fig. 4e), which kept increasing, and the qPCR result, in which the copy number per N. aeruginosum nuclear genome decreased from day 6 (Fig. 4h), was probably due to the ratio of N. aeruginosum cells with Chroomonas sp. nuclei, which decreased from day 6 because of the unequal inheritance of nuclei during N. aeruginosum cell division (Fig. 4d, g). In addition to the nuclear genome, qPCR showed that DNA content of mitochondrial (cox1), nucleomorph (gidA), and chloroplast (rpoB) genomes of Chroomonas sp. Dc01 also increased until day 6 and then decreased.
Transcriptome analyses showed that many contigs annotated as Chroomonas sp. nuclear genes, especially those involved in metabolism and translation, were upregulated after ingestion. However, the upregulation or downregulation of a particular gene in RNA-seq results indicates a respective increase or decrease in the ratio of the mRNA levels of the gene to the total mRNA of Chroomonas sp. Thus, upregulation or downregulation does not necessarily indicate a respective increase or decrease in the absolute level of mRNA (i.e., the mRNA level of a given gene per Chroomonas sp. nucleus). To investigate the relationship between the upregulated and downregulated genes and the increase in the ploidy of the Chroomonas sp. nucleus, we examined the change in mRNA levels of some genes and that of 18S rRNA (to assess the relative number of ribosomes) per Chroomonas sp. nucleus in the course of ingestion and polyploidization (Fig. 5). To this end, the starved N. aeruginosum cells were fed Chroomonas sp. for 9 h and, afterwards, free Chroomonas sp. cells were removed from the culture (day 0) before N. aeruginosum with kleptoplasts were cultured for 12 days (Fig. 5a). By the removal of free Chroomonas sp., ~60% of the N. aeruginosum cells ingested 1–14 Chroomonas sp. cells. Subsequently, N. aeruginosum cells that did not ingest Chroomonas sp. started to die, resulting in a decrease in the ratio of N. aeruginosum cells without Chroomonas sp. nuclei at day 6 (Fig. 5b, c). After the removal of free Chroomonas sp. from the culture at day 0, the number of Chroomonas sp. nuclei per N. aeruginosum cell also decreased because of the lack of nuclear division of Chroomonas sp. in N. aeruginosum cells and the unequal inheritance during N. aeruginosum cell division (Fig. 5b, c).
a Schematic diagrams showing the coculture of N. aeruginosum and Chroomonas sp. Dc01 used to investigate the change in mRNA and rRNA levels of Chroomonas sp. ingested by N. aeruginosum in (b–d). Chroomonas sp. Dc01 was removed from the coculture for N. aeruginosum cells to digest most of the kleptoplasts derived from Chroomonas sp. (~95% and 60% of N. aeruginosum cells had completely digested the nucleus and kleptoplast derived from Chroomonas sp., respectively). The starved N. aeruginosum cells were again fed Chroomonas sp. until ~60% of the cells had ingested Chroomonas sp. Then, the free-living Chroomonas sp. was removed from the coculture (day 0) and N. aeruginosum cells with kleptoplasts were cultured for 12 days under continuous light (10 µmol photons m−2 s−1). b Change in N. aeruginosum cell number after the removal of free Chroomonas sp. c Histogram showing the change in the ratio of N. aeruginosum cells possessing cryptomonad nuclei and the number of the nuclei per cell. d RT-qPCR showing the changes in mRNA levels per Chroomonas sp. nucleus of LHC1 (LIGHT HARVESTING COMPLEX 1), PSBM, PETC, FBA, APX, RPL10, ACTIN, and TUBULIN A mRNAs, and 18S rRNA, upon and after ingestion. The values were normalized with that of an internal control, which was equivalent to the culture volume, and the number of Chroomonas sp. nuclei per culture volume. The averaged value of free-living Chroomonas sp. was defined as “1.0.” The levels in the free-living Chroomonas sp. were determined in the culture immediately after the addition of Chroomonas sp. to the starved N. aeruginosum culture. The error bars represent standard deviation (four independent cultures). *P < 0.05; N.S., no significant difference (t-test).
