Abstract
If gut microbes influence host behavioral ecology in the short term, over evolutionary time, they could drive host niche differentiation. We explored this possibility by comparing the gut microbiota of Madagascar’s folivorous lemurs from Indriidae and Lepilemuridae. Occurring sympatrically in the eastern rainforest, our four, target species have different dietary specializations, including frugo-folivory (sifakas), young-leaf folivory (indri and woolly lemurs), and mature-leaf folivory (sportive lemurs). We collected fecal samples, from 2013 to 2017, and used amplicon sequencing, metagenomic sequencing, and nuclear magnetic resonance spectroscopy, respectively, to integrate analyses of gut microbiome structure and function with analysis of the colonic metabolome. The lemurs harbored species-specific microbiomes, metagenomes, and metabolomes that were tuned to their dietary specializations: Frugo-folivores had greater microbial and metagenomic diversity, and harbored generalist taxa. Mature-leaf folivores had greater individual microbiome variation, and taxa and metabolites putatively involved in cellulolysis. The consortia even differed between related, young-leaf specialists, with indri prioritizing metabolism of fiber and plant secondary compounds, and woolly lemurs prioritizing amino-acid cycling. Specialized gut microbiota and associated gastrointestinal morphologies enable folivores to variably tolerate resource fluctuation and support nutrient extraction from challenging resources (e.g., by metabolizing plant secondary compounds or recalcitrant fibers), perhaps ultimately facilitating host species’ diversity and specialized feeding ecologies.
Similar content being viewed by others
Introduction
Niche differentiation is fundamental to maintaining species diversity because resource partitioning facilitates sympatric coexistence [1]. Dietary partitioning is one axis by which sympatric species avoid competition, and is typically documented by cataloging diets to show minimal overlap [2,3,4], describing the morphological traits that facilitate dietary specialization [5, 6], or experimentally highlighting inter-species dynamics [7]. Gut microbiota—the consortia of microorganisms, including bacteria, archaea, fungi, and protists that inhabit animal gastrointestinal tracts—represent an underappreciated means of partitioning resources. Gut microbes are metabolically versatile and, via their role in digesting substrates otherwise unavailable to hosts, can help animals expand their dietary options. Scavengers, for example, may benefit from the microbial breakdown of carrion [8] that would be otherwise toxic; cichlids harbor algal farms to optimize phototropism [9]; bamboo specialists depend on microbial detoxification of cyanide [10]; and insectivores and krill specialists use bacterial chitinases to degrade exoskeletons and improve protein bioavailability [11,12,13]. Whereas researchers recognize microbially facilitated resource partitioning across host feeding strategies, we lack understanding about the role of gut microbiota in promoting niche differentiation within host feeding strategies.
Nowhere is there greater potential for gut microbiota to facilitate dietary niche differentiation than among folivores or leaf eaters (and herbivores, more generally). Folivores rely on their gut microbes to solve nutritional challenges and, in turn, gut microbes rely on their folivorous hosts to provide the substrates needed for metabolism. Gut microbes metabolize plant secondary compounds [10, 14] that would otherwise inhibit nutrient bioavailability for the host [15, 16]. They also ferment recalcitrant plant fibers into short-chain fatty acids [17] that can account for 30–50% of the host’s daily energy demands [18,19,20]. Comparative studies have highlighted diet-related differences in the composition and function of gut microbiota between folivores and non-folivores [21,22,23] and between related folivores across environments [24, 25]. In allopatric lineages that converged on folivory, gut microbiota better relate to host phylogeny and gut morphology than to dietary niche [21]; nevertheless, for sympatric species that differentiate their dietary niches within folivory, diet-related differences in gut microbiota rarely have been examined [26, 27].
The lemurs of Madagascar include 50 folivorous species that stem from different phylogenetic lineages, occupy different niches, and are routinely sympatric throughout diverse habitats. Madagascar’s extreme seasonal and climatic variability required that species adjust their feeding strategies and dietary selection to cope with fluctuating resources over ecological and evolutionary timescales [28], thereby contributing to the impressive diversification of folivores. Two extant families, Indriidae and Lepilemuridae, contain four genera and 45 species of leaf eaters: The former comprises the sifakas (Propithecus spp.), indri (Indri indri), and woolly lemurs (Avahi spp.); the latter comprises the sportive lemurs or lepilemurids (Lepilemur spp.) (Fig. 1a). Sifakas (3–6 kg) are frugo-folivores; in the rainforest, fruits, seeds, and flowers account for 40–80% of their diet [29, 30]. Indri (6–10 kg) are young-leaf specialists; immature foliage accounts for 50–90% of their diet [30, 31], but indri also consume seasonal fruits, seeds, flowers, and mature leaves. Woolly lemurs (0.6–1.2 kg) are young-leaf specialists; immature foliage accounts for up to 98% of their diet [32]. They will forage on flowers, but rarely eat mature leaves or fruit [33]. Lastly, sportive lemurs (0.5–1 kg) are mature-leaf specialists [34], but, barring woolly lemurs, will readily consume young leaves [35].
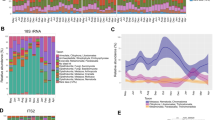
Depicted in a are the following: color-coded photographs of the lemur species, including sifakas (yellow), indri (light green), woolly lemurs (dark green), and sportive lemurs (teal); their respective geographic ranges [81], shown using the same color code, with the red dot pinpointing the study site; illustrations of their dietary composition, with the relative size of fruits, flowers, young (light green) and mature (dark green) leaves corresponding to their relative dietary proportion; and key features of their gastrointestinal morphology, including the cecum (gray), based on previous publications [36, 37, 39]. Depicted in b are pie charts showing lemur gut microbiome composition (i.e., genera that account for ≥1% of sequences, on average, across individuals, in minimally one host species), and a microbial key for which color families refer to microbial phyla and distinct shades refer to unique microbial genera. “Unassigned” refers to microbial genera that could not be phylogenetically assigned via online databases. “Other” refers to the sum of all microbial genera that failed to reach the 1% cutoff. Photographs provided by Nick Garbutt (nickgarbutt.com); illustrations provided by Sally Bornbusch.
