Abstract
Fungal endophytes can improve plant tolerance to abiotic stress. However, the role of these plant–fungal interactions in invasive species ecology and their management implications remain unclear. This study characterized the fungal endophyte communities of native and invasive lineages of Phragmites australis and assessed the role of dark septate endophytes (DSE) in salt tolerance of this species. We used Illumina sequencing to characterize root fungal endophytes of contiguous stands of native and invasive P. australis along a salinity gradient. DSE colonization was assessed throughout the growing season in the field, and effects of fungal inoculation on salinity tolerance were investigated using laboratory and greenhouse studies. Native and invasive lineages had distinct fungal endophyte communities that shifted across the salinity gradient. DSE colonization was greater in the invasive lineage and increased with salinity. Laboratory studies showed that DSE inoculation increased P. australis seedling survival under salt stress; and a greenhouse assay revealed that the invasive lineage had higher aboveground biomass under mesohaline conditions when inoculated with a DSE. We observed that P. australis can establish mutualistic associations with DSE when subjected to salt stress. This type of plant–fungal association merits further investigation in integrated management strategies of invasive species and restoration of native Phragmites.
Similar content being viewed by others
Introduction
Fungal endophytes establish mutualistic associations with most plant species, and can play a major role in plant ecology and community structure [1]. These endophytes can improve host nutrient uptake [2, 3], improve host defense against pathogens [4], modify trophic interactions [5,6,7], and improve host tolerance to abiotic stress [8, 9]. At the plant community level, they can affect plant diversity [10, 11] and can be important factors in plant invasion ecology [12, 13]. A better understanding of plant–microbe interactions can help improve various aspects of invasive species management [14]. Kowalski et al. [15] recently proposed a framework for a microbial-based control strategy of invasive species; the basis of this strategy is that greater understanding of key microbial association of invasive and native species can lead to new insights of invasive species’ success and improve management practices.
The aggressive expansion of the invasive European lineage of Phragmites australis is an issue in several regions of the United States. Management of this lineage has been costly. Despite agencies spending over $4.6 million/year [16], most eradication efforts are unsuccessful and focused on short-term results [15, 17]. Once established, invasive P. australis forms dense monotypic stands affecting native plant diversity [18,19,20], hydrology [21], and biogeochemistry [22, 23] in invaded areas. Expansion of this lineage has been common in brackish marshes [24, 25] and salt marshes, where it can significantly alter ecological functions [19, 26].
The native North American haplotype F of P. australis [27] is less salt tolerant than the invasive European haplotype M [28], and is therefore predominantly found in low-salinity habitats [29]. Both lineages share the same physiological mechanisms of salt tolerance, which are K+ accumulation in plant tissues and Na+ exclusion [30, 31]; but the growth potential of the invasive lineage has been considered key to its invasiveness at higher salinities [28]. Expansion of the invasive lineage into salt marshes has also been related to clonal integration [32] and temporary decreases in soil salinity [20, 33]. Benefits of microbial associations for salinity tolerance of P. australis have been theorized [34] but have not been assessed until this study.
In wetlands, one of the most common groups of root endophytes are dark septate endophytes (DSE). In these systems they are commonly found to coexist with mycorrhizal fungi and are more prevalent in monocotyledonous than dicotyledonous plant species [35, 36]. DSE are considered generalist root fungi and have been found to associate with over 600 plant species, including some that are non-mycorrhizal in various ecosystems [37, 38]. Based on the classification by Rodriguez et al. [1], these Class IV endophytes can be characterized as sterile or conidial, they have dark melanized hyphae and microsclerotia, and are likely to play an important role in plant ecophysiology. Several studies have found DSE colonization is common in plants exposed to abiotic stress [39,40,41], and experimental inoculation of plants with DSE has been reported to improve host tolerance to heavy metal contamination [42] and drought [43]. Some of the possible mechanisms by which DSE can affect host fitness include the production of bioactive compounds [42, 44], and increasing nutrient uptake by colonized hosts [2, 43, 45]. Considering the ubiquitous nature of DSE in wetland grass species and their ability to promote stress tolerance in various hosts, their associations with wetland plants, and potential functional roles merit further investigation. Specifically, their interactions with native and invasive plants like P. australis could be of interest to improve management of the invasive lineage as proposed by Kowalski et al. [15].
In this study we characterized the fungal endophyte communities of contiguous stands of native and invasive P. australis across a salinity gradient. We used next generation sequencing and microscopy to address the role of lineage and salinity in structuring root fungal communities over a growing season. In pursuing this objective, we identified salinity-driven DSE colonization patterns that led to a follow-up question: Can fungal endophytes improve salt tolerance of P. australis? We hypothesized that DSE mutualists played a role in stress tolerance of the invasive P. australis lineage, and used laboratory and greenhouse assays to test this prediction.
Materials and methods
Study sites and sampling
We selected three sites with contiguous stands of native and invasive P. australis along a salinity gradient in the Choptank River in eastern Maryland, USA (Fig. 1). The salinity regimes at these tidal wetland sites range from freshwater (<0.5%) to oligohaline (0.5–5%) [46]. During the summer of 2016, we collected rhizomes from visibly healthy native and invasive P. australis by excavating the plant and clipping rhizomes that had multiple lateral and fine roots. Four rhizomes were sampled from each stand from plants that were at least 5 m away from each other. Sampling was carried out approximately every 2 weeks between June and October, resulting in a total of 84 rhizomes of each lineage that were collected for analysis (168 total rhizomes-3 sites × 2 lineages × 7 time points × 4 rhizomes/plant). To monitor water level, we installed loggers (HOBO U20L-04, Bourne MA, USA) in stands of native and invasive P. australis at sites A and C, and water level was recorded every 5 min from July to October. We calculated the level of inundation for each stand based on the percent time that the water was above the soil surface over the 2-week period before each rhizome sampling date. Salinity was recorded at each site using a portable salinity meter (YSI, Yellow Springs OH, USA). We collected six soil samples at a depth of 25 cm from each site in July and analyzed their pH using a 1:5 soil:DI water slurry, soil organic matter (SOM) using loss-on-ignition (550 °C for 2 h) and then sent them to the Delaware Soil Testing Lab for analyses of percent nitrogen (%N) and carbon (%C) by combustion at 950 °C. We characterized root morphology of native and invasive P. australis based on three samples from each stand and measured lateral root density and length, and root hair density [47].