qRT-PCR showed that the 18S rRNA level per Chroomonas sp. nucleus remained almost constant upon ingestion and then increased in the N. aeruginosum cell. Of six genes that were found to be upregulated DEGs in the transcriptome analysis, mRNA levels of PSBM, PETC, LHC1, and RPL10 per Chroomonas sp. nucleus increased upon ingestion (i.e., free vs. hour 0 in Fig. 5d). After ingestion, mRNA levels of all the six genes continued increasing (Fig. 5d) in accordance with polyploidization of the ingested nucleus (Fig. 4). In contrast, mRNA levels of the downregulated DEGs ACTIN and TUBULIN A per Chroomonas sp. nucleus decreased upon ingestion and then increased afterward, although the magnitudes of their increase were smaller than those of PSBM, PETC, LHC1 FBA, APX, and RPL10 (Fig. 5d). Therefore, the increase or decrease of mRNA levels upon ingestion and the differential magnitudes of the increase during polyploidization of the ingested nucleus probably contributed to the upregulation or downregulation of genes in the transcriptome (Fig. 2).
The ingested cryptophyte nucleus allays high light-induced damage to the kleptoplasts and dinoflagellate cell
Above results suggest that the ingested nucleus supports the growth of the kleptoplast in N. aeruginosum cells. To elucidate any additional effects of the ingested Chroomonas sp. nucleus on the kleptoplast and/or dinoflagellate host, we changed the light intensity of the culture, which led to changes in photosynthetic activity and oxidative stress, and we examined the states of N. aeruginosum and the kleptoplast with or without the ingested nuclei.
We first compared the cellular growth rate in the culture of Chroomonas sp. alone with that in N. aeruginosum cells that had mostly retained Chroomonas sp. nuclei (immediately after free Chroomonas sp. cells were removed from the coculture with N. aeruginosum) under various light intensities: in the dark or in the light at 10, 50, 100, or 200 µmol photons m−2 s−1 (Fig. 6a, b). In the dark, neither free Chroomonas sp. nor N. aeruginosum that had ingested Chroomonas sp. grew (Fig. 6a, b), which shows that N. aeruginosum growth requires light and is based on photosynthesis by Chroomonas sp. The growth rate of Chroomonas sp. peaked at 50 µmol photons m−2 s−1 and decreased gradually as light intensity increased further (Fig. 6b; Supplementary Fig. 1). In contrast, the growth rate of N. aeruginosum with ingested Chroomonas sp. nuclei peaked at 10 µmol photons m−2 s−1 and substantially decreased at 50 and 100 µmol photons m−2 s−1; at 200 µmol photons m−2 s−1, the cell number of N. aeruginosum decreased further (Fig. 6b; Supplementary Fig. 1).
a To examine the cellular growth of N. aeruginosum with kleptoplasts, free Chroomonas sp. Dc01 was first removed from the coculture with illumination (10 µmol photons m−2 s−1) in order to inhibit N. aeruginosum from newly ingesting Chroomonas sp., and then the culture was exposed to the indicated light intensities. Changes in N. aeruginosum cell number for 5 days were determined. Free Chroomonas sp. Dc01 alone was simultaneously cultured in the same inorganic medium with illumination (10 µmol photons m−2 s−1) and then exposed to the indicated light intensities for 5 days. b Growth rates under the indicated light intensities. The growth rate (µ) was calculated as the difference between the natural logarithm of the cell number (N) at different time points (t, day 0–5) according to the following equation: µ = ln(Nt5/Nt0)/(t5−t0). The error bars represent standard deviation (three independent cultures). *P < 0.05; N.S., no significant difference (t-test). The changes in respective cell numbers are show in Supplementary Fig. 1. c To examine the effects of nuclei derived from Chroomonas sp. Dc01 on the growth/survival of N. aeruginosum in e and f, free Chroomonas sp. Dc01 was removed from the coculture with illumination (10 µmol photons m−2 s−1). N. aeruginosum with kleptoplasts were further cultured for 12 days until ~40% of the cells had lost nuclei derived from Chroomonas sp. Dc01. Then, from hour 0, the culture was kept at 10 µmol photons m−2 s−1 or subjected to 200 µmol photons m−2 s−1, and further cultured for 5 days. d Micrographs showing the change in the nucleus and kleptoplast derived from Chroomonas sp. in N. aeruginosum cells at 10 and 200 µmol photons m−2 s−1, respectively. Cells were stained with SYBR Green. Images of differential interference contrast (DIC), SYBR Green staining, and kleptoplast red fluorescence (Chl) are shown. The arrowhead indicates a nucleus derived from Chroomonas sp. For comparison, Chroomonas sp. Dc01 cells alone, which were cultured at 200 µmol photons m−2 s−1, are also shown. Scale bar = 10 μm. Changes in N. aeruginosum cell density (e) and ratio of N. aeruginosum cells possessing the nuclei derived from Chroomonas sp. Dc01 (f) at 10 and 200 µmol photons m−2 s−1. The error bars represent standard deviation (three independent cultures).
