Abstract
Chlorine disinfection to drinking water plays an important role in preventing and controlling waterborne disease outbreaks globally. Nevertheless, little is known about why it enriches the antibiotic resistance genes (ARGs) in bacteria after chlorination. Here, ARGs released from killed antibiotic-resistant bacteria (ARB), and culturable chlorine-injured bacteria produced in the chlorination process as the recipient, were investigated to determine their contribution to the horizontal transfer of ARGs during disinfection treatment. We discovered Escherichia coli, Salmonella aberdeen, Pseudomonas aeruginosa and Enterococcus faecalis showed diverse resistance to sodium hypochlorite, and transferable RP4 could be released from killed sensitive donor consistently. Meanwhile, the survival of chlorine-tolerant injured bacteria with enhanced cell membrane permeabilisation and a strong oxidative stress-response demonstrated that a physiologically competent cell could be transferred by RP4 with an improved transformation frequency of up to 550 times compared with the corresponding untreated bacteria. Furthermore, the water quality factors involving chemical oxygen demand (CODMn), ammonium nitrogen and metal ions (Ca2+ and K+) could significantly promote above transformation frequency of released RP4 into injured E. faecalis. Our findings demonstrated that the chlorination process promoted the horizontal transfer of plasmids by natural transformation, which resulted in the exchange of ARGs across bacterial genera and the emergence of new ARB, as well as the transfer of chlorine-injured opportunistic pathogen from non-ARB to ARB. Considering that the transfer elements were quite resistant to degradation through disinfection, this situation poses a potential risk to public health.
Similar content being viewed by others
Introduction
Antibiotic resistance has become a worldwide crisis. It is estimated that antibiotic resistance will cause as many as 10 million casualties annually by 2050 if no action is taken now [1,2,3,4]. Meanwhile, as increasing kinds of antibiotic resistance genes (ARGs) and antibiotic-resistant bacteria (ARB) are discovered in aquatic ecosystems [5,6,7], environmental ARGs that may promote the global transmission of ARB have been classified as emerging pollutants. Therefore, approaches to help disseminate ARGs in the environment are attracting global attention [8,9,10,11,12,13,14,15].
As a widely used tool to kill pathogens and ensure the microbiological safety of drinking water, chlorine disinfection has made a great contribution to preventing and controlling waterborne disease outbreaks [16,17,18]. Theoretically, it should help diminish, or even eliminate, ARB and ARG. However, the reported enrichment of the ARG level in the finished water after chlorination means a much higher proportion of either intracellular ARG (iARG) or extracellular ARG (eARG) in post-disinfection water than that in pre-disinfection water [19,20,21,22]. This realisation that chlorine disinfection enriches the ARG in bacteria and promotes the transmission of ARB and ARG in water poses a potential risk to public health. Interestingly, the reason for this and the factors that trigger antibiotic resistance dissemination during disinfection remain unclear.
Natural genetic transformation is a process in which a competent bacterial recipient takes up naked DNA and incorporates it into its own chromosome or converts it into an autonomous extra-chromosomal replicon [23]. As an important approach for spreading ARG, besides the competent bacterial recipient, a donor is required to provide the ARG for transfer. Considering that a large number of ARG carriers with biological activity would be released from the killed donors during the chlorination process [24], which would contribute to increasing eARG after disinfection [25], it has been hypothesised that the chlorination process would enhance iARG abundance by promoting the uptake of released ARG and genetic transformation across bacteria. Furthermore, culturable chlorine-injured bacteria may play roles in this process. Culturable chlorine-injured bacteria are viable but physiologically unhealthy populations tolerant to chlorine. These bacteria may account for as much as 90% of all indicator bacteria present after disinfection [26]. Due to suffering from reversible damage as a consequence of partial or inappropriate disinfection, they are undetectable using specific selective media, resulting in the underestimation of their presence, and health can be recovered only in certain circumstances, thereby posing hazards for water safety [13, 27]. Previously, we found temporal physiological persistence to antibiotics in culturable chlorine-injured Pseudomonas aeruginosa [28]. However, the contribution of cultivated chlorine-injured bacteria to genetic transformation during the treatment of disinfection remained unclear.
Herein, after the exposure of sodium hypochlorite (NaClO) to Escherichia coli, Salmonella aberdeen, P. aeruginosa and Enterococcus faecalis with plasmid RP4, the transferability of RP4 released from killed ARB (donor) and their genetic transformability to chlorine-injured bacteria (recipient) were observed. Then, the effects of physicochemical parameters, including pH, temperature, chemical oxygen demand (CODMn), ammonium nitrogen (NH4+–N) and metal ions (Ca2+ and K+) on the natural transformation frequency of released plasmid RP4 from killed bacteria into chlorine-injured E. faecalis, which are frequent causes of biofilm-associated opportunistic infections [29, 30], were further explored. For the first time, we showed that cultivated chlorine-tolerant injured non-ARB, which was a kind of competent cell with cell membrane permeabilisation and a strong oxidative stress-response, could uptake released RP4 from sensitive donors and then shift into ARB persistently with a higher frequency. This information will help in comprehending the reasons for the enrichment of the ARG level during the chlorination process and reveal the dissemination approaches of ARGs between bacteria during the disinfection process.