Root processing and endophyte sequencing
Lateral and fine roots were clipped and separated for different uses. Some were stored in 50% ethanol for staining and microscopy, and the rest were surfaced sterilized and either stored at −80 °C for Illumina sequencing, or used to isolate fungal endophytes. Surface sterilization was carried out as described in Ban et al. [48], with 99% ethanol for 1 m, 35% hydrogen peroxide for 5 m, 99% ethanol for 30 s, and washing three times in sterile DI water. We confirmed the success of the root sterilization by imprinting the roots on potato dextrose agar (PDA) and confirming no signs of growth after incubation.
To isolate root endophytes we clipped the ends of the roots and placed them on PDA with ampicillin (50 μg ml−1) and streptomycin (25 μg ml−1). Plates were incubated in the dark at 23 °C and after about 10 days fungi that emerged from the roots were transferred to new PDA plates. We characterized the isolated endophytes using Sanger sequencing. Endophyte recovery from the Choptank sites was low and we were only able to obtain one DSE from native and invasive Phragmites at Site B and one from Site G after plating four segments of eight root samples collected at each site. Therefore additional isolates were obtained using the same methods from ten root samples of invasive Phragmites that were located at a mesohaline site (8 ppt) on the Patuxent River (N38°32′20″, W76°40′3″). We extracted fungal DNA using a Zymo Quick DNA Fungal/Bacterial kit according to the manufacturer’s instructions. BigDye® Terminator v3.1 (ThermoFisher) was used for PCR amplification using the ITS1F/ITS4R and the EF1-728F/EF1-986R primer sets to amplify the internal transcribed spacer (ITS) region and alpha elongation factor (EF), respectively. EdgeBio cleanup plates were used to recover the cleaned sample, which was then vacuum concentrated using a speedvac, resuspended in 20 µl of HiDi formamide, and denatured for 2 m at 95 °C. We processed the resulting sequences using SeqScanner v.1.0 (ABI) to check quality, DNAStar to assemble contigs at 97% similarity, and BLAST (NCBI) to assign taxonomy. Sequences of the isolated endophytes were deposited in GenBank under accession numbers MT094851-MT094863.
Surface sterilized roots from two of our sampling dates (June 30th and August 24th) were used for DNA extractions and subsequent Illumina sequencing of the ITS1 region. We used a PowerPlant Pro DNA isolation kit (MoBio, Carlsbad CA, USA) for DNA extractions and followed the manufacturer’s instructions, except for the lysing step, which was carried out using a FastPrep®-24 (two 60 s cycles at 6 m s−1; MP Biomedicals, LLC, Solon OH, USA). We quantified the extracted DNA using a Qubit 2.0 fluorometer and diluted it to 5 ng μl−1 for PCR and amplicon sequencing. The ITS region was targeted using the primer + adapter for ITS1F (5′-TCG TCGGCAGCGTCAGATGTGTATAAGAGACAGCTTGGTCATTTAGAGGAAGTAA-3′) and ITS2 (5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGCTGCGTTCTTCAT CGATGC-3′) with Illumina adapters. The PCR reaction had 3.5 μl of DNA, 17.5 μl of ThermoScientificTM PhusionTM Flash High-Fidelity PCR Mastermix (Thermo Fisher Scientific), and 7 μl of each primer (1 ng/μl). PCR products were purified using AMPure XP beads (Beckman Coulter, Pasadena CA, USA) following the Illumina protocol (Part # 15044223 Rev. B, support.illumina.com), and indexed using the Illumina Nextera XT 96 index kit. Samples were pooled, and amplicon size of the library was checked using a Bioanalyzer 2100 (Agilent Technologies). We quantified the library using Q-PCR, and the final library was diluted to 12 pM, spiked with 30% PhiX (Illumina), and ran on an Illumina MiSeq using a 600-cycle v3 cartridge. Resulting sequences were deposited in the NCBI Sequence Read Archive (accession numbers: SAMN14121326-SAMN14121358)
Roots that were stored in 50% ethanol were used for fungal colonization assessment using microscopy. They were cleared by autoclaving in 10% KOH for 15 m, acidified in 1% HCl for 20 m, and stained with 0.05% trypan blue for 2 h to detect if arbuscular mycorrhizas were present together with DSE [49]. Roots were destained overnight in 50% glycerol and stored in lactoglycerol. Percent colonization of DSE was assessed by quantifying melanized DSE hyphae and microsclerotia using the grid intercept method [50], with 100 intersections per slide.
Laboratory and greenhouse experiments
We assessed the salt resistance of the isolated endophytes from the mesohaline site by growing them in PDA with 200, 400, and 600 mM NaCl. We then used all salt tolerant endophytes in a laboratory experiment to evaluate their effect on survival of P. australis under salt stress. Seeds were sterilized with 70% ethanol for 2 m, 10% bleach for 5 m, and three sterile DI water rinses.
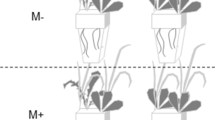
For the laboratory experiment, we germinated surface sterilized seeds of invasive P. australis on 1% agar. After germination, four seedlings were transferred to Magenta boxes containing solid Murashige and Skoog media with 100 mM NaCl. We then added either a disc of actively growing fungi, or a sterile PDA plug as a control next to each seedling. For the less salt-resistant native P. australis lineage, we inoculated the seeds prior to adding them to the Magenta Box because 100 mM of NaCl can induce a stress response for this lineage [28] and we speculated that the endophytes might improve host’s chances of surviving the transplant. We added the sterilized seeds to PDA plates with and without endophytes (Control) and after 24 h transferred them to 1% agar for germination. We then added the seedlings to Magenta boxes containing solid MS agar with 100 mM NaCl and ampicillin. We recorded the number of surviving seedlings of native and invasive P. australis after 2 months. Based on these results, we selected one of the endophytes for a greenhouse experiment to further evaluate its effect on salinity tolerance of invasive Phragmites. In addition, we stained a subset of the seedling roots (as described in the previous section), to confirm fungal colonization by DSE.