We also examined cellular growth/survival at either 10 or 200 µmol photons m−2 s−1 in an N. aeruginosum culture in which ~40% of the cells had lost Chroomonas sp. nuclei (Fig. 6c–f). At 10 µmol photons m−2 s−1, cells without Chroomonas sp. nuclei survived for 5 days (Fig. 6c–f). In contrast, at 200 µmol photons m−2 s−1, the number of cells decreased gradually, while the ratio of cells with Chroomonas sp. nuclei increased during 5 days (Fig. 6c–f). Thus, the cells that had lost Chroomonas sp. nuclei apparently died more quickly than those that had retained the nuclei.
By microscopic observation, we found that kleptoplasts gradually turned from green to yellow in N. aeruginosum cultures under 200 but not 10 µmol photons m−2 s−1 regardless of the presence or absence of Chroomonas sp. nuclei; in contrast, the chloroplast of free Chroomonas sp. remained green (Fig. 6d). The rate of cell death, i.e., the decrease in cell number, in N. aeruginosum was higher at 200 µmol photons m−2 s−1 than in the dark (i.e., without photosynthesis), which suggests that cell death under high light is not solely due to the reduced photosynthetic activity of the kleptoplast but also cellular damage in N. aeruginosum cells.
These results suggest that (1) high-intensity light conditions, specifically those in the range in which free Chroomonas sp. can maintain the integrity of chloroplasts and grow, cause lethal damage to kleptoplasts and N. aeruginosum cells and that (2) the ingested Chroomonas sp. nucleus increases the longevity of the kleptoplast and its host N. aeruginosum under high light conditions.
Discussion
Polyploidization and the loss of transcriptional regulation in endosymbionts are common in endosymbiotic evolution
Our previous study showed that N. aeruginosum enlarged the kleptoplast only during the retention of the symbiont nucleus [20]. Here, we have shown that the ingested Chroomonas sp. nucleus is transcriptionally active in N. aeruginosum. Following ingestion, contigs annotated as genes involved in metabolism, including photosynthesis and protein synthesis, were upregulated, while those involved in sensory systems and cellular activities, such as signal transduction and cell motility, were downregulated (Fig. 2; Supplementary Table 1). A similar transcriptomic change has been observed in the cryptomonad nucleus after ingestion by M. rubrum, although kleptoplasty was acquired and evolved in the ancestor of this ciliate, independently from that of Nusuttodinium spp. [18, 46, 47]. These results suggest that the cryptomonad cell is enslaved as a “machine” specialized for photosynthetic metabolism and protein synthesis that supports growth of the kleptoplast and dinoflagellate host.
Our results also showed that transcriptomic changes that followed illumination were almost abolished in the ingested cryptomonad nucleus. In the kleptoplasty of N. aeruginosum, the cryptomonad loses the plasma membrane after being ingested and its cytosol, therefore, becomes enclosed in the phagosome of the dinoflagellate host [19, 20]; presumably, this keeps the cryptomonad cytosol in a more stable condition than if it were exposed to a natural water environment across cell membrane. In addition, the eyespot and flagella of the cryptomonad disappear after it is ingested [19, 20, 48]. Thus, it is apparently reasonable that, after being ingested by the dinoflagellate, the cryptophyte genes involved in sensory systems and cell motility are downregulated and their transcriptomic responses are abolished.