Materials and methods
Bacterial strains and media
A list of the bacterial strains and plasmids used in this study is given in Supplementary Table S1. The culture conditions of the bacteria are shown in Supplementary Text 1.
Disinfection experiments
Microbial inactivation experiments were conducted in sterile 250-mL glass bottles containing 100 mL of bacterial suspension at a final concentration of 105–106 cfu/mL. The preparation of the bacterial suspension for the exposure of NaClO (Sigma–Aldrich, USA) is shown in Supplementary Text 2. NaClO was then added to all vials (except for the control vials), and the samples were mixed well in a shaker at 150 rpm. Ten mililitres samples was collected before and after NaClO contact with the bacteria suspension at various time points (i.e., 15 s, and 1, 2, 5, 10, 20 and 30 min). All the samples treated with NaClO were immediately neutralised with 0.1 mol/L of sodium thiosulphate and then analysed for residual chlorine using the N,N-diethyl-p-phenylenediamine method [31]. Each experiment was performed, in triplicate, in a temperature-controlled incubator. The details of the disinfection kinetic modelling of NaClO are given in Supplementary Text 3.
Numeration of injured bacteria
The number of viable injured bacteria in the treated water samples was determined, according to the previous method [5, 6, 13, 28]. Briefly, appropriate dilutions of bacterial samples treated with NaClO were spread on TSYA and corresponding selection plates, in triplicate (Supplementary Text 1). After overnight incubation at 37 °C, both the number of viable bacteria on the TSYA and uninjured bacteria on the selective medium were calculated. The difference value was determined to be the viable injured bacteria.
Detection of transferable RP4 by genetic transformation
Functional transferable RP4 was detected as positive if the samples could transform chemically competent E. coli DH5α successfully through the heat-shock method [32]. DNA released in the disinfected samples at different time points was concentrated, according to Supplementary Text 4. Then, 10 μL of concentrated DNA samples was added to 50 μL of chemically competent E. coli DH5α (Takara, Dalian, China), which was placed on ice for 30 min before a heat-shock at 42 °C for 90 s. A total of 500 μL of SOC medium was added to each tube and incubated in a 37 °C water bath for 1 h. The number of transformants was determined by spreading aliquots of cell suspension on SOB plates containing 50 mg/L kanamycin (Kan), 60 mg/L ampicillin (Amp) and 40 mg/L tetracycline (Tet). Each experiment was performed in triplicate.
Cell permeability assessment
The hydrolysis rate of the o-nitrophenyl-β-d-galactopyranoside (ONPG) by E. coli was measured to assess cell permeability [13] and the absorbance values at 420 nm were monitored. Each experiment was performed in triplicate. The details are shown in Supplementary Text 5 and 6.
Bacterial reactive oxygen species (ROS) and antioxidant systems measurement
Bacterial ROS were detected with a DCF-DA/H2DCFDA-cellular ROS detection assay kit (Abcam, UK) and flow cytometry (BD FACS Calibur, USA). Bacterial antioxidant systems, including catalase (CAT) activity, superoxide dismutase (SOD) activity and glutathione peroxidase (GSH-Px) activity, were assayed with commercial kits from the Nanjing Jiancheng Bioengineering Institute (Nanjing, China) and a microplate reader (Molecular Devices, USA). All assays were done in compliance with the manufacturer’s instructions and were performed in triplicate. The details of these procedures are shown in Supplementary Text 5, 7 and 8.
Assay of natural transformation induced by NaClO exposure
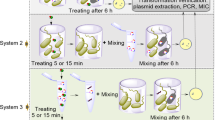
Considering the occurrence of conjugal transfer between the bacteria, two kinds of bacteria with different chlorine resistances were exposed to NaClO under the same conditions, separately, and then mixed in order to demonstrate the plasmid transformation induced by NaClO exposure. Briefly, water samples with 109 cfu/mL of E. coli, S. aberdeen and P. aeruginosa (carrier of RP4, donors) were treated with 6 mg/L NaClO for 20 min to kill most cells (Supplementary Table S2) and then neutralised by 0.1 mol/L of sodium thiosulfate and filtered (0.22-μm filter, Millipore, USA) to remove the entire residual bacteria. In parallel, 6.3 × 108 cfu/mL of chlorine-injured E. faecalis without RP4 (recipients) was prepared with 6 mg/L NaClO for 20 min (Supplementary Text 5). Then, 10 μL of the filtered solution was added to 100 μL of prepared injured bacteria and co-cultured at 37 °C for 60 min. Following this, the mixtures were spread on SOB medium containing 50 mg/L Kan, 60 mg/L Amp and 40 mg/L Tet. After overnight incubation at 37 °C, the colonies were counted. Each experiment was performed in triplicate.
