Abstract
Lakes receive large amounts of terrestrially derived dissolved organic matter (tDOM). However, little is known about how aquatic microbial communities interact with tDOM in lakes. Here, by performing microcosm experiments we investigated how microbial community responded to tDOM influx in six Tibetan lakes of different salinities (ranging from 1 to 358 g/l). In response to tDOM addition, microbial biomass increased while dissolved organic carbon (DOC) decreased. The amount of DOC decrease did not show any significant correlation with salinity. However, salinity influenced tDOM transformation, i.e., microbial communities from higher salinity lakes exhibited a stronger ability to utilize tDOM of high carbon numbers than those from lower salinity. Abundant taxa and copiotrophs were actively involved in tDOM transformation, suggesting their vital roles in lacustrine carbon cycle. Network analysis indicated that 66 operational taxonomic units (OTUs, affiliated with Alphaproteobacteria, Actinobacteria, Bacteroidia, Bacilli, Gammaproteobacteria, Halobacteria, Planctomycetacia, Rhodothermia, and Verrucomicrobiae) were associated with degradation of CHO compounds, while four bacterial OTUs (affiliated with Actinobacteria, Alphaproteobacteria, Bacteroidia and Gammaproteobacteria) were highly associated with the degradation of CHOS compounds. Network analysis further revealed that tDOM transformation may be a synergestic process, involving cooperation among multiple species. In summary, our study provides new insights into a microbial role in transforming tDOM in saline lakes and has important implications for understanding the carbon cycle in aquatic environments.
Similar content being viewed by others
Introduction
Saline lakes are globally widespread and occupy almost a half of total inland water surface area [1]. Generally, saline lakes are located in catchment basins, which receive large amounts of terrigenous materials, and thus contain high concentrations of dissolved organic carbon (DOC), largely due to their evaporative condensation effect [2, 3]. Therefore, saline lakes contribute significantly to the global carbon budget [4]. Within saline lakes, a considerable amount of DOC is originated from terrestrially derived dissolved organic matter (tDOM) from surrounding soils [5], and may undergo extensive transformations by lacustrine microbial communities [6]. However to date, little is known about the biogeochemical fate of tDOM in saline lakes, which is of great importance to the understanding of the global carbon cycle [7].
Traditionally, tDOM is considered to be refractory to biological utilization, because a large portion of tDOM molecules contain highly complex and aromatic chemical structures [8, 9]. Nevertheless, an increasing number of recent studies have indicated that tDOM can be utilized by aquatic heterotrophic microorganisms [10,11,12,13]. For example, tDOM can be utilized by aquatic bacterial communities in freshwater environments to produce new microbial biomass [11] or even mineralized to carbon dioxide (CO2), which is an important source for atmospheric CO2 [12, 14]. tDOM can also be transformed to recalcitrant DOM by microorganisms in oceans [15]. Therefore, unveiling the linkage between aquatic microbial community and tDOM is essential to comprehending carbon cycling in aquatic environments such as lakes.
The Qinghai–Tibetan Plateau (QTP) hosts thousands of lakes (including many saline and hypersaline lakes) with a salinity range from 0.1 to 426.3 g/l [16]. Previous studies have shown that tDOM occurs widely in the QTP lakes and is a very important organic carbon source for microbial communities in these lakes [17, 18]. In addition, previous studies have indicated that the QTP lakes are inhabited by a broad range of microbial species, and their taxonomic compositions become more divergent with increasing salinity difference [19,20,21,22,23,24,25,26,27]. However, little is known about the role of microbial taxa in potential transformation of tDOM in these high-elevation lakes. For example, little is known about which key microbial taxa are responsible for tDOM degradation in saline lakes, which tDOM compounds are transformed by those microbial taxa, and how such microbially catalyzed tDOM transformation responds to salinity change. To address these knowledge gaps, the linkage between aquatic microbial communities and the degradation potentials of tDOM in the QTP lakes of different salinity was investigated in this study by performing microcosm experiments and using a suite of analytical techniques.
Materials and methods
Sampling
Six Qinghai–Tibetan lakes with different salinities were selected for this study: Erhai Lake (EHL) is a freshwater lake; Qinghai Lake (QHL) and Tuosu Lake (TSL) are saline lakes; Gahai Lake (GHL), Xiaochaidan Lake (XCDL), and Chaka Lake (CKL) are hypersaline lakes [28]. A sampling cruise was carried out in May 2017. At inshore sites (~10–20 m away from shoreline) of each lake, salinity, pH, and temperature of surface water (~0–10 cm) were measured with portable meters (SANXIN, Shanghai, China). Water samples (~40 ml each) for measurements of total dissolved nitrogen (TDN) and total dissolved phosphorus (TDP) were collected after filtration through 0.22-μm Nuclepore filters (Whatman, UK). Subsequently, about 6-l surface water was collected from each lake into acid-washed and sterilized 2-l polycarbonate bottles (Nalgene, USA). To obtain tDOM, onshore soils around each lake were taken from topsoil layer (0–10 cm) without plant. All samples were kept cold and in the dark during transportation to the laboratory. Samples were stored at 4 °C in the laboratory until further processing.
Experimental set-up
To avoid exogenous contamination, all glassware used in the experiments was combusted at 450 °C for 6 h, and Whatman nuclepore filters or plastic instruments were sterilized by either autoclaving or ultraviolet irradiation before use.
Preparation of tDOM-containing media: to minimize any difference in tDOM source, six onshore soils around the lakes were equally mixed to make one composite soil sample. The composite soil sample was mixed (water:soil = 2:1, v–v) with lake water from each lake to obtain six water–soil mixtures. The mixtures were shaken on a rotary table for 1 h, and then placed on bench for 1 h in the dark. Subsequently, the resulting tDOM-containing supernatant (~3 l) in each mixture was filtrated through a 0.22-μm Whatman nuclepore membrane filter, and collected into a pre-combusted glass bottle. The resulting tDOM-containing filtrate was diluted using corresponding microbe-free lake water to maintain a similar level of tDOM, i.e., the DOC content of the tDOM-containing lake waters minus the DOC content of their corresponding lake water was ~12 mg/l. Microbe-free lake water was obtained by filtering lake water through a Whatman 0.22-μm nuclepore membrane filter. The resulting tDOM-containing lake water was finally used as a basic culture medium for subsequent microcosm experiment.