Although the responses of ingested nuclei to environmental changes have not yet been examined in other kleptoplastic organisms, the loss of the transcriptional response observed in our study is likely to be a common evolutionary trend in endosymbiotic enslavement of endosymbionts by eukaryotic hosts (Fig. 7). In the cryptophyte Guillardia theta, cell-cycle-regulated genes, which are specifically transcribed at certain cell-cycle stages in free-living eukaryotes, are constantly expressed from a nucleomorph (a descendant of a red algal endosymbiont) regardless of its replication or division cycle [49]. In the chlorarachniophyte Bigelowiella natans, the mRNA levels of almost all of the genes of the nucleomorph (a descendant of a green algal endosymbiont) remained constant throughout a 12-h-light and 12-h-dark cycle [50]. The loss or reduction of the transcriptional response to light is also applicable to endosymbiotic evolution of organelles from cyanobacterial endosymbionts. In contrast to cyanobacteria, chloroplast-encoded transcripts in the green alga Chlamydomonas reinhardtii exhibited little change following illumination; therefore, it was suggested that levels and activities of chloroplast-encoded proteins are instead regulated post-transcriptionally [51]. The rhizarian amoeba Paulinella chromatophora possesses cyanobacterium-derived photosynthetic organelles known as chromatophores, which were established much more recently than chloroplasts. As is the case in the chloroplast genome, mRNA levels of chromatophore-encoded genes change little following illumination, which suggests that the chromatophore has lost cyanobacterial transcriptional regulation of the response to light [52].
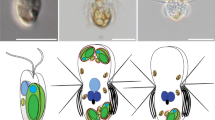
Predators start digesting algae immediately after phagocytic ingestion of algae (left). Kleptoplastic species retain the ingested algae for days to weeks in the cell before digesting them in some cases, including that of N. aeruginosum, with the nucleus of the algae (middle). The present study shows that the ingested nucleus is polyploidized in the host cell, which increases mRNA levels. The ingested nucleus almost loses transcriptional regulation. These changes were also common to the course of establishment of nucleomorph and chloroplast (also other types of plastids) as obligate endosymbionts in eukaryotes (right).
Following ingestion by the dinoflagellate, the ploidy of the cryptomonad nuclear and organellar genomes increased, which correlates with an increase in mRNA levels of the encoded genes (Fig. 4). Our previous study showed that nuclear division ceased in the cryptomonad after it was ingested by the dinoflagellate [19, 20]. In our present study, transcriptome analyses showed that, after ingestion, genes involved in nuclear DNA replication were upregulated, while genes encoding tubulin, which forms spindles for chromosome segregation and an M-phase-protein, CDC20, were downregulated (Fig. 2). These results suggest that the ingested nucleus is arrested in the S phase and continues to be replicated.
In a previous study, although ploidy was not examined, cryptomonad nuclei ingested by the ciliate M. rubrum were shown to have been enlarged by up to four times their original diameter after ingestion [53]. The nucleomorph genomes are diploid in the chlorarachniophyte B. natans and tetraploid in the cryptophyte G. theta, respectively, and it has been suggested that these genomes were polyploidized during endosymbiotic evolution [54]. Chloroplast and mitochondrial genomes also multiplied substantially during endosymbiotic evolution [54, 55]. In addition, the genome of symbiotic or parasitic bacteria such as Buchnera [56], Neisseria [57], Borrelia [58], and Epulopiscium [59] are polyploid. Thus, polyploidization of the genomes of endosymbionts/parasites is apparently a common trend in endosymbiotic/parasitic relationships (Fig. 7). Given that the dinoflagellate host is able to divide and inherit kleptoplasts during cell division and that the ingested nucleus is required to grow the kleptoplast [20], if the replicated chromosomes can be segregated and inherited (amitotically because of the loss of tubulin expression) by both daughter cells, the period of kleptoplast retention will be extended. Indeed, in the dinoflagellate Durinskia baltica, which contains a permanent diatom endosymbiont, the nucleus of the diatom undergoes amitosis and is inherited by daughter dinoflagellate cells during cell division [15]. Furthermore, the nucleomorph apparently divides amitotically [60,61,62].