In addition, to ensure that the bacteria growing on the SOB medium were transformers rather than any spontaneous mutations of the recipients or the contamination of donors, at least five colonies were spread on CATC plates of E. faecalis containing 50 mg/L Kan, 60 mg/L Amp and 40 mg/L Tet, and their plasmids were extracted to detect the specific TraG of RP4 by PCR. Detailed information about the RP4 detection by PCR is presented in Supplementary Table S3. The transformation frequency was calculated by dividing the number of transformants by the viable counts.
Statistical analysis
Statistical analyses were performed using IBM SPSS Statistics (version 20.0, Armonk, NY: IBM Corp). The transformation frequency and oxidative stress-response of various groups were analysed using the Student’s t test. The difference of the ONPG hydrolysis curves between E. coli suffering from chlorine injury or not was determined by one-way ANOVA test. For all tests, only data resulting in P values < 0.05 were regarded as statistically significant.
Results
ARB presented diverse resistance to chlorine disinfection during NaClO exposure
To observe the dynamics of ARB killing, the inactivation kinetics of four kinds of ARB—E. coli, P. aeruginosa, S. aberdeen and E. faecalis (with RP4)—were investigated during bench-scale inactivation experiments with NaClO, which were carried out in buffered disinfectant demand-free water at pH 7.2 and 20 °C. Fitted chlorine decay curves and bacterial inactivation kinetics are shown in Supplementary Figs. S1–S3. Chlorine decay constants k′ with different initial NaClO concentrations, and the estimated efficiency factor Hom (EFH) model parameters are summarised in Supplementary Table S4. These data showed that the EFH model was a good fit for the inactivation data of all observed ARB, with R2 above 0.97. The Ct values (Supplementary Table S5), that is, the product of the disinfectant concentration and contact time needed to achieve a defined reduction of target organisms, were calculated by fitting the EFH model for different levels of ARB inactivation by NaClO.
The inactivation curves of observed ARB (Fig. 1) were based on the Ct value calculated by fitting the EFH model. A similar lethal bacterial tendency can be found whether it was detected by TSYA or a selective medium, but the lethal rate detected with a selective medium was higher than that with TSYA due to its detection failure of chlorine-injured bacteria. For example, after the exposure of 8.4 mg/L min, 4.0-log killing of E. coli could be observed for the TSYA medium, while it was above 4.5-log killing for the Endo media. Importantly, the strains exhibited diverse resistant abilities to NaClO, although E. faecalis was the strongest. To kill 4-log E. faecalis fully, the Ct values reached 22.8 mg/L min, which was detected with TSYA medium. Otherwise, E. coli was demonstrated to be the most sensitive against NaClO, among the observed bacteria. The Ct value to kill 4-log E. coli fully was only 8.4 mg/L min. Significant differences between the Ct values to kill various bacteria fully demonstrated that it is possible that bacteria with strong resistance to chlorine, such as E. faecalis, can survive NaClO exposure even if sensitive E. coli are fully killed.
The baseline conditions were as follows: the initial concentration of bacteria was 105–106 cfu/mL (pH 7.2, 20 °C). Nt/No: number of viable bacteria detected by TSYA medium or selective medium at a given time/number of bacteria at zero time. Dash lines, fitted Ct values by the estimated efficiency factor Hom (EFH) model. Solid line, observed Ct values (n = 3; mean ± SD).
Killed ARB after NaClO exposure can donate transferable plasmids to surroundings
During the process of NaClO suffering, an increasing amount of killed ARB released the corresponding RP4 into the surroundings gradually. To test the transferability of RP4 released from ARB, ARB suspensions suffering from different doses of NaClO were filtered to remove ARB and then co-cultured with chemically competent E. coli DH5α. Table 1 summarises the Ct values for destroying the RP4 released from ARB at pH 7.2 and 20 °C. It showed that transformants could still be formed for the filtered water after disinfection even at Ct values of 25.8 mg/L min for P. aeruginosa and 39.8 mg/L min for E. faecalis when no viable ARB was detectable by the Cetrimide agar or CATC agar (Fig. 1). Therefore, transferable plasmids could be released from killed sensitive ARB, and much more NaClO was required to remove plasmids than was required to kill ARB (compare Table 1 and Fig. 1). Even if the ARB were killed, RP4, as an ARG carrier donated by the ARB present in water, was stable and remained transferable.