Preparation of inocula: for each lake, about 2-l water was filtrated through 0.22-μm Whatman nuclepore membrane filters. The resulting biomass-containing filters were immersed into corresponding microbe-free lake water (~200 ml), and manually stirred for 30 min. After the filters were removed, the resulting concentrated microbial inocula were used for subsequent microcosm experiments.
Microcosm experiments and sample collection: prior to the microcosm experiments, lake waters were pre-incubated at 15 °C in the dark for 7 days to remove any labile lake-derived DOM. Six microcosms (i.e., four experimental treatments and two controls) were prepared for each lake. Microcosm experiments were conducted in 1-l glass bottles containing 450 ml tDOM-containing basic medium (prepared as above). To each bottle, 50 ml of microbial inocula (prepared above) were added into four microcosms as experimental treatments, while two control bottles received 50 ml microbe-free lake water. At zero time point (i.e., the beginning of the experiment), water samples were taken from two experimental treatments of each lake for microbial counts (n = 2), DNA extraction (n = 2), DOC (n = 2), and FT-ICR-MS (n = 1) analyses. The other two experimental treatments and controls for each lake were sealed with air permeable and microbe-proofing films and then were incubated for 7 days at 15 °C in the dark. At the end of incubation (i.e., day 7), duplicate water samples from both treatments and abiotic controls of each lake were collected for microbial count (n = 4), DOC (n = 4), and FT-ICR-MS (n = 4) analyses. Detailed sampling procedures for microbial count, DOC, DNA extraction, and FT-ICR-MS analyses were described in the Supplementary materials.
Laboratory analyses
DOC concentrations were determined on a multi N/C 2100S analyzer (Analytik Jena, Germany). TDN and TDP were analyzed using published colorimetric methods [29, 30]. Acridine orange direct count was used to determine the total number of microbial cells in water samples according to the procedure described previously [31].
FT-ICR-MS spectrometry
DOM in the water samples was isolated using solid phase extraction (SPE) as described previously [32]. All the SPE-extracted DOM samples were analyzed on a 12 Tesla Bruker SolariX FT-ICR-MS spectrometer equipped with negative-mode electrospray ionization at Pacific Northwest National Laboratory, Richland, Washington, USA. Details concerning DOM isolation and FT-ICR-MS analyses were provided in the Supplementary materials.
DNA extraction, illumina sequencing, and sequence analysis
Total DNA was extracted from biomass-containing filters using the Fast DNA SPIN Kit for Soil (MP Biomedical, USA). The extracted DNA was amplified with a universal 16S rRNA gene primer set: 515F (5′-GTGYCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACNVGGGTWTCTAAT-3′) according to the PCR conditions described previously [33]. Amplicon sequencing were performed by using an Illumina-Miseq platform (paired-ends sequencing of 2 × 250 bp) [34]. Detailed procedures for PCR preparation and sequence processing were provided in the Supplementary Materials. All the raw sequences obtained from this study have been deposited at the NCBI Sequence Read Archive under the project PRJNA377204 with biosample accession number of SAMN10334138- SAMN10334161.
Statistical analyses
Unless otherwise indicated, all statistical analyses were carried out in the R program (http://cran.r-project.org/) implemented with various packages. Nonmetric multidimensional scaling (NMDS) was employed to evaluate the microbial community composition difference between the beginning and end of incubation, based on the Bray–Curtis dissimilarity using the “vegan” packages. The total number of molecular formulae was calculated according to specific categories (e.g., CHO, CHON, CHONP, CHONS, CHONSP, CHOP, CHOS, and CHOSP). Relative peak intensities (normalized to the sum of all peak intensities of identified molecular formulae per sample) were used to semi-quantitatively assess the changes in DOM molecular composition as a result of incubation. Principal coordinate analyses (PCoA) were conducted to illuminate the difference in DOM molecular compositions among different experimental samples using the “ape” package. The van Krevelen diagrams [35], in which the ratios of hydrogen to carbon (H/C) was plotted against the ratios of oxygen to carbon (O/C), were used to compare the DOM compositional difference between different samples and as a result of incubation. To test the significance of difference in microbial community and DOM molecular composition among samples, PERMANOVA analysis was performed based on Bray–Curtis dissimilarity with 9999 permutations.
Seven DOM molecular groups were defined according to the presence–absence of DOM formulae among the staring samples, treatments, and controls: (1) transformed DOM by biotic or abiotic process was indicated by the presence of DOM formulae only in the starting samples; (2) transformed DOM by abiotic process was indicated by the presence of DOM formulae in both the starting samples and biotic treatments but by the absence in abiotic controls; (3) transformed DOM by biotic activity was indicated by the presence of DOM formulae in both the starting samples and abiotic controls but by the absence in biotic treatment; (4) relatively stable DOM was indicated by DOM formulae that were shared among the starting samples, biotic treatments, and abiotic controls; (5) newly produced DOM by biotic activity was indicated by the presence of DOM formulae only in biotic treatments; (6) newly produced DOM by abiotic processes was indicated by the presence of DOM formulae only in abiotic controls; and (7) newly produced DOM by biotic or abiotic process was indicated by the presence of DOM formulae in both biotic treatments and abiotic controls but by the absence in the starting samples. These groups were hereafter called G1–G7 for brevity.
In order to explore the association between microbial OTUs and DOM molecular formulae, the network analysis was performed. Briefly, Spearman’s rank correlations were performed between relative abundance of microbial OTUs and relative peak intensity of DOM molecules for all experimental treatments at the end of microcosm experiments (a total of 12 samples) using the R package “Hmisc”. Rare OTUs (relative abundance <0.01%) in each sample were removed before correlation analysis. To be statistically significant, microbial OTUs and DOM molecular formulae that occurred simultaneously in at least six samples were included in the analysis. To correct false discovery rates, all p values generated by Spearman correlation analyses were adjusted using the Benjamini–Hochberg method [36]. Spearman’s correlation coefficient |R| >0.7 with p < 0.01 was considered as robust [37]. All the robust correlations were used to construct a network where nodes represented microbial OTUs or DOM molecular formulae, and edges indicated strong and significant correlations between nodes. Network visualization was conducted using the interactive platform Gephi (http://gephi.github.io/).