Comparison of the efficiency of kleptoplasty in the dinoflagellate N. aeruginosum and in other organisms
In comparison to the growth of the free-living cryptomonad, the growth rate of N. aeruginosum with kleptoplasts was saturated at lower light intensities, even when the nucleus of the cryptomonad had been retained (Fig. 6). Even under low light, genes involved in ROS dissipation were upregulated in the cryptomonad nucleus after its ingestion by the dinoflagellate (Supplemental Table 1). With high-intensity light in the range that did not affect the growth and integrity of the free-living cryptomonad’s chloroplast, we found that the kleptoplast was bleached. These results suggest that the dinoflagellate cell is unable to maximize the potential of the cryptomonad chloroplast. In a similar manner, in another dinoflagellate species that possesses a kleptoplast derived from a haptophyte, the photosynthetic activity of the haptophyte was reduced after it was ingested by the dinoflagellate [63].
In contrast, in M. rubrum, the photosynthetic activity of the cryptomonad per chlorophyll is maintained after ingestion; additionally, by enlargement and division of the kleptoplast in the M. rubrum cell, the photosynthetic activity per M. rubrum cell increases after the ingestion of the cryptomonad [64]. In a photosymbiont-harboring radiolarian species, an increase in the electron transfer rate of photosystem II in a haptophyte was observed after ingestion [65]. These organisms are planktonic and often form blooms around the surface of the sea where they are exposed to high light [66,67,68]. Thus, these acquired phototrophic organisms have likely acquired the ability to use kleptoplasts/photosymbionts under high light and thereby cope with photosynthetic oxidative stress. Indeed, M. rubrum has been shown to produce mycosporine-like amino acids [64], which function as UV radiation protectors and antioxidants [69]. In addition, a haptophyte ingested by a radiolarian has been shown to produce dimethylsulfoniopropionate, which also functions as an antioxidant, at levels more than 100-fold greater than those produced by the free-living algae [68, 70].
In contrast to M. rubrum and the radiolarian, N. aeruginosum is benthic [48]. Thus, it appears reasonable that N. aeruginosum is relatively weak to high light and photosynthetic oxidative stress; however, it is currently unclear whether it produces the antioxidants required to cope with oxidative stress in the light. P. chromatophora is also benthic, its chromatophore is bleached, and the cell dies even under moderate light [52]. Therefore, a correlation apparently exists between the habitat of organisms and their ability to use kleptoplasts or photosynthetic organelles; further studies are required to confirm this hypothesis.
Data availability
All of the RNA-seq raw data obtained in this study and the assembled contig sequences were deposited at the DDBJ Sequence Read Archive (accession codes BioProject PRJDB9674) and at the DDBJ Transcriptome Shotgun Assembly Database (accession codes ICPR01000001-ICPR01070969).
References
Johnson MD. The acquisition of phototrophy: adaptive strategies of hosting endosymbionts and organelles. Photosynth Res. 2011;107:117–32.
Stoecker DK, Johnson MD, De Vargas C, Not F. Acquired phototrophy in aquatic protists. Aquat Microb Ecol. 2009;57:279–310.
Keeling PJ. The number, speed, and impact of plastid endosymbioses in eukaryotic evolution. Annu Rev Plant Biol. 2013;64:583–607.
Archibald JM. Endosymbiosis and eukaryotic cell evolution. Curr Biol. 2015;25:R911–21.
Gustafson DE, Stoecker DK, Johnson MD, Van Heukelem WF, Sneider K. Cryptophyte algae are robbed of their organelles by the marine ciliate Mesodinium rubrum. Nature. 2000;405:1049–52.
Takano Y, Yamaguchi H, Inouye I, Moestrup O, Horiguchi T. Phylogeny of five species of Nusuttodinium gen. nov. (Dinophyceae), a genus of unarmoured kleptoplastidic dinoflagellates. Protist. 2014;165:759–78.
Koike K, Takishita K. Anucleated cryptophyte vestiges in the gonyaulacalean dinoflagellates Amylax buxus and Amylax triacantha (Dinophyceae). Phycol Res. 2008;56:301–11.
Gast RJ, Moran DM, Dennett MR, Caron DA. Kleptoplasty in an Antarctic dinoflagellate: caught in evolutionary transition? Environ Microbiol. 2007;9:39–45.
Hehenberger E, Gast RJ, Keeling PJ. A kleptoplastidic dinoflagellate and the tipping point between transient and fully integrated plastid endosymbiosis. Proc Natl Acad Sci USA. 2019;36:17934–42.