Survival of chlorine-injured bacteria produced throughout NaClO exposure showed physiological competence with increased membrane permeability and a strong oxidative stress-response
As a parallel, the production of chlorine-injured bacteria during the process of NaClO exposure was found continuously. From Fig. 2, the concentration of injured bacteria would peak at the 2-min exposure-time point, whose Ct values are in the range 2.0–4.2 mg/L min. Then, their concentration would decline gradually as the exposure dose increased. Otherwise, the percentage of injured bacteria in viable cells would increase sharply and all residual viable E. coli, P. aeruginosa, S. aberdeen, and 97% of viable E. faecalis, would become injured (below 100 cfu/mL) when exposure to NaClO reached 16.1, 19.3, 29.3 and 38.3 mg/L min, respectively. Therefore, during the chlorination process, viable bacteria became injured gradually and, ultimately, surviving populations with stronger tolerance to chlorine were injured.
To test if chlorine-injured bacteria were physiologically competent, RP4 was used to evaluate their transformability to chlorine-injured or CaCl2-prepared competent E. coli, P. aeruginosa, S. aberdeen and E. faecalis (Fig. 3, Supplementary Text 5). The data showed that RP4 could enter injured bacteria of all kinds of those mentioned above and survive in them successfully, presenting a more efficient transformation than corresponding CaCl2-prepared competent bacteria (p = 1.87 × 10−4, 1.01 × 10−4, 6.61 × 10−3, 1.88 × 10−3, respectively) and untreated ones (p = 1.0 × 10−4, 8.8 × 10−5, 3.3 × 10−3, 1.5 × 10−3, respectively). Above all, compared with the corresponding untreated bacteria, chlorine-injured E. faecalis improved the transformation frequency of RP4 by 550-fold, reaching 9.8 × 10−5, while it was only enhanced 21-fold for CaCl2-prepared competent bacteria, equating to a transformation frequency of 3.7 × 10−6.
RP4 (100 ng) was added to 100 µL of 108 cfu/mL chlorine-injured or competent bacteria prepared by the CaCl2 method, according to the heat-shock assay [32, 45]. Bacteria (108 cfu/mL) without exposure to NaClO were selected as controls. *p < 0.01, compared with untreated bacteria; #p < 0.01, compared with CaCl2-prepared competent bacteria.
An ONPG hydrolysis rate of 108 cfu/mL above chlorine-injured E. coli was exhibited (Fig. 4). Compared with the control (untreated E. coli), injured E. coli increased their hydrolysis rate of ONPG by a significant 3.1-fold (p = 0.0095). It indicated that the cell membrane had increased permeability in the chlorine-injured bacteria after the exposure to NaClO.
Furthermore, the oxidative stress-response in chlorine-injured E. coli, P. aeruginosa, S. aberdeen and E. faecalis was investigated (Fig. 5). It was found that the production of ROS increased in three injured bacteria (E. coli, P. aeruginosa and E. faecalis) up to 2.3-fold and their ROS levels were evident higher than those in the control. Meanwhile, to protect bacteria from ROS destruction, cellular antioxidant systems, including CAT activity, SOD activity, and GSH-Px activity, were activated, correspondingly. The SOD level increased dramatically up to 4.5-fold in the chlorine-injured bacteria (p < 0.01). CAT activity was only expressed in P. aeruginosa before chlorine exposure, but all four bacteria expressed it after chlorination, and S. aberdeen showed the maximal CAT activity of 8.3 U/108 cfu. No GSH-Px activity was expressed in all untreated bacteria, but chlorine-injured P. aeruginosa, S. aberdeen and E. faecalis had improved GSH-Px activity of up to 11.3 U/108 cfu. All these results indicate that a strong oxidative stress-response occurred in the injured bacteria suffering from NaClO exposure.
Bacterial suspensions (108 cfu/mL), chlorine-injured or not, were sonicated at 20 kHz for 10 min and then centrifuged at 5000 × g, 4 °C for 3 min. The levels of superoxide dismutase (SOD), catalase (CAT) and glutathione peroxidase (GSH-Px) in the supernatants were assayed. Reactive oxygen species (ROS) in 106 cfu/mL chlorine-injured bacteria or not were measured and the fold changes of average ROS between them were calculated.
Released RP4 from chlorine-killed ARB horizontally transferred to chlorine-injured bacteria by natural transformation more frequently under the same environmental circumstances
The RP4 released from killed sensitive E. coli, P. aeruginosa and S. aberdeen transformed into chlorine-resistant injury E. faecalis naturally, with frequencies of 8.8 × 10−6–1.7 × 10−5 (Fig. 6). In comparison to the control group, in which E. faecalis was untreated by NaClO, the transformation frequencies were improved in the range 37–134-fold (Supplementary Fig. S4).
Suspensions (109 cfu/mL) of E. coli, P. aeruginosa and S. aberdeen (with RP4) treated by 6 mg/L NaClO for 20 min, separately, were filtered to remove ARB and then co-cultured with 108 cfu/mL E. faecalis chlorine-injured or not for 60 min (1:10, v/v). Then, transformers on SOB medium containing 50 mg/L Kan, 60 mg/L Amp and 40 mg/L Tet were picked after overnight incubation at 37 °C.