Reflecting their dietary niches, these lemurs’ gastrointestinal tracts are similarly specialized (Fig. 1a). Lacking a large foregut, the indriids boast elongated intestines and sacculated ceca that together stretch to >14 times body length [36, 37]. Whereas sifakas have small intestines and ceca that are, respectively, nine and one times body length, indri have small intestines and vascularized ceca that are, respectively, seven and three times body length [30]. The tradeoff between intestinal and cecal capacity facilitates greater sugar and fat absorption from fruits in the sifakas’ long intestines, and greater fiber fermentation from leaves in the indri’s enormous cecum [30]. Woolly lemurs presumably have an intestinal tract that is similar in complexity to that of other indriids; however, their greater reliance on young leaves may be associated with even greater cecal volume than is present in indri. Sportive lemurs, conversely, have a short small intestine, but a voluminous, sac-like cecum that has a total capacity on par with that of woolly lemurs [16, 38, 39].
The gut microbiota of folivorous lemurs are also specialized, comprising taxa and metabolic pathways that are associated with fiber fermentation, short-chain fatty acid production, and plant secondary compound metabolism [22, 23, 40], and are distinct from one another [27, 41]. Across species, phylogenetic distances in gut microbiota are broadly correlated to phylogenetic relationships among their hosts, with little evidence of microbiome convergence between indriids and lepilemurids [41]. Nevertheless, within the indriid lineage, lemurs harbor gut microbiota that are more disparate across species, genera, and habitats than are those of non-folivorous lemurs [41, 42]. The gut microbiota in folivorous lemurs closely track environmental and dietary resource availability [40, 43]. Folivory in lemurs is thus associated with host-specific gut microbiota that are adapted to, and potentially unable to thrive without, their host’s preferred resources; however, comparisons across sympatric species would provide greater resolution for determining microbial contributions specific to resource partitioning.
For four, folivorous species living sympatrically in a Malagasy rainforest, each representing a genus (Table 1), we determined gut microbiome structure and function. We used amplicon sequencing to identify the presence and abundance of bacterial taxa, metagenomic sequencing to determine the metabolic pathways available to gut microbes, and spectroscopy analysis to reveal the colonic metabolome or metabolites excreted by lemurs. This multipronged approach simultaneously illuminates the microbes and metabolic strategies that may differentially contribute to digestion across lemurs. Under the hypothesis that gut microbiota facilitate resource partitioning, we predict that each host species will harbor structurally and functionally distinct microbiomes and metabolomes that reflect their dietary specialization and associated gastrointestinal morphology [27]. For example, sifakas may show greater signatures of fruit metabolism, but reduced signatures of plant secondary compound metabolism; sportive lemurs may show greater capacity for cellulose degradation; and indri and woolly lemurs may be intermediaries between these extremes. If host evolutionary history predominately shapes folivore gut microbiota, we would expect indriids to have similar consortia, distinct from those of sportive lemurs [21, 41]. If habitat occupation predominantly shapes folivore gut microbiota, regardless of dietary niche, we would expect similar gut microbiota across species [42].
Subjects and methods
Subjects
The lemurs included 67 adult or juvenile diademed sifakas (P. diadema), indri (I. indri), eastern woolly lemurs (A. laniger), and weasel sportive lemurs (L. mustelinus) (Table 1). Folivorous lemurs converge on their adult microbiota by 4–6 months of age [22]: The four juveniles in our study were estimated to be past this age threshold. The study populations live sympatrically in the Ambatovy Conservation Zone (Fig. 1a) and are part of a monitoring project for which captures occur biannually [44, 45]. We chose capture timing, split between the late dry (September–October) and late rainy (March) seasons, to minimize risk to pregnant females or neonates. Our procedures were approved by the Institutional Animal Care and Use Committee of Duke University (protocols: A028-14-02; A007-17-01), and by Madagascar’s Ministère de l’Environnement, de l’Ecologie et des Forêts (MEEF/SG/DGF/DAPT/SCBT.Re permits: 197/13; 85/14; 68/15; 38/16; 83/17).
Samples
During six capture missions from 2013 to 2017, we collected 89 fecal samples, directly post voiding or from the rectum using a fecal loop, and placed them at −20 °C within 1 h. We caught 47 subjects only once and recaptured 20 animals at least once, all within a 65 km2 area. We caught representatives of each species in both seasons. We used sterile equipment and consistent methods across lemurs and missions, and maintained storage conditions during transit to Duke University, where samples were stored at −80 °C.
Analytical procedures
Amplicon sequencing, bioinformatics, and statistics
We extracted DNA from feces using QIAGEN’s DNeasy PowerSoil Kit (Hilden, Germany). We shipped 12-µL aliquots (5–50 ng/µL) in two batches to Argonne National Laboratories (Lemont, IL) for amplicon sequencing of the V4 region of the 16S rRNA gene using the 515f-806r primer set, 151 × 151 bp paired-end sequencing, and Illumina’s MiSeq platform. Each batch included representatives from all species during both seasons. Amplicon sequences are available online (NCBI accession PRJNA606765).