The network modularity and module division were calculated through a method of fast greedy optimization [38] using the R package of igraph. Subsequently, the roles of the network nodes were assigned according to their within-module connectivity (Zi) and among-module connectivity (Pi) [39]. Four categories can be defined for each node: (1) peripheral nodes (Zi < 2.5, Pi < 0.62), (2) connectors (Zi < 2.5, Pi > 0.62), (3) module hubs (Zi > 2.5, Pi < 0.62), and (4) network hubs (Zi > 2.5, Pi > 0.62) [40, 41]. The network and module hubs, and connectors are generally proposed as keystone module members [40].
Results
Physicochemical conditions of the sampling lake waters
Salinity of lake water was 1–358 g/l, pH was 7.2–9.4, and water temperature was 9.3–16.1 °C (Table 1), TDN and TDP were 1.6–14.9 and 0.1–0.2 mg/l, respectively.
Variation of microbial abundance and community composition as a result of incubation
Microbial cell abundance increased considerably from the beginning of the incubation (i.e., day 0) to the end (i.e., day 7) (Table 2). The average amount of increase was 4.97 × 105, 3.11 × 105, 2.42 × 105, 2.70 × 105, 4.82 × 105, and 5.38 × 106 cells mL−1 for EHL, QHL, TSL, GHL, XCDL, and CKL, respectively. The hypersaline CKL sample had the highest amount of increase. Microbial cells in abiotic control samples showed no observable increase.
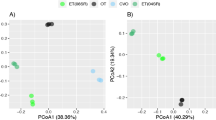
A total of 883,350 qualified sequences were obtained with an average of 36,806 per sample (Supplementary Table S1). The observed OTUs ranged from 115 to 299, and the Shannon index ranged from 1.9 to 3.9 (Supplementary Table S1). PERMONOVA analysis showed that microbial community composition was significantly distinct (R2 = 0.875, p < 0.001) among different lakes (Fig. 1). For example, Actinobacteria, Alphaproteobacteria, Bacteroidia, and Gammaproteobacteria dominated (relative abundance >10%) in the experimental samples of EHL, QHL, TSL, GHL, XCDL, while hypersaline lake CKL was dominated by Halobacteria, whose relative abundance was up to 80% (Fig. 1). Such a difference in microbial community composition among the lakes was further supported by NMDS analysis (Supplementary Fig. S1), which showed that all samples from the same lake were clustered together but distinct from those of other lakes, regardless of incubation time.
Microbial community composition exhibited a discernible shift as a result of incubation (Supplementary Fig. S1). However, PERMONOVA analysis did not show a significant (p > 0.05) difference in microbial community composition between the beginning and the end of incubation. We found that there were a total of 26 OTUs, whose average relative abundances increased by >1% from the beginning to the end of incubation in certain lake samples (Supplementary Table S2). The phylogenetic affiliations of these OTUs were shown in Supplementary Table S2. Interestingly, 16 out of these 26 OTUs were abundant (average relative abundance >1%) in both the beginning and the end of microcosms of certain lakes. For example, OTU599 (Alphaproteobacteria) was abundant in three saline lake samples of TSL, GHL, and XCDL (Supplementary Table S2). Notably, the relative abundances of four OTUs (i.e., OTU313, OTU375, OTU554, and OTU581) increased by >10 times from the beginning to the end of incubation in the TSL and XCDL samples (Supplementary Table S2): OTU313 and OTU375 in TSL samples belonging to Gammaproteobacteria increased from the average relative abundance of 0.05% and 0.08% in the beginning to 2.61% and 10.94% in the end, respectively. Likewise, in the XCDL samples, OTU554 (Alphaproteobacteria) and OTU581 (Gammaproteobacteria) increased from <0.01% and 0.05% in the beginning to 8.13% and 19.11% in the end, respectively (Supplementary Table S2). According to the definition established previously [42], these four OTUs could be considered as copiotroph-like OTUs, (i.e., OTUs with relative abundance <0.1% at day 0 but >1% at day 7).
DOC concentration and DOM molecular composition
The concentrations of DOC in the experimental treatments decreased from the beginning (i.e., day 0) to the end (i.e., day 7) of incubation, with the mean amount of decrease of 1.42, 4.87, 3.20, 2.61, 1.65, and 6.5 mg L−1 for the EHL, QHL, TSL, GHL, XCDL, and CKL samples, respectively (Table 3). The highest amount of DOC decrease was observed in the hypersaline CKL samples. The DOC concentrations in abiotic controls also experienced slight declines after incubation (Table 3). Therefore, the mean amounts of net DOC decrease derived from microbial activity were 1.09, 4.70, 2.85, 2.32, 1.48, and 6.0 mg L−1 for the EHL, QHL, TSL, GHL, XCDL, and CKL samples, respectively (Table 3), and such DOC decreases did not show any significant (p > 0.05) correlation with lake salinity.
The total number of DOM molecular formulae determined by FT-ICR-MS ranged from 2087 to 4229 per sample, with CHO, CHON, and CHOS compounds being dominant in all samples (Supplementary Table S3). PCoA showed the samples from different lakes contained distinct DOM molecular compositions, which was further supported by PERMONOVA analysis (R2 = 0.751, p < 0.001; Supplementary Fig. S2). Notably, although all DOM was initially eluted from the same composite soil sample, its composition was different even at time 0 (Supplementary Fig. S2). This difference could be caused by distinct DOM molecular composition in lake water that was used to elute tDOM from soil, or a salinity effect on tDOM elution from soil [43]. Furthermore, PCoA also indicated that there appeared to be recognizable differences among the starting samples, biotic treatments and abiotic controls for each lake (Supplementary Fig. S2), and this was also supported by the van Krevelen diagrams (Supplementary Fig. S3). However, such differences were not significant, as indicated by the PERMONOVA analysis (data not shown).
To discern the difference in DOM molecular composition among the starting samples, biotic treatments and abiotic controls of each lake, the numbers of DOM molecular formulae belonging to seven defined group (see the “Method” section) were calculated (Fig. 2a). Each group contained various numbers of DOM molecular formulae in different lakes, and G4 (relatively stable DOM) contained the majority of DOM molecular formulae (1243–2483) in all lakes (Fig. 2a). PERMONOVA analysis indicated that DOM molecular compositions in lake microcosms were distinct (R2 = 0.243, p < 0.001) among different groups (Fig. 2b). Furthermore, the relative abundances of molecular categories (e.g., CHO, CHON, and CHOS) in each group varied in lakes (Supplementary Fig. S4). The average carbon numbers of DOM molecular formulae were all >20 in most groups and in all lakes, expect for G4 whose average carbon numbers were all <20 in six lakes (Supplementary Fig. S5).