For the greenhouse experiment, sterile seeds of invasive P. australis were germinated on 1% agar at 14 h of light and a 30/18 °C diurnal temperature shift. We transferred seedlings into Magenta boxes with half-strength MS basal salt agar and ampicillin. After 3 weeks, we planted 23 seedlings into 2L pots containing a sterile mix of 2:1 Sungro potting soil and sand. Plant height was recorded at the beginning of the experiment and used as a covariate for analysis. One week after planting, we began the endophyte treatment by adding a disc of the selected fungal endophyte that was actively growing on PDA near the base of each plant, or a disc of sterile PDA media for control plants. A week later, we began the salt treatments by adding 100 mM of NaCl to irrigation water and gradually increasing additions by 100 mM weekly until the final treatment levels (Mesohaline = 200 mM and Polyhaline = 400 mM) were reached. This gave us a factorial design with three levels of salinity (Freshwater, Mesohaline, and Polyhaline) and two levels of endophyte (Endophyte and No Endophyte). We placed the pots into aluminum pans to collect drainage water, and plants were watered twice weekly with 1 L of tap water. We added NaCl weekly to the irrigation water to maintain salt treatments, and fertilized the plants biweekly by adding a quarter teaspoon of fertilizer to the water (Jack’s All Purpose 20% total N, 20% P2O5, and 20% K2O). After 2 months, plants were repotted into 4 L pots and we increased watering frequency to 3 times a week. Biweekly measurements included plant height, number of shoots, and salinity of the drainage and reservoir water using a portable salinity meter (YSI, Yellow Springs, Ohio). At the end of the experiment, we measured chlorophyll fluorescence as an indicator of stress using a PAM-2100 Chlorophyll Fluorometer (Walz, Effeltrich Germany) on the second collared leaf of two stems per pot. We recorded the quantum yield (Y) during the day, and the maximum quantum yield (Fv/Fm) at night. After 4 months of plant–fungal symbiosis plants were harvested and we recorded leaf number, leaf area (LI-COR LI-3100), total above and belowground dry biomass, number of shoots, lateral root length and density, and rhizome diameter. Total nitrogen and total carbon of leaf tissue was analyzed by combustion using a LECO CN628 analyzer (LECO, St. Joseph, MI, USA).
Data analyses
We used R v.1.0.153 [51] for all data analysis and figure drawings. Paired end sequences from Illumina were processed using the dada2 package [52] and taxonomy assigned using the UNITE database [53]. The phyloseq [54] and vegan [55] packages were then used for data analysis and ggplot2 [56] for plotting figures. Samples were rarefied to 14,705 sequences which provided overall good coverage based on rarefaction curves (Supplementary Fig. 1). Samples were then filtered based on abundance, and taxa with a prevalence of at least 7% were retained. Nonmetric multidimensional scaling (NMDS) based on a Bray–Curtis dissimilarity matrix was used to visualize endophyte community composition across sites and between lineages. Permutational multivariate ANOVA (PerMANOVA) was used to assess differences between communities, and homogeneity of group dispersion was checked using the vegan functions betadisper and permutest. When PerMANOVA was significant, we used pairwise comparisons to contrast the specific factors using the package RVAideMemoire [57]. Alpha diversity based on log-transformed observed and Fisher’s alpha index was evaluated using ANOVA (type III SS). Differential abundance of taxa between lineages and across sites was evaluated using the R packages Deseq2 [58] and mvabund [59].
Differences in DSE root colonization across dates and between sites for each lineages were assessed using two-way ANOVA (type III SS) and Tukey’s post hoc means comparisons test. Pearson correlation coefficients were calculated to determine the relationship of percent colonization with sampling date and salinity.
Greenhouse results were first analyzed using ANOVA (type III SS) to evaluate if initial height was a significant explanatory variable for each parameter. When it was, the data were analyzed as an ANCOVA using covariate-adjusted means with the package emmeans [60]. Planned pairwise contrasts with a Tukey adjustment were used to assess differences between endophyte treatments at each salinity level. When the initial height was not significant, it was removed from the model and data were analyzed as an ANOVA (type III SS). Variables were log-transformed to meet ANOVA assumptions when necessary.
Results
Site characteristics
In site A, where pH tended to be lower than at sites B and C, percent carbon (%C) and nitrogen (%N) were higher. There were also a few site specific differences between native and invasive stands, specifically the native stand soils had higher %C in sites B and C and overall higher %N at all sites; and invasive stands had higher %SOM in sites B and C. Root morphological characteristics did not differ significantly overall between lineages or across sites (Table 1).
Fungal root endophyte community analyses
After rarefaction and filtering of Illumina sequences there was a total of 165 amplicon sequence variants (ASVs). Only 83 (50%) of the ASVs could be assigned to the Genus level, and 55 (33.5%) to Species. Most fungal ASVs were present in both lineages (71%) and half of them were found at the three sites. The most abundant Orders were Lulworthiales, Agaricales, Pezizales, and Pleosporales (Supplementary Fig. 2), all of which contain taxa that have been identified as DSE [37, 61, 62]. The most abundant Genera within those orders were Lulworthia, Psathyrella, Conlarium, and Anguillospora respectively. Fungal endophytes communities did not differ between June and August (PerMANOVA, F41 = 0.99, p = 0.46), and were therefore combined for the rest of the analyses. When evaluating beta diversity between the sites and lineages, we found a significant interaction between factors (PerMANOVA, F37 = 2.2, p = 0.001). We then ran separate PerMANOVAs for each lineage and site to evaluate changes in community composition across the salinity gradient. Fungal endophytes associated with native P. australis in Site A (~0.7 ppt), differed from those at sites B (~1.2 ppt), and C (~3 ppt) (PerMANOVA, F20 = 2.49, p = 0.002). For invasive P. australis, fungal communities only differed between Site A and Site C (PerMANOVA, F17 = 2.79, p = 0.002). Contiguous stands of native and invasive P. australis had distinct endophyte communities in every site (Site A: F12 = 2.49, p = 0.01; Site B: F14 = 2.69, p = 0.007; Site C: F11 = 2.69, p = 0.02) (Fig. 2 NMDS). Alpha diversity did not differ between lineages or across sites (p > 0.05) (Supplementary Fig. 3). Differential taxa analysis between lineages revealed that only two of the seven differentially abundant fungi could be identified at the Genus level (Fig. 2), two others belonged to the Class Sordariomycetes and the rest could not be resolved beyond the Phylum level (two were Ascomycota and one Basidiomycota).