To further explore the essential water quality factors on the natural transformation of injured bacteria, the effects of physicochemical parameters, including pH, temperature, CODMn, NH4+–N and metal ions, on the above-stated transformation frequency were observed (Fig. 7, Supplementary Figs. S5 and S6, Supplementary Text 9). The transformation frequency of released RP4 from killed E. coli, P. aeruginosa and S. aberdeen into injured E. faecalis under various physicochemical conditions, revealed that CODMn, NH4+–N and metal ions (Ca2+ and K+) could promote the transformation frequency of released RP4 into injured bacteria (p < 0.05, n = 3, Supplementary Table S6). Above all, transformation frequencies had increased as the concentration of NH4+–N increased. When it was 5 mg/L, the whole transformation frequency of released plasmid RP4 from killed E. coli into injured E. faecalis reached around 7.9 × 10−4, which was upregulated 55-fold compared with the corresponding injured bacteria without NH4+–N exposure. Furthermore, the pH value of the water had a great effect on the transformation frequency of released RP4, which reached at most 2.8 × 10−5 at pH 7 when RP4 were released from E. coli. Then, it would decline with increasing or decreasing pH, reaching zero at pH 10. However, a similar transformation frequency of around 3.0 × 10−5 was found when the temperature was in the range 4–37 °C, and there were no significant differences between them (p > 0.05, Supplementary Table S6).
The baseline conditions were as follows: 109 cfu/mL E. coli suspension (with RP4) treated by 6 mg/L NaClO for 20 min were filtered to remove ARB and then co-cultured with 108 cfu/mL of chlorine-injured E. faecalis under different conditions of water quality parameters for 60 min (1:10, v/v). Then, transformers on SOB medium containing 50 mg/L Kan, 60 mg/L Amp and 40 mg/L Tet were picked after overnight incubation at 37 °C.
Discussion
Nowadays, antibiotic use, environmental temperature and urban wastewater treatment plants size have been realised as important factors related to resistance persistence and spread in the environment [33, 34]. However, this study is the first to find that chlorine disinfection, which has been used to eliminate pathogens and improve public health in past years [35], increased the frequency of natural transformation and inevitably promoted the horizontal transfer of ARG across bacterial genera via culturable chlorine-injured bacteria. In a sense, chlorine disinfection naturally accelerated the genetic exchange in or across bacterial genera, resulting in the enrichment of ARGs in bacteria after chlorination.
This phenomenon occurred because disinfection of drinking water can help release DNA, including various ARGs and mobile genetic elements (MGEs), such as plasmids, integrons and insertion sequences, from killed donors into the environment [24]. It is more difficult to remove all functional antibiotic-resistant plasmids or ARGs in the ARB than to kill the ARB itself [36, 37]. This study also showed much more NaClO was required to remove transferable plasmids released from killed sensitive ARB. Therefore, using NaClO to decrease bacteria viability is merely fulfilling a necessary demand to “kill” the bacteria. A much higher dose of NaClO should be utilised to destroy plasmids and further break down ARGs in the ARB. Considering that the MGEs involved in the horizontal transfer of ARGs are still quite recalcitrant to disinfection and retain their biological activity [37,38,39], released ARGs located on MGEs can easily spread via horizontal transfer among species, including human pathogens [40]. Roller et al. reported that even at 6-log inactivation of Haemophilus influenzae cells by chlorine dioxide (ClO2), the intracellular DNA was largely intact [41]. Here, we further demonstrated that although ARB cells had effectively died following chlorination, their plasmids still retained biological activity and could be available to other bacterial communities via transformation. Hence, when disinfection is performed just to inactivate bacteria or the disinfection dose is not enough, large amounts of functional plasmids or ARGs with transporters are released from the dead bacteria that provide a great opportunity to transfer ARB to other environmental bacteria by natural transformation.