Some microbially transformed DOM molecules (i.e., G3) were observed in the studied microcosms (Fig. 2a, b). Different types of DOM molecules were transformed by microbial activity in different lakes (Fig. 2b). For example, the CHON molecules in G3 were more abundant in the EHL and QHL samples of lower salinity than in the samples of higher salinity (i.e., TSL, GHL, XCDL, and CKL samples), whereas CHOP molecules showed an opposite trend (Supplementary Fig. S4). Furthermore, the numbers of carbon in the DOM molecules of G3 were significantly (Kruskal–Wallis rank sum test: chi-squared = 21.371, p < 0.001) lower in the microcosms with low salinity (i.e., EHL, QHL, and TSL) than in the microcosms with high salinity (i.e., GHL, XCDL, and CKL) (Supplementary Fig. S5).
Associations between microbial OTUs and DOM molecular formulae
Spearman correlation analysis showed that there were a total of 66 OTUs, whose relative abundances showed strong (|R| > 0.7, p < 0.01) correlations with the relative intensities of some DOM molecular formulae. The phylogenetic affiliations of these OTUs were shown in Supplementary Table S4. The DOM molecular formulae that were correlated with the abovementioned OTUs possessed various H/C and O/C ratios and the number of carbon (Fig. 3 and Supplementary Fig. S6). Most of these microbial OTUs were widely correlated with CHO molecules except for OTU137 (Gammaproteobacteria), OTU158 (Bacteroidia), OTU376 (Actinobacteria), and OTU445 (Alphaproteobacteria), which were correlated with both CHO and CHOS molecules (Supplementary Fig. S6). The carbon number of DOM formulae associated with microbial OTUs ranged from 6 to 27, and their distribution patterns changed with different OTUs (Supplementary Fig. S7).
Network analysis indicated that the abovementioned correlations between microbial OTUs and DOM molecular formulae formed a complex network, which consisted of 1716 nodes and 4638 edges (Fig. 3a). The network modularity was 0.53 (Fig. 3b), which suggests a good (modularity >0.4) modular structure [44]. The network was composed of eight modules, and each module contained different types and numbers of DOM molecular formulae and microbial OTUs (Fig. 3b). The numbers of DOM molecular formulae in Module 2 was the highest, followed by Module 1 and Module 4 in a decreasing order (Supplementary Fig. S8a). CHO formulae were widely distributed in all modules, and were dominant in Module 4 and Module 8; CHOS formulae were frequently present in Module 2, Module 3, and Module 5; CHON formulae were dominant in Module1 and Module 7. The numbers of microbial OTUs were highest in Module 4 and Module 1, while the lowest number was observed in Module 8 with only one OTU affiliated with Gammaproteobacteria (Supplementary Fig. S8b). Microbial OTUs affiliated with Gammaproteobacteria were widely distributed in most of the identified modules except for Module 5, while microbial OTUs belonging to Bacteroidia were frequently present in Module 4 (Supplementary Fig. S8b).
According to connectivity within and among modules, we identified four network hubs, 39 module hubs, 176 connectors, and 1497 peripheral nodes (Fig. 4 and Supplementary Table S5). It was noteworthy that all modules and network hubs were microbial OTUs, and most connectors were DOM formulae except for OTU46 (Bacteroidia) and OTU93 (Gammaproteobacteria) (Fig. 4 and Supplementary Table S5). The molecular characteristics (i.e., ratios of H/C and O/C) of these identified DOM connectors were shown in the van Krevelen diagrams, and they possessed a broad ranges of H/C and O/C ratios (Supplementary Fig. S9a). The DOM connectors mainly consisted of CHO, CHON, CHONS, and CHOS formulae, and they were dominated by CHO formulae, which accounted for 53% of the total DOM connectors (Supplementary Fig. S9b).
Discussion
Microorganisms play key roles in transformation of tDOM
Our data revealed that microbial activities decreased the concentration of tDOM in the studied treatments. The decreased amount of tDOM (by as much as 1.1–6.0 mg/l, in Table 3) may be transformed into microbial cellular carbon [45, 46] and/or mineralized to CO2 through microbial respiration [47, 48]. Assuming one microbial cell contains 21 fg (1 fg = 1 × 10−12 mg) carbon [49], the increased microbial cell numbers (Table 2) only accounted for <1% of the decreased tDOM (<0.01 mg L−1 vs. 1.1–6.0 mg L−1); thus a large fraction of the decreased tDOM (>99%) may have been mineralized to CO2. This result suggests that the input of tDOM to lakes may increase CO2 emission. Furthermore, the lack of correlation between salinity and the decreased amount of tDOM in the microcosms suggests that other environmental factors may have contributed to the observed tDOM degradation. In addition, microbial activities transformed some tDOM to novel DOM molecules, as evidenced by unique DOM molecules observed in biotic treatments but not in abiotic controls and starting samples (e.g., G5 in Fig. 2a, b). This observation is consistent with previous studies in showing that microbial activity can transform DOM into new products [15, 50,51,52,53].
Our combined molecular and physicochemical studies allowed us to identify the dominant microorganisms that may be responsible for such tDOM transformation. The relative abundance of abundant OTUs (i.e., relative abundance >1% both in the beginning and end of the experiment) increased in response to addition of tDOM (Supplementary Table S2), suggesting their vital roles in transformation and mineralization of tDOM. This observation may be ascribed to the fact that abundant taxa can usually utilize a broad spectrum of DOM compounds as substrates [54]. Furthermore, it is also notable that these tDOM-transforming taxa were phylogenetically diverse (i.e., Actinobacteria, Alphaproteobacteria, Bacteroidia, Gammaproteobacteria, Halobacteria, Verrucomicrobiae; as shown in Supplementary Table S2). In the meanwhile, distinct microbial taxa appeared to be responsible for the transformation of tDOM among different lakes, as evidenced by abundance increases of lake-specific OTUs after microcosm incubations (Supplementary Table S2). This observation could be due to the fact that the original microbial communities differed among the studied lakes with different salinity [19, 21, 23, 25, 55, 56].