DSE colonization
Percent DSE colonization was consistent overall throughout the growing season, and root colonization was significantly higher in the invasive than in the native lineage (F147 = 61.49, p < 0.01; Fig. 3a). There was a significant positive correlation between DSE colonization and salinity in the invasive lineage (r = 0.47, n = 82, p < 0.01), but no correlation in the native lineage (r = −0.037, n = 79, p = 0.75) (Fig. 3b). Inundation level had no apparent effect on the observed percent DSE colonization (Fig. 4).
The y-axis shows either the %DSE Colonization or the % inundation (percentage of time when the water was above the soil surface in the 14 days prior to sampling). The lines indicate the % inundation over time in loggers placed in Sites A (top) and C (bottom) in native and invasive Phragmites stands. The circles show the mean %DSE colonization for each lineage at the different sampling times, and the error bars show the standard error of the mean.
Endophyte isolation and Sanger sequencing
We isolated 15 fungal endophytes from invasive P. australis roots, which were categorized as DSE based on microscopic observation of DSE hyphae and microsclerotia in inoculated plant roots. Sequencing output based on the ITS region resulted in 12 contigs that predominantly matched to uncultured fungi (Supplementary Table 1). Based on ITS, only two of the isolates found matches to known cultured fungi—Phialocephala sp. (97.2% match) and Trematosphaeria hydrela (98.1%). Sequencing of the alpha-EF gene did not provide enough resolution for further differentiation of the isolates.
Endophyte salinity tolerance
Fourteen of the isolated endophytes showed growth on PDA with up to 600 mM NaCl (Supplementary Fig. 4); therefore, all of these were tested in Magenta box laboratory assays to evaluate their effect on seedling survival of both lineages. Seedlings of invasive and native P. australis inoculated with endophyte GG2D showed the highest survival relative to the control treatment and other endophytes tested (Table 2). Based on these results, endophyte GG2D was selected to further evaluate its effect on salt tolerance of P. australis in a greenhouse experiment. Taxonomic information on this fungi could not be retrieved from BLAST but it was found to closely match a root endophyte isolated from Persicaria amphibia which is also an aquatic macrophyte (Supplementary Table 1). When grown on PDA, the colony was round and greyish, and grew slowly reaching a diameter of 5.5 cm after 3 weeks at 22 °C. The sterile hyphae measured about 4–7 μm wide and were mostly pigmented, but hyaline hyphae were also observed (Supplementary Fig. 4).
Greenhouse assay
Endophyte inoculation increased aboveground biomass of invasive P. australis only at the mesohaline salinity treatment (ANCOVA, contrast t15 = 2.42, p = 0.029) (Fig. 5a). This was mainly driven by a significant increase in average stem height (ANOVA, F11 = 6.77, p = 0.039) (Fig. 5b) and leaf biomass (ANCOVA, contrast t15 = 2.58, p = 0.021) (Fig. 5c), and an increase in stem biomass (ANCOVA, contrast t15 = 2.1, p = 0.053) at that salinity level in inoculated plants. Other aboveground parameters, including number of stems, leaf count, and leaf area, did not differ significantly between inoculated and non-inoculated plants (Supplementary Table 2). There was also no effect of inoculation on belowground parameters including rhizome and lateral root biomass, rhizome diameter, and lateral root length and number (p > 0.05, Supplementary Table 2); but overall root-to-shoot ratio was lower in endophyte treatments across all salinity levels (p < 0.1) (Fig. 5d).
Bar plots showing means and standard error of the mean for the effects of dark septate endophyte inoculation of invasive P. australis at different salinity levels (Freshwater: no added NaCl, Mesohaline: 200 mM NaCl, and Polyhaline: 400 mM NaCl) on (a) aboveground biomass, (b) average stem height, (c), leaf biomass, and (d) root:shoot ratio. NS indicates p values greater than p = 0.1. Average stem height for freshwater treatments was not recorded because there were sometimes over one hundred per pot making it difficult to get an accurate count.
We also did not observe endophyte or salinity treatment effects on some of the other measured variables. Photosynthetic efficiency based on quantum yield (Y) and maximum quantum efficiency (Fv/Fm) did not differ between salinity treatments (ANOVA, Y: F16 = 1.3, p = 0.29; Fv/Fm: F16 = 0.37, p = 0.69), or between inoculated and not inoculated plants (ANOVA, Y: F16 = 0.1, p = 0.75; Fv/Fm: F16 = 1.02, p = 0.33) (Supplementary Table 2). Similarly, total carbon content in leaf tissue was not affected by salinity or fungal treatments (ANOVA, F15 = 0.28, p = 0.75; F15 = 0.08, p = 0.78; respectively) and total nitrogen did not differ between inoculated and non-inoculated plants (F15 = 0.64, p = 0.44). However, salinity did affect total leaf nitrogen content (ANOVA, F15 = 8.73, p = 0.003), and was highest in mesohaline conditions and lowest in freshwater (Supplementary Table 2).
Discussion
Native and invasive lineages of the common reed P. australis are colonized by distinct fungal endophytes that can improve the salt tolerance of these grasses. We observed an increase in DSE colonization with salinity in the invasive lineage, and greater overall colonization of this lineage relative to the native (Fig. 3a); we speculated this was due to a mutualistic association between DSE and the invasive lineage, likely related to salt tolerance. This warranted further investigation on the potential role of DSE in salinity tolerance of invasive P. australis, which has not been considered a relevant factor to explain P. australis expansion into saline areas so far.