We processed sequences using the Quantitative Insights Into Microbial Ecology (QIIME) package (version 2-2019.4; Supplementary material, S1) [46]. We followed the developers’ suggested workflow: We demultiplexed and denoised sequences from each run via DADA2 (i.e., filtered for quality, chimeras, and singletons), using default parameters. DADA2 clusters sequences into “amplicon sequence variants” (ASVs) based on 100% sequence identity. We removed one sample from an indri represented by <10,000 reads. Our final dataset comprised 88 samples and our shallowest depth was 19,679 sequences/sample.
We merged the ASV tables and representative sequences, and assigned taxonomy using the SILVA 132 99% classifier pre-trained for the V4 region of the 16S rRNA gene [47]. We removed chloroplast and mitochondrial sequences. We computed the relative abundance of genera per sample. We determined alpha diversity (using Observed ASVs, Shannon’s H index, and Faith’s Phylogenetic Diversity) and beta diversity (using unweighted and weighted UniFrac distances). We rarefied samples to 19,000 sequences at the time of metric computation.
To analyze alpha diversity, we computed linear mixed models (LMM) using the glmmADMB package (v 0.8.3.3) and R software (v 3.3.3) in RStudio (v 1.1.463) [48,49,50]. We used alpha-diversity metrics as the dependent variables, host species and season nested within species as the explanatory variables, and lemur nested within its social group as a random variable. To analyze beta diversity, we used the vegan package (v 2.4-5) [51] to perform permutational analysis of variance using unweighted and weighted UniFrac distances. We included host species, season nested within species, and individual nested within social group as explanatory variables, and tested that variable order did not influence our results. We reran all diversity analyses using sex as an additional explanatory variable, but failed to find any sex differences. We report findings from the simplified models. We likewise failed to find differences by age class.
We determined if microbial variability between conspecifics differed between species and seasons. We computed pairwise comparisons of UniFrac distances across lemur pairs, excluding comparisons in which the samples derived from the same lemur or from lemurs within the same social group. Using GraphPad Prism (version 8.0.2), we computed Kruskal–Wallis and Dunn’s post-hoc tests of conspecific UniFrac distances between host species, and repeated the analysis separately within and between seasons.
Metagenomic sequencing, bioinformatics, and statistics
We selected 12 samples from sifakas and indri (1 per subject) for metagenomic sequencing (Table 1), because more information is available about these species’ feeding ecology compared with woolly and sportive lemurs. We selected high-quality samples that were split evenly between seasons. We shipped aliquots (>1 µg of total DNA/sample) to the New York Genome Center for sequencing [23]. All samples underwent TruSeq PCR-Free preparation and were sequenced in one lane of Illumina’s HiSeq 2500 platform, targeting 2 × 125 bp reads. We generated minimally 20 million reads/sample. Metagenomic sequences are available online (NCBI accession PRJNA606765).
Sequences were processed via established methods [23]. Sequences were filtered for host DNA (0.1–0.6% of sequences/sample) by comparison to the genomes of the Coquerel’s sifaka (P. coquereli: NCBI accession PRJNA281642) and gray mouse lemur (Microcebus murinus: NCBI accession PRJNA19967) and for plant sequences from viridiplantae (0.2–0.4% of sequences/sample). We determined functional composition using HUMAnN2 (v0.5.0) [52] and the UniRef database, which provide the relative abundance of metabolic pathways based on the abundance of the genes catalyzing each pathway’s component reactions. We retained for analysis metabolic pathways that were present in minimally two samples.
Our statistical analyses of metabolic pathways mirrored those applied to ASVs. We calculated the number of pathways per sample, and computed a LMM, using species and season nested within species as explanatory variables, and social group as a random term. We calculated Bray–Curtis dissimilarity scores in pathway profiles across sample pairs, and computed Wilcoxon tests to assess if metagenomic variation differed within and between species. We performed permutational analysis of variance using Bray–Curtis metrics, using host species and season nested within species as explanatory variables. We used Linear discriminant analysis Effect Size (LEfSe) [53] with the Benjamini–Hochberg correction factor [54] to determine which pathways were enriched in the species’ metagenomes.
Metabolomic profiling and statistics
We selected 36 samples across species (Table 1) for colonic metabolomics profiling via nuclear magnetic resonance (NMR) spectroscopy [23, 43]. We mixed feces (~0.05 g) with 600 µL PBS prepared in D2O, pH 7.5. We centrifuged homogenized samples through 0.22 µm and 30 kDa MWCO filters. We added 60 µL of an internal standard (500 µM 3-(trimethylsilyl)-1-propanesulfonic acid-d6, sodium salt (DSS-d6)), and stored samples at −20 °C until analysis. We acquired spectral data using a 700 MHz NMR spectrometer and a standard 1D-NOESY Preset experiment. We analyzed the data using the Advanced Chemistry Development Spectrus Processor software (v 2016.1) and the Chenomex NMR Suite (v 8.2). We determined metabolite concentrations by quantitative fitting of each spectrum. The final concentrations were normalized based on sample weight. The values for total phenolic compounds were determined by spectral integration over the region from 6.8 to 8.5 ppm. Samples from one woolly lemur and one sportive lemur that were contaminated with propylene glycol were removed from analysis: Our final sample size for metabolomics profiling was 34 samples.
We performed a principle component analysis (PCA) in JMP (v 14.0.) on the identified metabolites. We retained the top four principle components (PCs) with eigenvalues >1 that accounted for >1% of the variation. We used these PCs as variables in a linear discriminant analysis (LDA) to test for species differences. We repeated PCAs/LDAs within each season. We computed LMMs for a subset of metabolites, including the short-chain fatty acids, branched-chain amino acids, and plant secondary compound derivatives. We entered metabolites as the dependent variables, host species and season as the explanatory variables, and social group as a random term. The few samples analyzed from sportive lemurs precluded testing for seasonal differences in metabolite concentrations within species. Metabolite concentrations are presented in the Supplementary material, S2.