It is interesting to observe that copiotrophs (e.g., OTUs belonging to genera of Vibrio and Photobacterium in the Gammaproteobacteria) also contributed to the transformation of the added tDOM. Copiotrophs are frequently observed in the nutrient-rich environments and are opportunistic r-strategists for growth, and thus they prefer labile substrates to support their fast growth [42, 57]. So copiotrophs are frequently observed in the microcosms amended with labile DOM (e.g., glucose) [42, 58,59,60]. In this study, the relative abundances of certain copiotrophic OTUs showed a dramatic increase (>10 times) in response to the addition of tDOM during microcosm incubations of two saline lake (TSL and XCDL) samples (Supplementary Table S2). Such response of copiotrophs can be ascribed to the possible presence of certain labile DOM (e.g., protein and sugars) eluted from the sampled terrestrial soils [9, 61]. Therefore, it was possible to observe that tDOM stimulated the growth of copiotrophs in saline and hypersaline lakes.
Potential interactions between microbial taxa and DOM
To further illuminate the relation between DOM transformation and specific microbial species responsible for such transformation, correlation analyses between microbial taxa and DOM molecules were performed to illuminate this link. Note that a significant correlation between microbial species and DOM molecules does not necessarily suggest that such microbial species actually transformed or were capable of transforming the DOM molecules [11]. However, such correlation analyses at least provide some direction for future investigation on microbial degradation of specific DOM compounds.
Correlation analyses in this study revealed that microorganisms may contribute to the transformation of DOM compounds with a broad range of H/C and O/C ratios, and carbon number. Different microbial OTUs were associated with the transformation of distinct DOM compounds (Supplementary Fig. S6). This can be ascribed to the fact that different microbial species possess different functional enzymes, which degrade different DOM compounds [11, 62]. In other words, some microbial species may produce some specific functional enzymes, which can degrade specific type of DOM compounds [63, 64]. For example, OTU137 (Gammaproteobacteria), OTU158 (Bacteroidia), OTU376 (Actinobacteria), and OTU445 (Alphaproteobacteria) are highly associated with the degradation of CHOS compounds (Supplementary Fig. S6). In addition to DOM types, some microbial taxa may be able to degrade DOM compounds with distinct ranges of carbon number [11]. For example, OTU205 (Rhodothermia) and OTU464 (Gammaproteobacteria) were positively correlated with compounds with low carbon number (~<15), and negatively correlated with compounds with high carbon number (~>15); whereas OTU346 (Verrucomicrobia), OTU376 (Actinobacteria), and OTU463 (Gammaproteobacteria) were positively correlated with DOM formulae of high carbon number, and negatively correlated with DOM formulae of low carbon number (Supplementary Fig. S7).
Remarkably, the abovementioned correlations between certain microbial taxa and DOM formulae formed a strong modular network with eight modules (Fig. 3b). Previous studies have indicated that a module within one network can be considered as a functional unit, which may perform an identical ecological task, and the involved microbial taxa within that module are highly connected with each other [40, 65]. Therefore, we speculated that microbial taxa within certain module may cooperate to degrade some specific types of DOM (e.g., refractory tDOM), and specific DOM compounds may serve as substrates. Indeed, one previous study suggested that one single species cannot completely degrade most large molecular weight DOM compounds [66], implying that it is necessary for multiple species to cooperate for tDOM degradation. Furthermore, different modules contained distinct DOM molecular composition (Supplementary Fig. S8A), suggesting different modules may contribute to different DOM transformations [67].
It is also important to mention the identification of keystone module members (i.e., module and network hubs, connectors) due to their high connectivity within or among modules and key roles in the network [39, 40]. In the present study, correlations between microbial OTUs and DOM formulae were used to construct the network, and many microbial OTUs were largely correlated with DOM formulae (Fig. 3a). Accordingly, it is not surprising that those microbial OTUs were prone to be identified as modules or network hubs. However, the identification of keystone module members may highlight the key roles of those microbial taxa in tDOM transformation. Strikingly, a large number of DOM connectors were identified in the studied network (Supplementary Fig. 9A, B). From ecological perspectives, connectors in the network are proposed as the bridges linking different modules [40, 41]. Accordingly, it can be speculated that those DOM connectors may be key intermediate substrates or products from microbial degradation of tDOM. However, FT-ICR-MS cannot predict the chemical structure of each formula, so it is difficult to know the actual characteristics of those DOM compounds. Furthermore, the above speculation needs validation by studying microbial functions and DOM structure.
Different DOM transformation in the lake microcosms of different salinity
In our incubation experiments, the DOM in the original lake waters were difficult to remove, thus any observed DOM transformation should be a combination of both tDOM and original lake DOM. However, little bias should have been introduced when comparing the change of DOM composition before and after incubation because (1) any labile DOM in the original lake waters should have been consumed before incubations, and (2) the tDOM used in lake microcosm experiment were adjusted to an equal level before incubations.
Our results indicated that both biotic and abiotic processes contributed to tDOM transformation, because some newly produced DOM were detected in both biotic treatments (i.e., G5) and abiotic controls (i.e., G6) (Fig. 2a, b). This result corroborated previous findings that certain DOM compounds in the environments could be degraded either abiotically (such as photo- or thermal degradation) or biotically (e.g., microbial activity) [68]. Nonetheless, a large portion of DOM compounds appeared to be stable during the experimental incubations. For example, more than 50% of DOM compounds were shared among the initial samples, biotic treatments, and abiotic controls (G4 in Fig. 2a). A likely reason is that most tDOM may be resistant to biological or nonbiological degradation and can exist for thousands of years [69], but the tDOM in our microcosm experiments were only incubated for only 7 days.
Despite the fact that tDOM is largely refractory to biological degradation, there are some DOM compounds that can be transformed by microbial activity, for example DOM compounds in G3 (Fig. 2a). It is notable that among those DOM compounds that are transformed by microbes, microbial communities in the microcosms with high salinity seemed to exhibit a stronger ability to transform DOM compounds of higher carbon numbers than those in the microcosms with low salinity (i.e., DOM formulae of G3 in Supplementary Fig. S5). We speculated that halophilic/halotolerant microbes in lakes of high salinity have to exploit a broader range of carbon sources due to high energy costs to deal with salinity stress relative to those in lakes of low salinity [70], and thus they may have developed an ability to degrade organic matter with a higher carbon number and more complex structures. Indeed, many halotolerant/halophilic prokaryotes have been indicated to possess specific enzymes that have a high efficiency to degrade recalcitrant organic matter (e.g., lignin, cellulose, and chitin) [71, 72].