The “habitat adapted hypothesis” [8] suggests that plants may associate with endophytes to improve their tolerance to environmental stress, and these endophytes can confer similar stress tolerance to genetically distant plants. This has been commonly reported for Class II endophytes that can benefit a host under a specific stress, and induce a similar response in closely related hosts [1, 8, 63]. Similarly, in our study Class IV DSE isolated from roots of invasive P. australis improved salt tolerance of both the invasive and native lineage (Table 2). This suggests that DSE mutualisms may be an additional mechanism of salt tolerance for P. australis that might enhance the invasion of the European lineage. On the other hand, these mutualisms could also be useful in restoration of the native lineage if inoculation improves its survival in areas susceptible to saltwater intrusions [28].
DSE associations can range from parasitic to mutualistic, but are predicted to be primarily the latter in plants under abiotic stress [64]. Accordingly, our greenhouse study showed that invasive P. australis did not appear to benefit from inoculation under freshwater conditions; but had higher aboveground biomass at mesohaline salinity (Fig. 5). These results highlight the importance of environmental drivers of plant–fungal symbiosis, as has been seen with mycorrhizae and other endophytic fungi [1, 65]; and can help explain why there are such mixed results in the literature concerning the effects of DSE on their hosts [66, 67].
An increase in photosynthetic efficiency is one of the mechanisms by which DSE could enhance plant tolerance to abiotic stress [68, 69]. In our study DSE inoculation had no effect on quantum yield (Y) or maximum quantum yield (Fv/Fm), and did not affect C or N content in leaves; so growth promotion was likely related to other factors. For example the reduction of reactive oxygen species by fungal melanins could promote plant tolerance to various types of abiotic stress including high salinity [70]; and melanin isolated from a DSE was reported to have high antioxidant activity [71]. Endophyte colonization could also affect plant–water relationships and promote salt tolerance through other mechanisms. In the mycorrhizal fungal symbiosis hyphae can directly uptake water into the plant, induce changes in gene expression relevant to osmotic stress, and increase the production of osmolytes [72]. Although these mechanisms of salt tolerance have not been specifically studied in DSE yet, production of trehalose and mannitol has been reported in these endophytes [2] and accumulation of these osmolytes could reduce salinity stress in colonized plants [70].
Fungal inoculation led to a marginally significant decrease in root:shoot ratio relative to non-inoculated controls (Fig. 5), as inoculated plants generally favored above over belowground growth. This is often observed in mycorrhizal plants where extensive hyphal networks promote water and nutrient uptake [73, 74], but a meta-analysis on DSE found no influence of inoculation on plant root:shoot ratio, and an overall increase in both root and shoot biomass [66]. We only identified the latter in our greenhouse assay and suggest that preferential allocation of C aboveground was not a result of an imposed abiotic stress, but rather a response of the host to inoculation.
In eutrophic wetlands, like our study system, greater aboveground biomass could translate to a significant competitive advantage. Given the abundance of nitrogen [75] and phosphorus in these wetlands [76], belowground competition for these resources is relaxed, and aboveground competition might have a greater role in plant community structure [77, 78]. Other factors to consider would be soil characteristics, disturbance, and the plant’s own physiological adaptations which can be key to determine the outcome of plant competition and contribute to plant invasions in wetlands [20, 28]. Based on our greenhouse findings, we propose DSE could play a role in expansion and establishment of invasive P. australis into brackish marshes by increasing its competitive ability.
Characterizing microbial communities of the native and invasive lineages, and identifying relevant microbial associations can help improve management of P. australis [14, 15]. Our study characterized fungal endophyte communities of contiguous stands of native and invasive P. australis and showed that community composition was lineage and site specific (Fig. 2), even though most taxa were present in both lineages and half of them were found at all sites. These results differ from those reported by Bickford et al. [79] who found no differences in root fungal endophytes between P. australis lineages in the Great Lakes, USA. However, soil saturation was a relevant environmental factor in that study, whereas water level did not appear to play a role in endophyte colonization or community structure in our study sites (Fig. 4). Differential abundance analysis showed that the genera Conlarium and Lulworthia were associated with native and invasive Phragmites respectively (Fig. 2). Conlarium sp. are DSE that have been found to be symbiotic in sugarcane roots [80] and Lulworthia sp. are DSE commonly isolated from marine and coastal systems where they are known to associate with roots of sea grasses and salt marsh plant species [62, 81]; the ecological roles of these endophytes are still unknown.
Dark septate endophyte colonization has been reported to vary seasonally, showing a decrease at the end of the growing season in alpine plant communities and in a tall grass prairie [82, 83]. In our study DSE colonization was prevalent in both lineages throughout the growing season (Fig. 3b), and likely underestimated because hyaline hyphae, which are harder to detect with our staining method, were not quantified [40]. The high prevalence of DSE and lack of evident disease symptoms in colonized plants, suggest a relevant and yet unexplored, role of these endophytes in our study system even at low salinities.
Our study focused on the effects of DSE on salt tolerance, and specifically looked at NaCl as a stress factor; future research could further address the ecological role of DSE in wetland plants by looking at their effects on sulfide tolerance for example. This would be particularly relevant in freshwater wetlands where saltwater intrusion is already affecting coastal biogeochemistry and plant community composition [84, 85]. Increased sulfide concentrations can be toxic for aquatic macrophytes and can limit their growth by reducing nitrogen uptake [86]. Given that DSE might help plants incorporate nitrogen [45, 66], it would be interesting to know if they can improve P. australis’ tolerance to sulfide toxicity. Sulfide could also have a negative effect on seedlings that colonize sites after invasive P. australis removal [87], so beneficial effects for plant restoration using DSE should also be evaluated. In conclusion, P. australis could benefit from DSE colonization when exposed to salt stress. Therefore, the role of fungal mutualists, particularly in a context of sea-level rise, is worth considering in future studies of invasion ecology, species management, and restoration of native plants.
References
Rodriguez RJ Jr, JFW, Arnold AE, Redman RS. Fungal endophytes: diversity and functional roles. New Phytol. 2009;182:314–30.