Okamoto N, Inouye I. Hatena arenicola gen. et sp. nov., a katablepharid undergoing probable plastid acquisition. Protist. 2006;157:401–19.
Lekieffre C, Jauffrais T, Geslin E, Jesus B, Bernhard JM, Giovani ME, et al. Inorganic carbon and nitrogen assimilation in cellular compartments of a benthic kleptoplastic foraminifer. Sci Rep. 2018;8:1–12.
Serôdio J, Cruz S, Cartaxana P, Calado R. Photophysiology of kleptoplasts: photosynthetic use of light by chloroplasts living in animal cells. Philos Trans R Soc B Biol Sci. 2014;369:20130242.
Van Steenkiste NWL, Stephenson I, Herranz M, Husnik F, Keeling PJ, Leander BS. A new case of kleptoplasty in animals: marine flatworms steal functional plastids from diatoms. Sci Adv. 2019;5:eaaw4337.
Yamada N, Bolton JJ, Trobajo R, Mann DG, Dąbek P, Witkowski A, et al. Discovery of a kleptoplastic ‘dinotom’ dinoflagellate and the unique nuclear dynamics of converting kleptoplastids to permanent plastids. Sci Rep. 2019;9:1–13.
Tippit DH, Pickett Heaps JD. Apparent amitosis in the binucleate dinoflagellate Peridinium balticum. J Cell Sci. 1976;21:273–89.
Douglas S, Zauner S, Fraunholz M, Beaton M, Penny S, Deng L-T, et al. The highly reduced genome of an enslaved algal nucleus. Nature. 2001;410:1091–6.
Gilson PR, Su V, Slamovits CH, Reith ME, Keeling PJ, Mcfadden GI. Complete nucleotide sequence of the chlorarachniophyte nucleomorph: nature’s smallest nucleus. Proc Natl Acad Sci USA. 2006;103:9566–71.
Johnson MD, Oldach D, Delwiche CF, Stoecker DK. Retention of transcriptionally active cryptophyte nuclei by the ciliate Myrionecta rubra. Nature. 2007;445:426–8.
Onuma R, Horiguchi T. Morphological transition in kleptochloroplasts after ingestion in the dinoflagellates Amphidinium poecilochroum and Gymnodinium aeruginosum (Dinophyceae). Protist. 2013;164:622–42.
Onuma R, Horiguchi T. Kleptochloroplast enlargement, karyoklepty and the distribution of the cryptomonad nucleus in Nusuttodinium (=Gymnodinium) aeruginosum (Dinophyceae). Protist. 2015;166:177–95.
Onuma R, Horiguchi T. Specificity of Chroomonas (Cryptophyceae) as a source of kleptochloroplast for Nusuttodinium aeruginosum (Dinophyceae). Phycol Res. 2016;64:35–43.
Larsen J. An ultrastructural study of Amphidinium poecilochroum (Dinophyceae), a phagotrophic dinoflagellate feeding on small species of cryptophytes. Phycologia. 1988;27:366–77.
Yamaguchi H, Nakayama T, Kai A, Inouye I. Taxonomy and phylogeny of a new kleptoplastidal dinoflagellate, Gymnodinium myriopyrenoides sp. nov. (Gymnodiniales, Dinophyceae), and its cryptophyte symbiont. Protist. 2011;162:650–67.
Onuma R, Watanabe K, Horiguchi T. Pellucidodinium psammophilum gen. & sp. nov. and Nusuttodinium desymbiontum sp. nov. (Dinophyceae), two novel heterotrophs closely related to kleptochloroplastidic dinoflagellates. Phycologia. 2015;54:192–209.
Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–12.
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–52.
Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–2.
Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–59.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–9.
Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2.
Ihaka R, Gentleman RR. A language for data analysis and graphics. J Comp Graph Stat. 1996;5:299–314.
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009;26:139–40.
Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999;27:29–34.
Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:182–5.
Sun J, Nishiyama T, Shimizu K, Kadota K. TCC: An R package for comparing tag count data with robust normalization strategies. BMC Bioinform. 2013;14:219.
Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–5.
Fujiwara T, Kanesaki Y, Hirooka S, Era A, Sumiya N, Yoshikawa H, et al. A nitrogen source-dependent inducible and repressible gene expression system in the red alga Cyanidioschyzon merolae. Front Plant Sci. 2015;6:1–10.