Microbial communities in microcosms with high salinity may selectively consume nitrogen (N)- and phosphors (P)-containing organic compounds. For example, the microbially transformed DOM molecules in the microcosms with high salinity showed a higher relative abundance of N- and a lower relative abundance of P-containing formulae than those in the microcosms with low salinity (G3 in Supplementary Fig. S4). Such difference may be due to different requirements for C/N/P in lakes of different salinity. Microbial biomass in different lakes tend to have distinct C:N:P stoichiometry [73], so microbes in different lakes selectively assimilate C/N/P from organic/inorganic substrates in the environment to keep their stoichiometry at equilibrium [74,75,76]. However, the underlying reasons still await further investigation.
In addition, it is surprising that the CKL samples with the highest salinity (358 g l−1) showed the highest increase of cell abundance and the highest DOC decrease during the incubations (Tables 2 and 3). This observation is inconsistent with the general principle that salinity decreases microbial growth and metabolism [70]. Such inconsistency may be explained by a possibility that the microbial population in the studied CKL microcosms adopt a high-efficiency strategy for resisting salinity stress, and they have adapted to grow on tDOM. Indeed, four microbial OTUs whose relative abundances increased by >1% from the beginning to the end of the incubation were all affiliated with Halobacteriales in the CKL samples (Supplementary Table S2). The Halobacteriales have been suggested to adopt the “salt-in” strategy that is energetically cheap and thus highly efficient to balance salinity pressure in the environment [70]. Moreover, Halobacteriales are capable of growth on cellulose and chitin [72], which are important components of the terrigenous organic carbon [9]. Therefore, it may be reasonable to observe that the highest microbial cell increase and DOC concentration decrease in the studied CKL microcosms during incubations with addition of tDOM.
In summary, our findings demonstrated that tDOM can be transformed by microbial populations from different saline lakes, and both abundant microbes and copiotrophs contributed to the transformation of tDOM. Microbial communities in the microcosms with different salinities exhibited different preference and capability in tDOM transformation: microbial populations in the microcosms with high salinity showed a stronger capability to degrade high carbon number DOM compounds than their counterpart in the microcosms with low salinity. Multiple microbial taxa may cooperate with each other to degrade certain kinds of tDOM compounds in the studied microcosms. Taken together, this study expands our understanding of microbial roles in tDOM degradation in saline lake ecosystems.
References
Wetzel RG. Limnology: lake and river ecosystems, 3rd ed. San Diego, CA: Academic Press; 2001. p. 15–40.
Song K, Wen Z, Shang Y, Yang H, Lyu L, Liu G, et al. Quantification of dissolved organic carbon (DOC) storage in lakes and reservoirs of mainland China. J Environ Manag. 2018;217:391–402.
Wen Z, Song K, Shang Y, Zhao Y, Fang C, Lyu L. Differences in the distribution and optical properties of DOM between fresh and saline lakes in a semi-arid area of Northern China. Aquat Sci. 2018;80:22.
Duarte CM, Prairie YT, Montes C, Cole JJ, Striegl R, Melack J, et al. CO2 emissions from saline lakes: a global estimate of a surprisingly large flux. J Geophys Res: Biogeosci. 2008;113:G04041.
Jansson M, Persson L, De Roos AM, Jones RI, Tranvik LJ. Terrestrial carbon and intraspecific size-variation shape lake ecosystems. Trends Ecol Evol. 2007;22:316–22.
Tranvik LJ, Downing JA, Cotner JB, Loiselle SA, Striegl RG, Ballatore TJ, et al. Lakes and reservoirs as regulators of carbon cycling and climate. Limnol Oceanogr. 2009;54:2298–314.
Cole JJ, Prairie YT, Caraco NF, McDowell WH, Tranvik LJ, Striegl RG, et al. Plumbing the global carbon cycle: integrating inland waters into the terrestrial carbon budget. Ecosystems. 2007;10:172–85.
Kellerman AM, Dittmar T, Kothawala DN, Tranvik LJ. Chemodiversity of dissolved organic matter in lakes driven by climate and hydrology. Nat Commun. 2014;5:3804.
Nebbioso A, Piccolo A. Molecular characterization of dissolved organic matter (DOM): a critical review. Anal Bioanal Chem. 2013;405:109–24.
Lapierre J-F, Guillemette F, Berggren M, del Giorgio PA. Increases in terrestrially derived carbon stimulate organic carbon processing and CO2 emissions in boreal aquatic ecosystems. Nat Commun. 2013;4:2972.
Logue JB, Stedmon CA, Kellerman AM, Nielsen NJ, Andersson AF, Laudon H, et al. Experimental insights into the importance of aquatic bacterial community composition to the degradation of dissolved organic matter. ISME J. 2015;10:533.
McCallister SL, del Giorgio PA. Evidence for the respiration of ancient terrestrial organic C in northern temperate lakes and streams. Proc Natl Acad Sci USA. 2012;109:16963–8.
Ward ND, Keil RG, Medeiros PM, Brito DC, Cunha AC, Dittmar T, et al. Degradation of terrestrially derived macromolecules in the Amazon River. Nat Geosci. 2013;6:530–3.
Gudasz C, Bastviken D, Steger K, Premke K, Sobek S, Tranvik LJ. Temperature-controlled organic carbon mineralization in lake sediments. Nature. 2010;466:478–81.
Jiao N, Herndl GJ, Hansell DA, Benner R, Kattner G, Wilhelm SW, et al. Microbial production of recalcitrant dissolved organic matter: long-term carbon storage in the global ocean. Nat Rev Microbiol. 2010;8:593–9.
Zheng M. An introduction to saline lakes on the Qinghai-Tibet plateau, 1st ed. Dordrecht: Kluwer Academic Publisher; 1997. p. 1–17.
Song K, Shang Y, Wen Z, Jacinthe P-A, Liu G, Lyu L, et al. Characterization of CDOM in saline and freshwater lakes across China using spectroscopic analysis. Water Res. 2019;150:403–17.
Spencer RGM, Guo W, Raymond PA, Dittmar T, Hood E, Fellman J, et al. Source and biolability of ancient dissolved organic matter in glacier and lake ecosystems on the Tibetan Plateau. Geochim Cosmochim Acta. 2014;142:64–74.
Jiang H, Dong H, Yu B, Liu X, Li Y, Ji S, et al. Microbial response to salinity change in Lake Chaka, a hypersaline lake on Tibetan plateau. Environ Microbiol. 2007;9:2603–21.