Usuki F, Narisawa K. A mutualistic symbiosis between a dark septate endophytic fungus, Heteroconium chaetospira, and a nonmycorrhizal plant, Chinese cabbage. Mycologia. 2007;99:175–84.
Yadav V, Kumar M, Deep DK, Kumar H, Sharma R, Tripathi T, et al. A phosphate transporter from the root endophytic fungus Piriformospora indica plays a role in phosphate transport to the host plant. J Biol Chem. 2010;285:26532–44.
Busby PE, Ridout M, Newcombe G. Fungal endophytes: modifiers of plant disease. Plant Mol Biol. 2016;90:645–55.
Clay K. Interactions among fungal endophytes, grasses and herbivores. Res Popul Ecol. 1996;38:191–201.
Omacini M, Chaneton EJ, Ghersa CM, Müller CB. Symbiotic fungal endophytes control insect host-parasite interaction webs. Nature. 2001;409:78–81.
Bultman TL, Bell GD. Interaction between fungal endophytes and environmental stressors influences plant resistance to insects. Oikos. 2003;103:182–90.
Rodriguez RJ, Henson J, Volkenburgh EV, Hoy M, Wright L, Beckwith F, et al. Stress tolerance in plants via habitat-adapted symbiosis. ISME J. 2008;2:404.
Gill SS, Gill R, Trivedi DK, Anjum NA, Sharma KK, Ansari MW, et al. Piriformospora indica: potential and Significance in plant stress tolerance. Front Microbiol. 2016;7:1–20.
Clay K, Holah J. Fungal endophyte symbiosis and plant diversity in successional fields. Science. 1999;285:1742–4.
Afkhami ME, Strauss SY. Native fungal endophytes suppress an exotic dominant and increase plant diversity over small and large spatial scales. Ecology. 2016;97:1159–69.
Uchitel A, Omacini M, Chaneton EJ. Inherited fungal symbionts enhance establishment of an invasive annual grass across successional habitats. Oecologia. 2011;165:465–75.
Aschehoug ET, Metlen KL, Callaway RM, Newcombe G. Fungal endophytes directly increase the competitive effects of an invasive forb. Ecology. 2012;93:3–8.
Dickie IA, Bufford JL, Cobb RC, Desprez‐Loustau M-L, Grelet G, Hulme PE, et al. The emerging science of linked plant–fungal invasions. New Phytol. 2017;215:1314–32.
Kowalski KP, Bacon C, Bickford W, Braun H, Clay K, Leduc-Lapierre M, et al. Advancing the science of microbial symbiosis to support invasive species management: a case study on Phragmites in the Great Lakes. Front Microbiol. 2015;6:1–14.
Martin LJ, Blossey B. The runaway weed: costs and failures of phragmites australis management in the USA. Estuaries Coasts. 2013;36:626–32.
Hazelton ELG, Mozdzer TJ, Burdick DM, Kettenring KM, Whigham DF. Phragmites australis management in the United States: 40 years of methods and outcomes. AoB Plants. 2014;6:1–19.
Farnsworth EJ, Meyerson LA. Species composition and inter-annual dynamics of a freshwater tidal plant community following removal of the invasive grass, Phragmites Australis. Biol Invasions. 1999;1:115–27.
Meyerson LA, Saltonstall K, Windham L, Kiviat E, Findlay S. A comparison of Phragmites Australisin freshwater and brackish marsh environments in North America. Wetl Ecol Manag. 2000;8:89–103.
Silliman BR, Bertness MD. Shoreline development drives invasion of Phragmites Australis and the loss of plant diversity on New England salt marshes. Conserv Biol. 2004;18:1424–34.
Rooth JE, Stevenson JC. Sediment deposition patterns in Phragmites australiscommunities: implications for coastal areas threatened by rising sea-level. Wetl Ecol Manag. 2000;8:173–83.
Mozdzer TJ, Megonigal JP. Increased methane emissions by an introduced Phragmites Australis lineage under global change. Wetlands. 2013;33:609–15.
Bernal B, Megonigal JP, Mozdzer TJ. An invasive wetland grass primes deep soil carbon pools. Glob Change Biol. 2017;23:2104–16.
Rice D, Rooth J, Stevenson JC. Colonization and expansion of Phragmites australis in upper Chesapeake Bay tidal marshes. Wetlands. 2000;20:280.
Packett CR, Chambers RM. Distribution and nutrient status of haplotypes of the marsh grass Phragmites australis along the Rappahannock River in Virginia. Estuaries Coasts. 2006;29:1222–5.
Able KW, Hagan SM, Brown SA. Mechanisms of marsh habitat alteration due toPhragmites: response of young-of-the-year mummichog (Fundulus heteroclitus) to treatment for Phragmites removal. Estuaries. 2003;26:484–94.
Saltonstall K. Cryptic invasion by a non-native genotype of the common reed, Phragmites australis, into North America. Proc Natl Acad Sci USA. 2002;99:2445–9.
Vasquez EA, Glenn EP, Brown JJ, Guntenspergen GR, Nelson SG. Salt tolerance underlies the cryptic invasion of North American salt marshes by an introduced haplotype of the common reed Phragmites australis (Poaceae). Mar Ecol Prog Ser. 2005;298:1–8.
Meadows RE, Saltonstall K. Distribution of native and introduced Phragmites australis in freshwater and oligohaline tidal marshes of the Delmarva peninsula and southern New Jersey1. tbot. 2007;134:99–107.
Glenn EP. Relationship between cation accumulation and water content of salt‐tolerant grasses and a sedge. Plant Cell Environ. 1987;10:205–12.
Lissner J, Schierup H-H. Effects of salinity on the growth of Phragmites australis. Aquat Bot. 1997;55:247–60.
Amsberry L, Baker MA, Ewanchuk PJ, Bertness MD. Clonal integration and the expansion of Phragmites Australis. Ecol Appl. 2000;10:1110–8.
Bart D, Hartman JM. The role of large rhizome dispersal and low salinity windows in the establishment of common reed, phragmites australis, in salt marshes: new links to human activities. Estuaries. 2003;26:436–43.