Results
Gut microbiome structure
Each lemur species harbored a structurally unique gut microbiome (Fig. 1b). The consortia of sportive lemurs were the most distinct; those of indri and woolly lemurs were the most similar (Supplementary material, S3). The lemurs’ consortia were dominated by the Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria phyla. Bacteroidetes were most abundant in sifakas (62.8%), intermediately abundant in indri (47.7%) and woolly lemurs (38.8%), and least abundant in sportive lemurs (14.1%). In contrast, Firmicutes were most abundant in sportive lemurs (60.0%), intermediately abundant in woolly lemurs (41.4%), and least abundant in sifakas (28.2%) and indri (19.7%). Indri had the greatest proportion of Proteobacteria (20.5%), whereas sportive lemurs had the greatest proportion of Actinobacteria (15.1%).
Thirty-five microbial genera accounted for ≥1% of the gut microbiome in minimally one host species. Two of these taxa—unassigned genera within the Prevotellaceae and the Lachnospiraceae families—were present in large proportions (i.e., ≥1%) in all host species. Otherwise, sifaka microbiomes were dominated by the unassigned Prevotellaceae genus (17.9%) and another genus within this family (UCG-001; 16.1%); indri microbiomes were dominated by the unassigned Prevotellaceae genus (24.4%), Prevotella 1 (18.1%), and Anaerobiospirillum from the Succinivibrionaceae family (18.6%); woolly lemur microbiomes were dominated by a genus within the Erysipelotrichaceae family (UCG-004; 24.7%) and Bacteroides (15.3%); and sportive lemur microbiomes were dominated by the unassigned Lachnospiraceae genus (33.8%), Oribacterium (17.2%), and Enterorhabdus from the Eggerthellaceae family (14.8%).
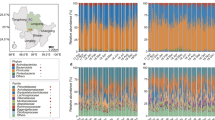
Sifaka gut microbiomes had the greatest alpha diversity, as revealed by Observed ASVs (LMM: all zs > 14.58, all ps < 0.001; Fig. 2a), Shannon’s H index (LMM: all zs > 11.52, all ps < 0.001; Fig. 2b) and Faith’s Phylogenetic Diversity (LMM: all zs > 11.86, all ps < 0.001; Fig. 2c). Relative to the dry season, the rainy season was associated with more Observed ASVs in sifakas (LMM: z = 2.62, p = 0.009), greater Shannon’s H index scores in sportive lemurs (LMM: z = 2.68, p = 0.007), and potentially greater Faith’s Phylogenetic Diversity scores in indri (LMM: z = 1.82, p = 0.068).
Depicted are measures of alpha diversity, including a Observed ASVs, b Shannon’s H index, and c Faith’s Phylogenetic Diversity, and measures of beta diversity, including unweighted UniFrac distances graphed (d–f) in Principle Coordinate (PC) space and g as pairwise comparisons across individuals within species. ***p < 0.001.
Analysis of beta diversity confirmed that microbiome structure varied by species, as captured by unweighted (PERMANOVA: R2 = 0.97, df = 3, p < 0.001; Fig. 2d–f) and weighted (PERMANOVA: R2 = 0.96; df = 3, p < 0.001; Supplementary material, S3) UniFrac distances. Within host species, season was associated with unweighted (PERMANOVA: R2 = 0.002, df = 3, p = 0.012), but not weighted (PERMANOVA: R2 = 0.002, df = 3, p = 0.249), UniFrac distances. Overall, season explained 0.2% of the variation in our dataset. Within-species variability differed between host species (unweighted UniFrac: Kruskal–Wallis H = 345.9, p < 0.001; Fig. 2g; weighted UniFrac: Kruskal–Wallis H = 230.8, p < 0.001; Supplementary material, S3): Post-hoc tests of unweighted UniFrac distances showed that, across individuals, sifakas harbored the most homogenized consortia (all ps < 0.001), whereas sportive lemurs harbored the most heterogeneous consortia (all ps < 0.007). Findings related to within-species differences between species held true when pairwise comparisons were examined exclusively within or between seasons (Supplementary material, S3), such that variable seasonal differences between species could not explain the results.
Gut microbiome function
We identified 578 metabolic pathways in the metagenomes of sifakas and indri. Compared to indri, the sifakas’ metagenomes had more pathways (LMM: z = 6.44, p < 0.001; Fig. 3a). For indri, more pathways were associated with the dry season than the rainy season (LMM: z = 4.09, p < 0.001), but sifakas showed no seasonal differences in pathway numbers (LMM: z = 1.26, p = 0.21). Analysis of Bray–Curtis distances indicated an effect of species (PERMANOVA: R2 = 0.45, df = 1, p = 0.001; Fig. 3b) and season within species (PERMANOVA: R2 = 0.26, df = 2, p = 0.027) on pathway composition. Wilcoxon tests of pairwise comparisons confirmed that there was greater metagenomic variability between sifaka-indri pairs than between paired conspecifics (sifakas: U = 229, p < 0.001; indri: U = 12, p < 0.001; Fig. 3c, see Supplementary material S4).
Depicted are measures of a alpha diversity, specifically the number of metabolic pathways, and b,c beta diversity, including Bray–Curtis dissimilarity scores graphed b in a multidimensional scaling plot and c as pairwise comparisons across individuals within and between species. d–f The abundances of individual metabolic pathways illustrate species’ tradeoffs in the microbial d metabolism of plant sugars, e fermentation, and f degradation of plant secondary compounds. Specific pathway names refer to those in the metaCyc database. *p < 0.05; ***p < 0.001.