Jiang H, Dong CZ, Huang Q, Wang G, Fang B, Zhang C, et al. Actinobacterial diversity in microbial mats of five hot springs in central and central-eastern Tibet, China. Geomicrobiol J. 2012;29:520–7.
Liu Y, Yao T, Jiao N, Zhu L, Hu A, Liu X, et al. Salinity impact on bacterial community composition in five high-altitude lakes from the Tibetan plateau, Western China. Geomicrobiol J. 2013;30:462–9.
Wang J, Yang D, Zhang Y, Shen J, Van Der Gast C, Hahn MW, et al. Do patterns of bacterial diversity along salinity gradients differ from those observed for macroorganisms? PLoS ONE. 2011;6:e27597.
Wu QL, Zwart G, Schauer M, Kamst-van Agterveld MP, Hahn MW. Bacterioplankton community composition along a salinity gradient of sixteen high-mountain lakes located on the Tibetan Plateau, China. Appl Environ Microbiol. 2006;72:5478–85.
Xing P, Hahn MW, Wu QL. Low taxon richness of bacterioplankton in high-altitude lakes of the eastern Tibetan Plateau, with a predominance of Bacteroidetes and Synechococcus spp. Appl Environ Microbiol. 2009;75:7017–25.
Yang J, Ma LA, Jiang H, Wu G, Dong H. Salinity shapes microbial diversity and community structure in surface sediments of the Qinghai-Tibetan Lakes. Sci Rep. 2016;6:25078.
Yang J, Jiang H, Wu G, Liu W. Phylum-level archaeal distributions in the sediments of Chinese lakes with a large range of salinity. Geomicrobiol J. 2018;35:404–10.
Zhong Z-P, Liu Y, Miao L-L, Wang F, Chu L-M, Wang J-L, et al. Prokaryotic community structure driven by salinity and ionic concentrations in plateau lakes of the Tibetan Plateau. Appl Environ Microbiol. 2016;82:1846–58.
Yang J, Jiang H, Dong H, Wang H, Wu G, Hou W, et al. amoA-encoding archaea and thaumarchaeol in the lakes on the northeastern Qinghai-Tibetan Plateau, China. Front Microbiol. 2013;4:329.
Neal C, Neal M, Wickham H. Phosphate measurement in natural waters: two examples of analytical problems associated with silica interference using phosphomolybdic acid methodologies. Sci Total Environ. 2000;251–2:511–22.
Willis RB, Montgomery ME, Allen PR. Improved method for manual, colorimetric determination of total Kjeldahl nitrogen using salicylate. J Agric Food Chem. 1996;44:1804–7.
Jiang H, Dong H, Zhang G, Yu B, Chapman LR, Fields MW. Microbial diversity in water and sediment of Lake Chaka, an athalassohaline lake in northwestern China. Appl Environ Microbiol. 2006;72:3832–45.
Dittmar T, Koch B, Hertkorn N, Kattner G. A simple and efficient method for the solid-phase extraction of dissolved organic matter (SPE-DOM) from seawater. Limnol Oceanogr Meth. 2008;6:230–5.
Walters W, Hyde ER, Berg-Lyons D, Ackermann G, Humphrey G, Parada A, et al. Improved bacterial 16S rRNA gene (V4 and V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. mSystems. 2015;1:e00009–00015.
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–4.
Krevelen DW. Graphical-statistical method for the study of structure and reaction processes of coal. Fuel. 1961;29:269–83.
Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B (Methodol). 1995;57:289–300.
Steinhauser D, Krall L, Müssig C, Büssis D, Usadel B. Correlation networks. In: Junker BH, Schreiber F, editors. Analysis of biological networks. Hoboken, New Jersey, USA: John Wiley & Sons; 2008. p. 305–33.
Clauset A, Newman MEJ, Moore C. Finding community structure in very large networks. Phys Rev E. 2004;70:066111.
Guimerà R, Nunes Amaral LA. Functional cartography of complex metabolic networks. Nature. 2005;433:895–900.
Deng Y, Jiang Y-H, Yang Y, He Z, Luo F, Zhou J. Molecular ecological network analyses. BMC Bioinforma. 2012;13:113.
Zhou J, Deng Y, Luo F, He Z, Yang Y. Phylogenetic molecular ecological network of soil microbial communities in response to elevated CO2. MBio. 2011;2:e00122–00111.
Nelson CE, Carlson CA. Tracking differential incorporation of dissolved organic carbon types among diverse lineages of Sargasso Sea bacterioplankton. Environ Microbiol. 2012;14:1500–16.
Painter SC, Lapworth DJ, Woodward EMS, Kroeger S, Evans CD, Mayor DJ, et al. Terrestrial dissolved organic matter distribution in the North Sea. Sci Total Environ. 2018;630:630–47.
Newman MEJ. Modularity and community structure in networks. Proc Natl Acad Sci USA. 2006;103:8577–82.
Boyer J, Dailey S, Gibson P, Rogers M, Mir-Gonzalez D. The role of dissolved organic matter bioavailability in promoting phytoplankton blooms in Florida Bay. Hydrobiologia. 2006;569:71–85.
Raymond PA, Bauer JE. Bacterial consumption of DOC during transport through a temperate estuary. Aquat Micro Ecol. 2000;22:1–12.
del Giorgio PA, Duarte CM. Respiration in the open ocean. Nature. 2002;420:379–84.
Sharma AK, Becker JW, Ottesen EA, Bryant JA, Duhamel S, Karl DM, et al. Distinct dissolved organic matter sources induce rapid transcriptional responses in coexisting populations of Prochlorococcus, Pelagibacter and the OM60 clade. Environ Microbiol. 2014;16:2815–30.
Magnabosco C, Lin LH, Dong H, Bomberg M, Ghiorse W, Stan-Lotter H, et al. The biomass and biodiversity of the continental subsurface. Nat Geosci. 2018;11:707–17.
Bugg TDH, Ahmad M, Hardiman EM, Rahmanpour R. Pathways for degradation of lignin in bacteria and fungi. Nat Prod Rep. 2011a;28:1883–96.
Bugg TDH, Ahmad M, Hardiman EM, Singh R. The emerging role for bacteria in lignin degradation and bio-product formation. Curr Opin Biotechnol. 2011b;22:394–400.
Medeiros PM, Seidel M, Gifford SM, Ballantyne F, Dittmar T, Whitman WB, et al. Microbially-mediated transformations of estuarine dissolved organic matter. Front Mar Sci. 2017;4:69.