Soares MA, Li H-Y, Kowalski KP, Bergen M, Torres MS, White JF. Evaluation of the functional roles of fungal endophytes of Phragmites australis from high saline and low saline habitats. Biol Invasions. 2016;18:2689–702.
Weishampel PA, Bedford BL. Wetland dicots and monocots differ in colonization by arbuscular mycorrhizal fungi and dark septate endophytes. Mycorrhiza. 2006;16:495–502.
Kandalepas D, Stevens KJ, Shaffer GP, Platt WJ. How abundant are root-colonizing fungi in Southeastern Louisiana’s degraded marshes? Wetlands. 2010;30:189–99.
Jumpponen A, Trappe JM. Dark septate endophytes: a review of facultative biotrophic root‐colonizing fungi. New Phytol. 1998;140:295–310.
Mandyam K, Jumpponen A. Seeking the elusive function of the root-colonising dark septate endophytic fungi. Stud Mycol. 2005;53:173–89.
Read DJ, Wandter KH. Observations on the mycorrhizal status of some alpine plant communities. New Phytol. 1981;88:341–52.
Barrow J, Aaltonen R. Evaluation of the internal colonization of Atriplex canescens (Pursh) Nutt. roots by dark septate fungi and the influence of host physiological activity. Mycorrhiza. 2001;11:199–205.
Rains KC, Nadkarni NM, Bledsoe CS. Epiphytic and terrestrial mycorrhizas in a lower montane Costa Rican cloud forest. Mycorrhiza. 2003;13:257–64.
Wang J, Li T, Liu G, Smith JM, Zhao Z. Unraveling the role of dark septate endophyte (DSE) colonizing maize (Zea mays) under cadmium stress: physiological, cytological and genic aspects. Sci Rep. 2016;6:1–12.
Li X, He X, Hou L, Ren Y, Wang S, Su F. Dark septate endophytes isolated from a xerophyte plant promote the growth of Ammopiptanthus mongolicus under drought condition. Sci Rep. 2018;8:1–11.
Zhu L, Li T, Wang C, Zhang X, Xu L, Xu R, et al. The effects of dark septate endophyte (DSE) inoculation on tomato seedlings under Zn and Cd stress. Environ Sci Pollut Res. 2018;25:35232–41.
Hill PW, Broughton R, Bougoure J, Havelange W, Newsham KK, Grant H, et al. Angiosperm symbioses with non-mycorrhizal fungal partners enhance N acquisition from ancient organic matter in a warming maritime Antarctic. Ecol Lett. 2019;22:2111–9.
U.S. Fish and Wildlife Service., Cowardin L. Classification of wetlands and deepwater habitats of the United States. Washington D.C.: Fish and Wildlife Service Office of Biological Services U.S. Dept. of the Interior; 1995.
Holdredge C, Bertness MD, Wettberg EV, Silliman BR. Nutrient enrichment enhances hidden differences in phenotype to drive a cryptic plant invasion. Oikos. 2010;119:1776–84.
Ban Y, Tang M, Chen H, Xu Z, Zhang H, Yang Y. The response of dark septate endophytes (DSE) to heavy metals in pure culture. PLoS ONE. 2012;7:1–11.
Burke DJ. Effects of Alliaria petiolata (garlic mustard; Brassicaceae) on mycorrhizal colonization and community structure in three herbaceous plants in a mixed deciduous forest. Am J Bot. 2008;95:1416–25.
McGonigle TP, Miller MH, Evans DG, Fairchild GL, Swan JA. A new method which gives an objective measure of colonization of roots by vesicular-arbuscular mycorrhizal fungi. New Phytol. 1990;115:495–501.
R Core Team. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2018. https://www.R-project.org/.
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–3.
Nilsson R. The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2019;47:D259–64.
McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 2013;8:e61217.
Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, et al. vegan: community ecology Package. 2019. https://CRAN.R-project.org/package=vegan.
Wickham H. ggplot2: elegant graphics for data analysis. New York: Springer-Verlag; 2016. http://ggplot2.org.
Hervé M. RVAideMemoire: testing and plotting procedures for biostatistics. 2019. https://CRAN.R-project.org/package=RVAideMemoire.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550.
Wang Y, Naumann U, Eddelbubettel D, Wilshire J. mvabund: statistical methods for analysing multivariate abundance data. Methods in Ecol Evol. 2012;3:471–4.
Lenth R. emmeans: estimated marginal means, aka least-squares means. 2019. https://CRAN.R-project.org/package=emmeans.
Xie L, Chen Y-L, Long Y-Y, Zhang Y, Liao S-T, Liu B, et al. Three new species of Conlarium from sugarcane rhizosphere in southern China. MycoKeys. 2019;56:1–11.
Vohník M, Borovec O, Župan Ivan, Kolařík M, Sudová R. Fungal root symbionts of the seagrass Posidonia oceanica in the central Adriatic Sea revealed by microscopy, culturing and 454-pyrosequencing. Mar Ecol Prog Ser. 2017;583:107–20.
Redman RS, Kim YO, Woodward CJDA, Greer C, Espino L, Doty SL, et al. Increased fitness of rice plants to abiotic stress via habitat adapted symbiosis: a strategy for mitigating impacts of climate change. PLOS ONE. 2011;6:e14823.
Mandyam KG, Jumpponen A. Mutualism–parasitism paradigm synthesized from results of root-endophyte models. Front Microbiol. 2015;5:1–15.
Hoeksema JD, Chaudhary VB, Gehring CA, Johnson NC, Karst J, Koide RT, et al. A meta‐analysis of context‐dependency in plant response to inoculation with mycorrhizal fungi. Ecol Lett. 2010;13:394–407.
Newsham KK. A meta-analysis of plant responses to dark septate root endophytes. New Phytol. 2011;190:783–93.
Mayerhofer MS, Kernaghan G, Harper KA. The effects of fungal root endophytes on plant growth: a meta-analysis. Mycorrhiza. 2013;23:119–28.
Ban Y, Xu Z, Yang Y, Zhang H, Chen H, Tang M. Effect of dark septate endophytic fungus gaeumannomyces cylindrosporus on plant growth, photosynthesis and Pb tolerance of maize (Zea mays L.). Pedosphere. 2017;27:283–92.