Another important reason explaining the enrichment of ARGs in bacteria after chlorination is that the challenge of disinfection in a factory cannot fully kill all bacteria. It induces survivors with a strong resistance to disinfection to enter a kind of physiologically damaged state in which they can easily capture environmental DNA, including various ARGs and MGEs. A large amount of DNA is released from sensitive bacteria surrounding these injured bacteria, during disinfection, which makes horizontal transfer happen more frequently. In this study, we demonstrated different extents of chlorine resistance among bacterial genera, which then yielded injured bacteria originating from higher resistance populations and, also, it was found that culturable chlorine-injured bacteria can frequently be transformed by antibiotic-resistant plasmids released by killed sensitive ARB. The transformation frequency of these culturable chlorine-injured bacteria increased >550-fold compared with untreated bacteria, resulting in a much higher proportion of iARG in post-disinfection water than that in pre-disinfection water [19]. Generally, competent E. coli cells are often used in bacterial transformation owing to their high permeability for bioorganic macromolecules like plasmid DNA [42]. Therefore, the mechanism for increased transformation here can be explained by the fact that the chlorine-injured bacteria also became competent, making plasmid uptake easier, due to the increased cell membrane permeability found in chlorine-injured E. coli in this study. Furthermore, the phenomenon of bacterial co-selection of disinfection resistance and antibiotic resistance in tap water found by Khan et al. [16]. might be explained by chlorine-resistant injured bacteria shifting from non-ARB to ARB by natural transformation. In addition, using a chlorine dosage that is lethal to E. coli but sub-lethal to E. faecalis led to the possibility of producing injured E. faecalis, which can uptake antibiotic-resistant plasmids released by dead ARB, resulting in the dissemination of antibiotics resistance across the bacterial genus. Due to Enterococci causing biofilm-associated opportunistic infections [30], such as wounds and infective endocarditis, promoting the natural transformation of released plasmids into chlorine-injured E. faecalis by the chlorination process may pose a potential risk to public health. Furthermore, the above results indicated that it was not ideal to use E. coli to determine the sanitary quality of drinking water, particularly for judging faecal contamination or predicting the possible presence of waterborne pathogens [13, 43], considering that it would result in insufficient bacterial inactivation and the production of culturable chlorine-injured bacteria that show more resistance to chlorination than E. coli. These organisms can survive chlorine exposure and enhance ARG transfer across bacterial genera via natural transformation.
Finally, environmental contaminants, CODMn, NH4+–N and metal ions (Ca2+ and K+), can enhance the transformation frequency of released plasmid RP4 from killed ARB into culturable chlorine-injured bacteria. In general, the cell competency drastically decreased in the case of planktonic cells when organic nutrients were richly available for cell growth [15]. However, chlorine-injured bacteria still can be induced to become competent in the presence of organic material in the environment, indicating they may also enter into a state of nutritional starvation similar to competent E. coil cells existing in the state of biofilm [15]. Ca2+ is one of the most typical competence-inducing factors. They are generally considered to induce the formation of pore-like structures on the cell surface for the passing of intact double-stranded DNA [14]. Alkali cations e.g. Na+, K+ in the presence of PEG6000 were also found all effective for the transformation of E. coli [7]. Liu et al. found that the change of total concentration of observed iARGs correlated strongly and positively with the NH4+–N concentration, during chlorination [19]. Zhang et al. found that a higher concentration of NH4+–N (15 mg/L) resulted in lower ARGs removal [44]. Therefore, external environmental factors, one of the DNA-uptake mechanisms for bacteria[14], may play important roles in horizontal transfer of ARGs resulting from the chlorination process. It indicated that controlling water quality before disinfection is an effective method to control ARG pollution.
Virtually, the promotion of conjugal transfer between bacteria may also contribute the enrichment of the iARG level in the finished water after chlorination. To determine the effect of chlorine on total transformation events, we also performed an experiment to treat co-cultures of donor (E. coli with RP4, Kan-, Amp- and Tet-resistant) and recipient cells (E. faecalis, nalidixic acid-resistant) using NaClO (Supplementary Text 10). The results showed (Supplementary Table S7) the number of transformers in viable E. faecalis (Kan-, Amp-, Tet- and nalidixic acid resistant) increased 8.3–8.7 folds even after 6–7 mg/L NaClO exposure, comparing with the control group (no chlorine). However, considering no genetic transformation should be occurred under the circumstance of the control (mixture of two kinds of bacteria), conjugal transfer was supposed to occur between E. coli and E. faecalis in the control. As a result, the increasing number of transformers after the chlorine exposure to mixed bacteria may also come from the promotion of conjugal transfer between bacteria, besides the genetic transformation.
In summary, chlorine-tolerant injured bacteria that are physiologically competent cells present higher plasmid transformation frequency than the corresponding untreated bacteria. Since the transferable plasmids released from killed sensitive ARB have a consistent resistance to degradation through disinfection, the chlorination process can promote the horizontal transfer of the released plasmid into chlorine-injured bacteria through natural transformation and lead to the enrichment of ARGs in viable bacteria. This process was enhanced by environmental contaminants, CODMn, NH4+–N and metal ions. These results indicate that chlorine disinfection naturally accelerates gene exchange in or between bacterial genera, so that the chlorine-injured opportunistic pathogens can be transferred from non-ARB to ARB through natural transformation during chlorination, which poses a potential risk to disseminate antibiotic resistance in water.
References
Pruden A, Pei R, Storteboom H, Carlson KH. Antibiotic resistance genes as emerging contaminants: studies in northern Colorado. Environ Sci Technol. 2006;40:7445–50.
O’Neill J. Antimicrobial resistance: tackling a crisis for the health and wealth of nations. London: The Review on Antimicrobial Resistance; 2015.