LEfSe identified 145 pathways that were differentially enriched in the species’ metagenomes, representing tradeoffs in fiber and sugar metabolism, short-chain fatty acid production, and plant secondary compound degradation. We present several illustrative results: First, the sifakas showed enrichment for one rhamnose-degradation pathway (RhamCat pwy), whereas the indri showed enrichment for seven galactose-degradation pathways, including the Leloir pathway (Log(LDA) > 2.58, ps < 0.05 for both comparisons; Fig. 3d). Second, the sifakas showed enrichment for heterolactic fermentation, whereas the indri showed enrichment for mixed-acid fermentation (Log(LDA) > 2.15, ps < 0.05 for both comparisons; Fig. 3e). Third, the sifakas lacked enrichment for any pathways associated with plant secondary compound metabolism, whereas the indri showed enrichment for 11 aromatic-compound degradation pathways, e.g., p-cymeme (pwy-5226) and cinnamate (pwy-6690) degradation (Log(LDA) > 2.78, ps < 0.05 for both comparisons; Fig. 3f). Lastly, there were other metagenomic differences between the species, including tradeoffs in vitamin, amino acid, and fatty-acid cycling, and enrichment for the citric acid cycle in indri (Supplementary material, S5).
Colonic metabolome
We identified 18 metabolites in the lemurs’ feces. PC/LDA revealed that metabolome composition varied by species (Wilks’ λ = 0.22, p < 0.001, Fig. 4a); however, seven of 34 samples were misclassified, with analyses misassigning woolly lemurs and sportive lemurs or misassigning sifakas and indri. Repeating the analyses within seasons confirmed species differences in both seasons (Supplementary material, S6).
Depicted are a a Linear Discriminant plot of the principal components calculated across all species and metabolites. b–j The concentrations of specific metabolites illustrate species’ tradeoffs in the colonic abundances of short-chain fatty acids, including b acetate, c propionate, and d butyrate, of branched chain amino acids, including e isoleucine, f leucine, and g valine, of plant secondary compound (PSC) derivatives, including h total phenolic abundance and i phloretic acid, and of j trimethylamine. §p < 0.10; *p < 0.05; **p < 0.01; ***p < 0.001.
In general, indri and sifakas had the greatest metabolomic concentrations of short-chain fatty acids (Supplementary material, S6), including acetate (indri > sifakas, LMM: z = 2.34, p = 0.019; indri, sifakas, or sportive lemurs > woolly lemurs, LMM: all zs > 2.20, all ps < 0.03; Fig. 4b), propionate (indri or sifakas > woolly lemurs or sportive lemurs, LMM: all zs > 2.30, all ps < 0.03; Fig. 4c), and butyrate (indri > other species, LMM: all zs > 2.70, all ps < 0.007; sifakas > woolly lemurs, LMM: z = 3.32, p = 0.009; Fig. 4d). In contrast, woolly lemurs and sportive lemurs generally had the greatest metabolomic concentrations of branched-chain amino acids (Supplementary material, S6), including isoleucine (sportive lemurs > other species, LMM: all zs > 2.87, all ps < 0.004; Fig. 4e), leucine (woolly lemurs or sportive lemurs > sifakas or indri, LMM: all zs > 2.13, all ps < 0.04; Fig. 4f), and valine (woolly lemurs > sifakas or indri, LMM: both zs > 2.0, both ps < 0.05; Fig. 4g).
Again, indri and sifakas generally had the greatest metabolomic concentrations of plant secondary compound derivatives (Supplementary material, S6), including total phenolic compounds (sifakas or indri > woolly lemurs or sportive lemurs, LMM: all zs > 3.08, all ps < 0.002; indri ≥ sifakas, LMM: z = 1.67, p = 0.094; Fig. 4h) and phloretic acid (indri > woolly lemurs, LMM: z = 2.78, p = 0.006; indri ≥ sportive lemurs, LMM: z = 1.82, p = 0.069; indri ≥ sifakas, LMM: z = 1.95, p = 0.051; Fig. 4i). Lastly, trimethylamine was present in most samples from sifakas and sportive lemurs, but was minimally present in samples from indri and woolly lemurs (Fig. 4j). Ethanol was variably present in the metabolomes of all species, but displayed a 5.83–34.65 fold increase, on average, in the sifakas’ metabolome compared to the metabolomes of the other species.
Discussion
By comparing gut microbiota across folivorous lemurs, we show that ecologically similar, sympatric hosts harbor strikingly different gut microbiomes, metagenomes, and metabolomes, in a manner that reflects the hosts’ dietary niches and associated gastrointestinal morphologies. The frugo-folivores had the most diverse microbial communities, but showed the least individual variability. Their microbiota were enriched for bacteria, metabolic pathways, and metabolites associated with simple-fiber metabolism. Conversely, the consortia of mature-leaf specialists were simple in structure and enriched for taxa and metabolites putatively associated with cellulolysis. The gut microbiota of the two young-leaf specialists appeared intermediate between these extremes, but were nevertheless distinct: Indri showed a strong reliance on microbially facilitated fiber and plant secondary compound degradation, whereas woolly lemurs may prioritize amino acid cycling. We suggest that the gut microbiota of these species differentially help their hosts withstand ingested fibers or plant secondary compounds, balance fruit and foliage fermentation, or cope with fluctuating resources in hypervariable environments (see [28]). Our results support the proposal that animal microbiota can influence host behavior [55], in this case, facilitating different folivorous strategies in sympatric species. Such findings illuminate how microbes can promote the evolution of folivory and the maintenance of macro-species diversity.