He Y, Yang Z, Li M, Jiang M, Zhan F, Zu Y, et al. Effects of a dark septate endophyte (DSE) on growth, cadmium content, and physiology in maize under cadmium stress. Environ Sci Pollut Res. 2017;24:18494–504.
Qin Y, Druzhinina IS, Pan X, Yuan Z. Microbially mediated plant salt tolerance and microbiome-based solutions for saline agriculture. Biotechnol Adv. 2016;34:1245–59.
Zhan F, He Y, Zu Y, Li T, Zhao Z. Characterization of melanin isolated from a dark septate endophyte (DSE), Exophiala pisciphila. World J Microbiol Biotechnol. 2011;27:2483–9.
Santander C, Aroca R, Ruiz-Lozano JM, Olave J, Cartes P, Borie F, et al. Arbuscular mycorrhiza effects on plant performance under osmotic stress. Mycorrhiza. 2017;27:639–57.
Kothari SK, Marschner H, George E. Effect of VA mycorrhizal fungi and rhizosphere microorganisms on root and shoot morphology, growth and water relations in maize. New Phytol. 1990;116:303–11.
Neumann, E. George, E. Nutrient Uptake: The Arbuscular Mycorrhiza Fungal Symbiosis as a Plant Nutrient Acquisition Strategy. In: Koltai H and Kapulnik Y (eds). Arbuscular Mycorrhizas: Physiology and Function. 2nd edn. (Springer Science+Business Media B.V., 2010) pp 137–67.
Yarwood SA, Baldwin AH, Mateu MG, Buyer JS. Archaeal rhizosphere communities differ between the native and invasive lineages of the wetland plant Phragmites australis (common reed) in a Chesapeake Bay subestuary. Biol Invasions. 2016;18:2717–28.
Dorfman DS, Mabrouk A, Bauer L, Nelson DM, Clement C, Claflin L. Choptank Ecological Assessment: Digital Atlas: Baseline Status Report. NOAA Technical Memorandum NOS NCCOS 213. 2016. Silver Spring, MD. pp 173.
Emery NC, Ewanchuk PJ, Bertness MD. Competition and salt-marsh plant zonation: stress tolerators may be dominant competitors. Ecology. 2001;82:2471–85.
Maurer DA, Zedler JB. Differential invasion of a wetland grass explained by tests of nutrients and light availability on establishment and clonal growth. Oecologia. 2002;131:279–88.
Bickford WA, Goldberg DE, Kowalski KP, Zak DR. Root endophytes and invasiveness: no difference between native and non-native Phragmites in the Great Lakes Region. Ecosphere. 2018;9:1–14.
Xie L, Chen Y-L, Long Y-Y, Zhang Y, Liao S-T, Liu B, et al. Three new species of Conlarium from sugarcane rhizosphere in southern China. MycoKeys. 2019;56:1–11.
Kandalepas D, Blum MJ, Van Bael SA. Shifts in symbiotic endophyte communities of a foundational salt marsh grass following oil exposure from the deepwater horizon oil spill. PLoS ONE. 2015;10:1–18.
Ruotsalainen A, Väre H, Vestberg M. Seasonality of root fungal colonization in low-alpine herbs. Mycorrhiza. 2002;12:29–36.
Mandyam K, Jumpponen A. Seasonal and temporal dynamics of arbuscular mycorrhizal and dark septate endophytic fungi in a tallgrass prairie ecosystem are minimally affected by nitrogen enrichment. Mycorrhiza. 2008;18:145–55.
Herbert ER, Boon P, Burgin AJ, Neubauer SC, Franklin RB, Ardón M, et al. A global perspective on wetland salinization: ecological consequences of a growing threat to freshwater wetlands. Ecosphere. 2015;6:art206.
Tully KL, Weissman D, Wyner WJ, Miller J, Jordan T. Soils in transition: saltwater intrusion alters soil chemistry in agricultural fields. Biogeochemistry. 2019;142:339–56.
Koch MS, Mendelssohn IA, McKee KL. Mechanism for the hydrogen sulfide-induced growth limitation in wetland macrophytes. Limnology and Oceanography. 1990;35:399–408.
Seliskar DM, Smart KE, Higashikubo BT, Gallagher JL. Seedling sulfide sensitivity among plant species colonizing Phragmites-infested wetlands. Wetlands. 2004;24:426–33.
Acknowledgements
We would like to thank the many helpers we had in the laboratory, field, and greenhouse: Zach Berry, Jessie King, Nicku Keshavarz, Zack Bernstein, Henry Wald, Amy Kuritzky, Diane Leason, Eni Baballari, Lindsay Wood, Brian Scott, and Dietrich Epp Schmidt. Also Ryan Kepler for his assistance with phylogenetic analysis of the Sanger sequences. We also wish to thank the three anonymous reviewers that provided constructive comments to improve this paper. Part of this work was supported by the Maryland Native Plant Society.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gonzalez Mateu, M., Baldwin, A.H., Maul, J.E. et al. Dark septate endophyte improves salt tolerance of native and invasive lineages of Phragmites australis. ISME J 14, 1943–1954 (2020). https://doi.org/10.1038/s41396-020-0654-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-020-0654-y
This article is cited by
-
Interplay of biotic and abiotic factors shapes tree seedling growth and root-associated microbial communities
Communications Biology (2024)
-
Endophytic fungi are able to induce tolerance to salt stress in date palm seedlings (Phoenix dactylifera L.)
Brazilian Journal of Microbiology (2024)
-
Towards a Better Understanding of Melanins from Dark Septate Endophytes (DSEs): Their Variability, Synthesis Pathways and Biological Roles
Journal of Soil Science and Plant Nutrition (2024)
-
Polydomus karssenii gen. nov. sp. nov. is a dark septate endophyte with a bifunctional lifestyle parasitising eggs of plant parasitic cyst nematodes (Heterodera spp.)
IMA Fungus (2023)
-
Elucidating the Strategies for Isolation of Endophytic Fungi and Their Functional Attributes for the Regulation of Plant Growth and Resilience to Stress
Journal of Plant Growth Regulation (2023)