Watkins RR, Bonomo RA. Overview: global and local impact of antibiotic resistance. Infect Dis Clin N Am. 2016;30:313–22.
Zhu YG, Gillings M, Simonet P, Stekel D, Banwart S, Penuelas J. Microbial mass movements. Science. 2017;357:1099–100.
Fricker CR, Eldred BJ. The effect of sodium azide concentration on the recovery of Enterococci from water. J Water Health. 2014;12:264–8.
Wang X, Devlieghere F, Geeraerd A, Uyttendaele M. Thermal inactivation and sublethal injury kinetics of Salmonella enterica and Listeria monocytogenes in broth versus agar surface. Int J Food Microbiol. 2017;243:70–7.
Ano T, Shoda M. Ultra-rapid transformation of Escherichia coli by an alkali cation. Biosci Biotechnol Biochem. 1992;56:1505.
Baquero F, Martinez JL, Canton R. Antibiotics and antibiotic resistance in water environments. Curr Opin Biotechnol. 2008;19:260–5.
Marti E, Variatza E, Balcazar JL. The role of aquatic ecosystems as reservoirs of antibiotic resistance. Trends Microbiol. 2014;22:36–41.
Karkman A, Do TT, Walsh F, Virta MPJ. Antibiotic-resistance genes in waste water. Trends Microbiol. 2018;26:220–8.
Czekalski N, Gascon Diez E, Burgmann H. Wastewater as a point source of antibiotic-resistance genes in the sediment of a freshwater lake. ISME J 2014;8:1381–90.
Yan L, Liu D, Wang XH, Wang Y, Zhang B, Wang M, et al. Bacterial plasmid-mediated quinolone resistance genes in aquatic environments in China. Sci Rep. 2017;7:40610.
Li J, Liu L, Yang D, Liu WL, Shen ZQ, Qu HM, et al. Culture-dependent enumeration methods failed to simultaneously detect disinfectant-injured and genetically modified Escherichia coli in drinking water. Environ Sci Process Impacts. 2017;19:720–6.
Hasegawa H, Suzuki E, Maeda S. Horizontal plasmid transfer by transformation in Escherichia coli: Environmental factors and possible mechanisms. Front Microbiol. 2018;9:2365.
Nishioka M, Mashayekhan S, Onishi K, Taya M. The biofilm environment offers a possible condition for inducing the competency of DNA recipient cells through nutritional starvation. Biocontrol Sci. 2007;12:115–8.
Khan S, Beattie TK, Knapp CW. Relationship between antibiotic- and disinfectant-resistance profiles in bacteria harvested from tap water. Chemosphere. 2016;152:132–41.
McCurry DL, Krasner SW, von Gunten U, Mitch WA. Determinants of disinfectant pretreatment efficacy for nitrosamine control in chloraminated drinking water. Water Res. 2015;84:161–70.
Nwachcuku N, Gerba CP. Emerging waterborne pathogens: can we kill them all? Curr Opin Biotechnol. 2004;15:175–80.
Liu SS, Qu HM, Yang D, Hu H, Liu WL, Qiu ZG, et al. Chlorine disinfection increases both intracellular and extracellular antibiotic resistance genes in a full-scale wastewater treatment plant. Water Res. 2018;136:131–6.
Shi P, Jia S, Zhang XX, Zhang T, Cheng S, Li A. Metagenomic insights into chlorination effects on microbial antibiotic resistance in drinking water. Water Res. 2013;47:111–20.
Zheng DW, Chen JL, Zhu JY, Rong L, Li B, Lei Q, et al. Highly integrated nano-platform for breaking the barrier between chemotherapy and immunotherapy. Nano Lett. 2016;16:4341–7.
Xi C, Zhang Y, Marrs CF, Ye W, Simon C, Foxman B, et al. Prevalence of antibiotic resistance in drinking water treatment and distribution systems. Appl Environ Microbiol. 2009;75:5714–8.
Baur B, Hanselmann K, Schlimme W, Jenni B. Genetic transformation in freshwater: Escherichia coli is able to develop natural competence. Appl Environ Microbiol. 1996;62:3673–8.
Dodd MC. Potential impacts of disinfection processes on elimination and deactivation of antibiotic resistance genes during water and wastewater treatment. J Environ Monit. 2012;14:1754–71.
Hao H, Shi DY, Yang D, Yang ZW, Qiu ZG, Liu WL, et al. Profiling of intracellular and extracellular antibiotic resistance genes in tap water. J Hazard Mater. 2019;365:340–5.
Bissonnette G, Jezeski J, McFeters G, Stuart D. Influence of environmental stress on enumeration of indicator bacteria from natural waters. Appl Microbiol. 1975;29:186–94.
McFeters GA, LeChevallier MW, Singh A, Kippin JS. Health significance and occurrence of injured bacteria in drinking water. Water Sci Technol. 1986;18:227–31.