The frugo-folivorous sifakas had gut microbiota that reflected their metabolic balance of fruit and foliage fermentation. The dominant bacteria in their consortia included non-cellulolytic fiber specialists, like Prevotellaceae members [56], fat metabolizers, like Rikenellaceae members [57], and complex-fiber specialists from the Clostridiales order [58] and Lachnospiraceae family [59]. The greater alpha diversity in the sifakas’ microbiota indicate that fruit consumption is associated with a relatively even distribution of generalist taxa common to all hosts [27, 42]. Whereas the greater number of metabolic pathways in the sifakas’ metagenomes, compared with those of indri, perhaps facilitates the sifakas’ more varied diet, the similar concentrations of short-chain fatty acids excreted by sifakas and indri perhaps point to a shared reliance on microbial fermentation of diverse substrates [60]. Although fruit consumption has clear energetic advantages, the ethanol and trimethylamine in the sifakas’ metabolomes, respectively associated with fruit over-ripening [61] and microbial decomposition [62], may point to the negative consequences of sugar fermentation occurring in a gastrointestinal system that is optimized for foliage digestion during lean seasons.
Indri, as young-leaf specialists, harbored large proportions of Prevotellaceae, and Clostridiales members from the Ruminococcaceae family, that likely reflect host reliance on fruits, flowers, and young leaves. Nevertheless, their gut microbiota also comprised abundant Proteobacteria, from the Anaerobiospirillum genus and Gammaproteobacteria class. The function of these Proteobacteria requires explanation, because they are typically associated with oxygen tolerance [63] and shorter gastrointestinal systems [42], and only rarely with folivory [26]. The metagenomic and metabolomic results indicate that these taxa may play a role in fiber metabolism: Anaerobiospirillum can use galactose [64], a sugar common in non-cellulolytic plant fibers, for energy. The metagenomes of indri (compared with sifakas) were enriched for pathways associated with galactose catabolism. Indri metagenomes were likewise enriched for mixed-acid fermentation pathways, reflecting a strategy used by Gammaproteobacteria to produce the short-chain fatty acid, acetate [65]. Acetate was more concentrated in the metabolomes of indri than in the metabolomes of the other indriids. These results may indicate that indri rely on the fibrolytic capacity of specialized Proteobacteria.
Compared with the other species, indri had signatures of greater plant secondary compound degradation. Both the sifakas and indri had relatively raised concentrations of total phenolic compounds, a finding that is consistent with diets similarly rich in secondary compounds [30]. Because leaf tannins in Malagasy rainforests tend to be most concentrated in the late afternoon [66], perhaps the diurnal indri and sifakas ingest more plant secondary compounds than do the nocturnal species. Yet, for indri, the abundance of metabolomic phloretic acid is revelatory about the dynamics between plant secondary compounds and gut microbes. Phloretic acid is produced by bacteria during the detoxification of p-coumaric acid [67], a hydroxycinnamic acid common in plant tissues [68]. Whereas Ruminococcaceae members degrade p-coumaric acid into phloretic acid [67, 69], Gammaproteobacteria also use hydroxycinnamic acids for energy [70]. Crucially, the metagenomes of indri, compared with those of sifakas, were enriched for one metabolic pathway that Gammaproteobacteria use to degrade cinnamates and hydroxycinnamates [70]. Indri may rely simultaneously on Firmicutes and Proteobacteria for fiber and plant secondary compound metabolism, suggesting functional redundancy to ensure that, even following the extinction of one phylum, the lemurs’ folivorous diets could remain sustainable.
Despite sharing dietary specialization and gastrointestinal morphology with indri, the woolly lemurs had distinct gut microbiota, characterized by different fibrolytic bacteria, like Fibrobacter [71], different Gammaproteobacteria, like Succinatimonas and Eschericia-Shigella, and by bacteria associated with fats and proteins, like Bacteroides and Erysipelotrichaceae members [72, 73]. Compared with indri and sifakas, woolly lemurs perhaps prioritize plant protein metabolism. This suggestion is supported by the lower metabolome concentrations of short-chain fatty acids, but greater concentrations of branched-chain amino acids, which are essential nutrients that must be acquired by consumption or microbial action. The nocturnal woolly lemurs may be particularly reliant on gut microbes for amino acid synthesis, because leaf-protein concentrations tend to wane overnight in Malagasy rainforests [66]. The similarly nocturnal sportive lemurs excreted concentrated branched-chain amino acids, raising the possibility that niche differentiation via activity pattern might influence the relationship between host diet and microbially facilitated nutrition. Alternately, perhaps woolly lemurs better absorb enteric short-chain fatty acids, leading to their reduced excretion. Future research could clarify how the gastrointestinal tract of woolly lemurs compares with those of other indriids, and potentially either limits short-chain fatty acid production or promotes their absorption.
The gut microbiota of sportive lemurs, the mature-leaf specialists, were simplest in structure, but most variable across individuals. Perhaps a diet of tough foliage and a simple gastrointestinal system are associated with a relatively uneven distribution of a few, specialist taxa. Overall, sportive lemurs had consortia that appeared to be optimized for microbial cellulolysis. Their gut microbiota were dominated by two Clostridiales taxa from the Lachnospiraceae family that together comprised >50% of sequences. These bacteria are known for their cellulolytic capacity [58]. The concentrated acetate, but limited propionate and butyrate, in the sportive lemurs’ metabolomes may reflect cellulose fermentation [74]. The third most abundant bacterial taxon in the sportive lemurs’ consortia, Enterorhabdus from the Eggerthellaceae family, is perhaps implicated in plant secondary compound metabolism. Some Eggerthellaceae taxa produce equol [75], a metabolite synthesized during the breakdown of the flavonoid daidzein [76]. Flavonoids are thought to promote host health, potentially by regulating gut microbiota [77]. Daidzein is produced almost exclusively by Fabaceae members, a plant family well represented in Madagascar [78]. Perhaps sportive lemurs preferentially feed on Fabaceae leaves and rely on Eggerthellaceae members for secondary compound detoxification. Continued study of medicinal plants and microbial metabolism by Madagascar’s folivores could have biomedical potential [79].