Pollegioni L, Tonin F, Rosini E. Lignin-degrading enzymes. FEBS J. 2015;282:1190–213.
Hambright KD, Beyer JE, Easton JD, Zamor RM, Easton AC, Hallidayschult TC. The niche of an invasive marine microbe in a subtropical freshwater impoundment. ISME J. 2015;9:256–64.
Foti MJ, Sorokin DY, Zacharova EE, Pimenov NV, Kuenen JG, Muyzer G. Bacterial diversity and activity along a salinity gradient in soda lakes of the Kulunda Steppe (Altai, Russia). Extremophiles. 2008;12:133–45.
Logares R, Lindstrom ES, Langenheder S, Logue JB, Paterson H, Laybourn-Parry J, et al. Biogeography of bacterial communities exposed to progressive long-term environmental change. ISME J. 2013;7:937–48.
Hunt DE, David LA, Gevers D, Preheim SP, Alm EJ, Polz MF. Resource partitioning and sympatric differentiation among closely related bacterioplankton. Science. 2008;320:1081–5.
McCarren J, Becker JW, Repeta DJ, Shi Y, Young CR, Malmstrom RR, et al. Microbial community transcriptomes reveal microbes and metabolic pathways associated with dissolved organic matter turnover in the sea. Proc Natl Acad Sci USA. 2010;107:16420–7.
Mills MM, Moore CM, Langlois R, Milne A, Achterberg E, Nachtigall K, et al. Nitrogen and phosphorus co-limitation of bacterial productivity and growth in the oligotrophic subtropical North Atlantic. Limnol Oceanogr. 2008;53:824–34.
Carlson CA, Giovannoni SJ, Hansell DA, Goldberg SJ, Parsons R, Vergin K. Interactions among dissolved organic carbon, microbial processes, and community structure in the mesopelagic zone of the northwestern Sargasso Sea. Limnol Oceanogr. 2004;49:1073–83.
Herlemann DPR, Manecki M, Meeske C, Pollehne F, Labrenz M, Schulz-Bull D, et al. Uncoupling of bacterial and terrigenous dissolved organic matter dynamics in decomposition experiments. PLoS ONE. 2014;9:e93945.
Zimmerman AE, Martiny AC, Allison SD. Microdiversity of extracellular enzyme genes among sequenced prokaryotic genomes. ISME J. 2013;7:1187.
Cottrell MT, Kirchman DL. Natural assemblages of marine Proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl Environ Microbiol. 2000;66:1692–7.
Gómez-Consarnau L, Lindh MV, Gasol JM, Pinhassi J. Structuring of bacterioplankton communities by specific dissolved organic carbon compounds. Environ Microbiol. 2012;14:2361–78.
Luo F, Zhong J, Yang Y, Zhou J. Application of random matrix theory to microarray data for discovering functional gene modules. Phys Rev E. 2006;73:031924.
Horemans B, Vandermaesen J, Smolders E, Springael D. Cooperative dissolved organic carbon assimilation by a linuron-degrading bacterial consortium. FEMS Microbiol Ecol. 2013;84:35–46.
Zhou J, Deng Y, Luo F, He Z, Tu Q, Zhi X. Functional molecular ecological networks. MBio. 2010;1:e00169–00110.
Zark M, Dittmar T. Universal molecular structures in natural dissolved organic matter. Nat Commun. 2018;9:3178.
Osterholz H, Niggemann J, Giebel H-A, Simon M, Dittmar T. Inefficient microbial production of refractory dissolved organic matter in the ocean. Nat Commun. 2015;6:7422.
Oren A. Thermodynamic limits to microbial life at high salt concentrations. Environ Microbiol. 2011;13:1908–23.
Cortes-Tolalpa L, Norder J, van Elsas JD, Falcao Salles J. Halotolerant microbial consortia able to degrade highly recalcitrant plant biomass substrate. Appl Microbiol Biotechnol. 2018;102:2913–27.
Sorokin DY, Toshchakov SV, Kolganova T, Kublanov IV. Halo(natrono)archaea isolated from hypersaline lakes utilize cellulose and chitin as growth substrates. Front Microbiol. 2015;6:942.
Godwin CM, Cotner JB. Aquatic heterotrophic bacteria have highly flexible phosphorus content and biomass stoichiometry. ISME J. 2015;9:2324.
Elser JJ, Sterner RW, Gorokhova E, Fagan WF, Markow TA, Cotner JB, et al. Biological stoichiometry from genes to ecosystems. Ecol Lett. 2000;3:540–50.
Manzoni S, Trofymow JA, Jackson RB, Porporato A. Stoichiometric controls on carbon, nitrogen, and phosphorus dynamics in decomposing litter. Ecol Monogr. 2010;80:89–106.
Spohn M. Element cycling as driven by stoichiometric homeostasis of soil microorganisms. Basic Appl Ecol. 2016;17:471–8.
Acknowledgements
This research was supported by grants from the National Natural Science Foundation of China (Grant Nos. 91751206, 41521001, 41972317, 41572328, and 41630103), the 111 Program (State Administration of Foreign Experts Affairs & the Ministry of Education of China, grant B18049), and Fundamental Research Funds for the Central Universities, China University of Geosciences (Wuhan), and State Key Laboratory of Biogeology and Environmental Geology, CUG (No. GBL11805). Portions of this work was performed in the W. R. Wiley Environmental Molecular Sciences Laboratory (EMSL), a DOE Office of Science User Facility sponsored by the Office of Biological and Environmental Research. We are grateful to anonymous reviewers for their constructive comments, which significantly improved the quality of the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Yang, J., Jiang, H., Liu, W. et al. Potential utilization of terrestrially derived dissolved organic matter by aquatic microbial communities in saline lakes. ISME J 14, 2313–2324 (2020). https://doi.org/10.1038/s41396-020-0689-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-020-0689-0
This article is cited by
-
Potential transformation of organic matter by microbes in cryoconite, Tibetan Plateau
Science China Earth Sciences (2024)
-
Photo-produced aromatic compounds stimulate microbial degradation of dissolved organic carbon in thermokarst lakes
Nature Communications (2023)
-
Spatial pattern and co-occurrence network of microbial community in response to extreme environment of salt lakes on the Qinghai-Tibet Plateau
Environmental Science and Pollution Research (2022)
-
Different distribution patterns of microorganisms between aquaculture pond sediment and water
Journal of Microbiology (2021)