Jeffrey SW, Humphrey GF. New spectrophotometric equations for determining chlorophylls a, b, c1 and c2 in higher plants, algae and natural phytoplankton. Biochem und Physiol der Pflanz. 1975;167:191–4.
Sabarinath S, Bharti S, Khanna-Chopra R. Superoxide dismutase and abiotic stress tolerance. Physiol Mol Biol Plants. 2005;11:187–98.
Ishikawa T, Shigeoka S. Recent advances in ascorbate biosynthesis and the physiological significance of ascorbate peroxidase in photosynthesizing organisms. Biosci Biotechnol Biochem. 2008;72:1143–54.
Huang J, Zhong L. Thioredoxin reductase. Adv Top Sci Technol China. 2012;8:41–64.
Yu H. Regulation of APC-Cdc20 by the spindle checkpoint. Curr Opin Cell Biol. 2002;14:706–14.
Stegmeier F, Amon A. Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annu Rev Genet. 2004;38:203–32.
Yu K, Imamura S, Minoda A, Kan T. External light conditions and internal cell cycle phases coordinate accumulation of chloroplast and mitochondrial transcripts in the red alga Cyanidioschyzon merolae. DNA Res. 2012;19:289–303.
Zones JM, Blaby IK, Merchant SS, Umen JG. High-resolution profiling of a synchronized diurnal transcriptome from Chlamydomonas reinhardtii reveals continuous cell and metabolic differentiation. Plant Cell. 2015;27:2743–69.
Lasek-Nesselquist E, Wisecaver JH, Hackett JD, Johnson MD. Insights into transcriptional changes that accompany organelle sequestration from the stolen nucleus of Mesodinium rubrum. BMC Genomics. 2015;16:805.
Kim GH, Han JH, Kim B, Han JW, Nam SW, Shin W, et al. Cryptophyte gene regulation in the kleptoplastidic, karyokleptic ciliate Mesodinium rubrum. Harmful Algae. 2016;52:23–33.
Schnepf E, Winter S, Mollenhauer D. Gymnodinium aeruginosum (Dinophyta): a blue-green dinoflagellate with a vestigial, anucleate, cryptophycean endosymbiont. Plant Syst Evol. 1989;164:75–91.
Onuma R, Mishra N, Miyagishima SY. Regulation of chloroplast and nucleomorph replication by the cell cycle in the cryptophyte Guillardia theta. Sci Rep. 2017;7:1–12.
Suzuki S, Ishida KI, Hirakawa Y. Diurnal transcriptional regulation of endosymbiotically derived genes in the chlorarachniophyte Bigelowiella natans. Genome Biol Evol. 2016;8:2672–82.
Yagi Y, Shiina T. Recent advances in the study of chloroplast gene expression and its evolution. Front Plant Sci. 2014;5:1–7.
Zhang R, Nowack ECM, Price DC, Bhattacharya D, Grossman AR. Impact of light intensity and quality on chromatophore and nuclear gene expression in Paulinella chromatophora, an amoeba with nascent photosynthetic organelles. Plant J. 2017;90:221–34.
Kim M, Drumm K, Daugbjerg N, Hansen PJ. Dynamics of sequestered cryptophyte nuclei in Mesodinium rubrum during starvation and refeeding. Front Microbiol. 2017;8:1–14.
Hirakawa Y, Ishida KI. Polyploidy of endosymbiotically derived genomes in complex algae. Genome Biol Evol. 2014;6:974–80.
Bendich AJ. Why do chloroplasts and mitochondria contain so many copies of their genome? BioEssays. 1987;6:279–82.
Komaki K, Ishikawa H. Intracellular bacterial symbionts of aphids possess many genomic copies per bacterium. J Mol Evol. 1999;48:717–22.
Tobiason DM, Seifert HS. The obligate human pathogen, Neisseria gonorrhoeae, is polyploid. PLoS Biol. 2006;4:1069–78.
Kitten T, Barbour AG. The relapsing fever agent Borrelia hermsii has multiple copies of its chromosome and linear plasmids. Genetics. 1992;132:311–24.