Alternately, the variability of the sportive lemurs’ gut microbiota may owe to their feeding flexibility. In the absence of competitive woolly lemurs [35], these mature-leaf specialists adopt a diet rich in immature foliage. It would be interesting to compare the gut microbiota of woolly lemurs and sportive lemurs throughout their extant ranges, including when living in sympatry versus in allopatry. It would likewise be interesting to compare gut microbiota in sifakas, woolly lemurs, and sportive lemurs living sympatrically in dry forests, where they experience greater seasonality than do their peers in the rainforest (see [27]). In other systems, correlational evidence for microbially facilitated resource partitioning could derive from comparison across sympatric hosts that depend on microbial metabolism for digestion. Experimental evidence will be critical, as our methods did not permit tracking the metabolism of dietary items by specific microbes, nor did they fully differentiate the metabolic action of hosts from that of their microbes. Necessary evidence might be obtained using germ-free models that could be colonized by the consortia of diverse hosts and exposed to variable diets [80]. Such studies would further establish that microbes provide hosts with the enzymatic potential for extracting nutrients from an array of resources and for adapting to variable environmental circumstances.
Previous work, focused across primate lineages evolving under different conditions and on different continents, has shown that convergence on folivory does not underlie convergence on a singular gut microbiome [21, 41]. Instead, host phylogenetic and morphological constraints better underlie microbial composition, especially when examined at broad scales. We have shown that related species of folivorous lemurs, characterized by different gastrointestinal morphologies, yet living sympatrically, each depend on the metabolic capacity of a unique set of microbes, some of which are highly specialized. We therefore suggest that folivory encompasses a greater diversity of feeding strategies than is generally recognized and that, within the constraints imposed by host phylogenetic placement, gut microbes can play a role in facilitating finer grained resource partitioning. The dietary plant variation experienced by folivores across niches, habitats, and continents have seemingly shaped host-microbiota coadaptation in lineage-specific ways [21, 42], such that, over evolutionary time, hosts and their microbes have become equally specialized and inter-dependent. That these lemurs’ specialized, host-specific diets were mirrored by equally specialized, host-specific gut microbiota may have conservation implications. Among Indriidae and Lepilemuridae, 82% of species are listed as endangered or critically endangered [81]. Although numerous factors contribute to extinction [82, 83], host-microbiota co-specialization might underlie a reduced resilience to dietary perturbation and a limited ability to buffer the consequences of the Anthropocene [84, 85].
References
Schoener TW. Resource partitioning in ecological communities. Science 1974;185:27–39.
De León LF, Podos J, Gardezi T, Herrel A, Hendry AP. Darwin’s finches and their diet niches: the sympatric coexistence of imperfect generalists. J Evol Biol. 2014;27:1093–104.
Kartzinel TR, Chen PA, Coverdale TC, Erickson DL, Kress WJ, Kuzmina ML, et al. DNA metabarcoding illuminates dietary niche partitioning by African large herbivores. Proc Natl Acad Sci. 2015;112:8019–24.
Winemiller KO. Ontogenetic diet shifts and resource partitioning among piscivorous fishes in the Venezuelan ilanos. Environ Biol Fish. 1989;26:177–99.
Lack D. Darwin’s finches. Cambridge: Cambridge University Press; 1947.
Pöysä H. Morphology-mediated niche organization in a guild of dabbling ducks. Ornis Scand. 1983;14:317–26.
Inouye DW. Resource partitioning in bumblebees: experimental studies of foraging behavior. Ecology 1978;59:672–8.
Roggenbuck M, Schnell IB, Blom N, Bælum J, Bertelsen F, Sicheritz-Pontén T, et al. The microbiome of New World vultures. Nat Commun. 2014;5:5498.
Hata H, Tanabe AS, Yamamoto S, Toju H, Kohda M, Hori M. Diet disparity among sympatric herbivorous cichlids in the same ectomorphs in Lake Tanganyika: amplicon pyrosequences on algal farms and stomach contents. BMC Biol. 2014;12:90.
Zhu L, Yang Z, Yao R, Xu L, Chen H, Gu X, et al. Potential mechanism of detoxification of cyanide compounds by gut microbiomes of bamboo-eating pandas. mSphere 2018;3:e00229–18.
Delsuc F, Metcalf JL, Parfrey LW, Song SJ, González A, Knight R. Convergence of gut microbiomes in myrmecophagous mammals. Mol Ecol. 2014;23:1301–17.
Mårtensson P-E, Nordøy ES, Blix AS. Digestibility of krill (Euphausia superba and Thysanoessa sp.) in minke whales (Balaenoptera acutorostrata) and crabeater seals (Lobodon carcinophagus). Br J Nutr. 1994;72:713–6.
Whitaker JO, Dannelly HK, Prentice DA. Chitinase in insectivorous bats. J Mammol. 2004;85:15–8.
Dearing DM, Kohl KD. Beyond fermentation: other important services provided to endothermic herbivores by their gut microbiota. Integr Comp Biol. 2017;57:723–31.