Hou AM, Yang D, Miao J, Shi DY, Yin J, Yang ZW, et al. Chlorine injury enhances antibiotic resistance in Pseudomonas aeruginosa through over expression of drug efflux pumps. Water Res. 2019;156:366–71.
Ch’ng JH, Chong KKL, Lam LN, Wong JJ, Kline KA. Biofilm-associated infection by Enterococci. Nat Rev Microbiol. 2019;17:82–94.
Kao PHN, Kline KA. Dr. Jekyll and Mr. Hide: how Enterococcus faecalis subverts the host immune response to cause infection. J Mol Biol. 2019;431:2932–45.
American Public Health Association (APHA). Standard methods for the examination of water and wastewater. 20th ed. Washington DC: APHA; 1998.
Froger A, Hall JE. Transformation of plasmid DNA into E. coli using the heat shock method. J Vis Exp. 2007:253.
Martinez JL. Environmental pollution by antibiotics and by antibiotic resistance determinants. Environ Pollut. 2009;157:2893–902.
MacFadden DR, McGough SF, Fisman D, Santillana M, Brownstein JS. Antibiotic resistance increases with local temperature. Nat Clim Chang. 2018;8:510–4.
Murphy EA, Post GB, Buckley BT, Lippincott RL, Robson MG. Future challenges to protecting public health from drinking-water contaminants. Annu Rev Public Health. 2012;33:209–24.
Stange C, Sidhu JPS, Toze S, Tiehm A. Comparative removal of antibiotic resistance genes during chlorination, ozonation, and UV treatment. Int J Hyg Environ Health. 2019;222:541–8.
He H, Zhou P, Shimabuku KK, Fang X, Li S, Lee Y, et al. Degradation and deactivation of bacterial antibiotic resistance genes during exposure to free chlorine, monochloramine, chlorine dioxide, ozone, ultraviolet light, and hydroxyl radical. Environ Sci Technol. 2019;53:2013–26.
McKinney CW, Pruden A. Ultraviolet disinfection of antibiotic resistant bacteria and their antibiotic resistance genes in water and wastewater. Environ Sci Technol. 2012;46:13393–400.
Yoon Y, Chung HJ, Wen Di DY, Dodd MC, Hur HG, Lee Y. Inactivation efficiency of plasmid-encoded antibiotic resistance genes during water treatment with chlorine, UV, and UV/H2O2. Water Res. 2017;123:783–93.
Rizzo L, Manaia C, Merlin C, Schwartz T, Dagot C, Ploy MC, et al. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: a review. Sci Total Environ. 2013;447:345–60.
Roller VO SD, Kawata K. Mode of bacterial inactivation by chlorine dioxide. Water Res. 1980;14:635–41.
Lim Y, Su CH, Liao YC, Lee SY. Impedimetric analysis on the mass transfer properties of intact and competent E. coli cells. Biochim Biophys Acta Biomembr. 2019;1861:9–16.
Leclerc H, Schwartzbrod L, Dei-Cas E. Microbial agents associated with waterborne diseases. Crit Rev Microbiol. 2002;28:371–409.
Zhang Y, Zhuang Y, Geng J, Ren H, Zhang Y, Ding L, et al. Inactivation of antibiotic resistance genes in municipal wastewater effluent by chlorination and sequential UV/chlorination disinfection. Sci Total Environ. 2015;512-513:125–32.
Seidman CE, Struhl K, Sheen J, Jessen T. Introduction of plasmid DNA into cells. Curr Protoc Mol Biol. 2001;37:1.8.1–1.8.10.
Acknowledgements
This work is supported by the grant from the National Natural Science Foundation of China (Grant No. 81972994 and 41831287). Min Jin would like to acknowledge Miss Hai-yan Wu and Miss Tian-jiao Chen for her contributions during the media preparation.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jin, M., Liu, L., Wang, Dn. et al. Chlorine disinfection promotes the exchange of antibiotic resistance genes across bacterial genera by natural transformation. ISME J 14, 1847–1856 (2020). https://doi.org/10.1038/s41396-020-0656-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-020-0656-9
This article is cited by
-
Phenotypic and molecular characterization of β-lactamase-producing Klebsiella species among children discharged from hospital in Western Kenya
BMC Microbiology (2024)
-
Enhancing microbiologically influenced corrosion protection of carbon steels with silanized epoxy-biocide hybrid coatings
Environmental Science and Pollution Research (2024)
-
Microplastic pollution interaction with disinfectant resistance genes: research progress, environmental impacts, and potential threats
Environmental Science and Pollution Research (2024)
-
Long-term exposure to food-grade disinfectants causes cross-resistance to antibiotics in Salmonella enterica serovar Typhimurium strains with different antibiograms and sequence types
Antimicrobial Resistance & Infection Control (2023)
-
Mechanisms of emerging resistance associated with non-antibiotic antimicrobial agents: a state-of-the-art review
The Journal of Antibiotics (2023)