Bresler V, Montgomery WL, Fishelson L, Pollak PE. Gigantism in a bacterium, Epulopiscium fishelsoni, correlates with complex patterns in arrangement, quantity, and segregation of DNA. J Bacteriol. 1998;180:5601–11.
Morrall S, Greenwood AD. Ultrastructure of nucleomorph division in species of Cryptophyceae and its evolutionary implications. J Cell Sci. 1982;54:311–28.
Sato T, Nagasato C, Hara Y, Motomura T. Cell cycle and nucleomorph division in Pyrenomonas helgolandii (cryptophyta). Protist. 2014;165:113–22.
Ludwig M, Gibbs SP. Evidence that the nucleomorphs of Chlorarachnion reptans (Chlorarachniophyceae) are vestigial nuclei: morphology, division and DNA‐DAPI fluorescence. J Phycol. 1989;25:385–94.
Stamatakis K, Vayenos D, Kotakis C, Gast RJ, Papageorgiou GC. The extraordinary longevity of kleptoplasts derived from the Ross Sea haptophyte Phaeocystis antarctica within dinoflagellate host cells relates to the diminished role of the oxygen-evolving Photosystem II and to supplementary light harvesting by mycosporine-like amino acids. Biochim Biophys Acta—Bioenerg. 2017;1858:189–95.
Johnson MD, Tengs T, Oldach D, Stoecker DK. Sequestration, performance, and functional control of cryptophyte plastids in the ciliate Myrionecta rubra (Ciliophora). J Phycol. 2006;1246:1235–46.
Decelle J, Stryhanyuk H, Gallet B, Veronesi G, Schmidt M, Balzano S, et al. Algal remodeling in a ubiquitous planktonic photosymbiosis. Curr Biol. 2019;29:968–78.e4.
Taylor FJR, Blackbourn DJ, Blackbourn J. The red-water ciliate Mesodinium rubrum and its “incomplete symbionts”: a review including new ultrastructural observations. J Fish Res Board Can. 1971;28:391–407.
Lindholm T. Mesodinium rubrum - a unique photosynthetic ciliate. Adv Aquat Microbiol. 1985;3:1–48.
Decelle J, Probert I, Bittner L, Desdevises Y, Colin S, De Vargas C, et al. An original mode of symbiosis in open ocean plankton. Proc Natl Acad Sci USA. 2012;109:18000–5.
Oren A, Gunde-Cimerman N. Mycosporines and mycosporine-like amino acids: UV protectants or multipurpose secondary metabolites? FEMS Microbiol Lett. 2007;269:1–10.
Gutierrez-Rodriguez A, Pillet L, Biard T, Said-Ahmad W, Amrani A, Simó R, et al. Dimethylated sulfur compounds in symbiotic protists: a potentially significant source for marine DMS(P). Limnol Oceanogr. 2017;62:1139–54.
Acknowledgements
We thank members of the S.-y.M. laboratory for their technical advice and support. Computations were partially performed on the National Institute of Genetics (NIG) supercomputer at the Research Organization of Information and Systems of the NIG. This study was supported by Ministry of Education, Culture, Sports, Science and Technology of Japan grants to S-yM (17H01446) and to RO (15H06834, 17K15168, and 18J01089), and by the Ministry of Education, Culture, Sports, Science and Technology-supported Program for Strategic Research Foundation at Private Universities to YK and HY (2013–2017 S1311017).
Author information
Authors and Affiliations
Contributions
RO and S-yM. conceived of and designed the research. RO established the axenic strains of the dinoflagellate and cryptophytes and performed culture experiments. RO, YK, and HY performed high-throughput sequencing. RO, SH, TF, and S-yM analyzed the RNA-seq data. RO and S-yM wrote the paper.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Onuma, R., Hirooka, S., Kanesaki, Y. et al. Changes in the transcriptome, ploidy, and optimal light intensity of a cryptomonad upon integration into a kleptoplastic dinoflagellate. ISME J 14, 2407–2423 (2020). https://doi.org/10.1038/s41396-020-0693-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-020-0693-4
This article is cited by
-
Taming the perils of photosynthesis by eukaryotes: constraints on endosymbiotic evolution in aquatic ecosystems
Communications Biology (2023)
-
Limits to the cellular control of sequestered cryptophyte prey in the marine ciliate Mesodinium rubrum
The ISME Journal (2021)