Iason G. The role of plant secondary metabolites in mammalian herbivory: ecological perspectives. Proc Nutr Soc. 2005;64:123–31.
Lambert JE. Primate digestion: interactions among anatomy, physiology, and feeding ecology. Evol Anthropol. 1998;7:8–20.
Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat Rev Microbiol. 2008;6:121–31.
Barboza PS, Hume ID. Hindgut fermentation in the wombats: two marsupial grazers. J Comp Physiol B. 1992;162:561–6.
Clayton JB, Gomez A, Amato K, Knights D, Travis DA, Blekhman R, et al. The gut microbiome of nonhuman primates: lessons in ecology and evolution. Am J Primatol. 2018;80:e22867.
Popovich DG, Jenkins D, Kendall C, Dierenfeld ES, Carroll RW, Tariq N, et al. The western lowland gorilla diet has implications for the health of humans and other hominoids. J Nutr. 1997;127:2000–5.
Amato KR, Sanders JG, Song SJ, Nute M, Metcalf JL, Thompson LR, et al. Evolutionary trends in host physiology outweigh dietary niche in structuring primate gut microbiomes. ISME J. 2019;13:576–87.
McKenney EA, Rodrigo A, Yoder AD. Patterns of gut bacterial colonization in three primate species. PloS ONE 2015;10:e0124618.
McKenney E, O’Connell TM, Rodrigo A, Yoder AD. Feeding strategy shapes gut metagenomic enrichment and functional specialization in captive lemurs. Gut Microbes 2017;9:202–17.
Amato KR, Martinez-Mota R, Righini N, Raguet-Schofield M, Corcione FP, Marini E, et al. Phylogenetic and ecological factors impact the gut microbiota of two Neotropical primate species. Oecologia 2016;180:717–33.
Gomez A, Rothman JM, Petrzelkova K, Yeoman CJ, Vlckova K, Umaña JD, et al. Temporal variation selects for diet-microbe co-metabolic traits in the gut of Gorilla spp. ISME J. 2016;10:514–26.
Dill-McFarland KA, Weimer PJ, Pauli JN, Peery MZ, Suen G. Diet specialization selects for an unusual and simplified gut microbiota in two- and three-toed sloths. Environ Microbiol. 2016;16:1391–402.
Perofsky AC, Lewis RJ, Meyers LA. Terrestriality and bacterial transfer: a comparative study of gut microbiomes in sympatric Malagasy mammals. ISME J. 2019;13:50–63.
Dewar RE, Richard AF. Evolution in the hypervariable environment of Madagascar. Proc Natl Acad Sci. 2007;104:13723–7.
Irwin MT. Ecologically enigmatic lemurs: the sifakas of the eastern forests (Propithecus candidus, P. diadema, P. edwardsi, P. perrieri and P. tattersalli). In: Gould L, Sauther M, editors. Lemurs: ecology and adaptation. New York: Springer; 2006. p. 305–26.
Powyzk JA, Mowry CB. Dietary and feeding differences between sympatric Propithecus diadema diadema and Indri indri. Int J Primatol. 2003;24:1143–62.
Britt A, Randriamandratonirina NJ, Glasscock KD, Iambana BR. Diet and feeding behavior of Indri indri in a low-altitude rain forest. Folia Primatol. 2002;73:225–39.
Faulkner AL, Lehman SR. Feeding patterns in a small-bodied nocturnal folivore (Avahi laniger) and the influence of leaf chemistry: a preliminary study. Folia Primatol. 2006;77:218–27.
Ganzhorn JU, Abraham JP, Razanahoera-Rakotomalala M. Some aspects of the natural history and food selection of Avahi laniger. Primates 1985;26:452–63.
Hladik CM, Charles-Dominique P. The behavior and ecology of the sportive lemur (Lepilemur mustelinus) in relation to its dietary peculiarities. In: Chivers D, Herbert J, editors. Prosimian biology. London: Duckworth; 1974. p. 23–7.
Ganzhorn JU. Flexibility and constraints of Lepilemur ecology. In: Kappeler PM, Ganzhorn JU, editors. Lemur social systems and their ecological basis. New York: Plenum Press; 1993. p. 153–65.
Campbell JL, Eisemann JH, Williams CV, Glenn KM. Description of the gastrointestinal tract of five lemur species: Propithecus tattersalli, Propithecus verreauxi coquereli, Varecia variegata, Hapalemur griseus, and Lemur catta. Am J Primatol. 2000;52:133–42.
Milne-Edwards A, Grandidier A. Histoire naturelle des mammifères, Tome I, Texte I. In: Grandidier A, editor. Histoire physique, naturelle et politique de Madagascar, Vol. 6. Les indrisinés, Imprimerie Nationale: Paris; 1875.
Charles-Dominique P, Hladik CM. Le Lepilemur du sud de Madagscar: ecologie, alimentation et vie sociale. La Terre et la Vie. 1971;25:3–66.
Chivers DJ, Hladik CM. Morphology of the gastrointestinal tract in primates: comparisons with other mammals in relation to diet. J Morphol. 1980;166:337–86.
Springer A, Fichtel C, Al‐Ghalith GA, Koch F, Amato KR, Clayton JB, et al. Patterns of seasonality and group membership characterize the gut microbiota in a longitudinal study of wild Verreaux’s sifakas (Propithecus verreauxi). Ecol Evol. 2017;7:5732–45.
Greene LK, Bornbusch SL, McKenney EA, Harris RL, Gorvetzian SR, Yoder AD, et al. The importance of scale in comparative microbiome research: new insights from the gut and glands of captive and wild lemurs. Am J Primatol. 2019;81:e22974.
Greene LK, Clayton JB, Rothman RS, Semel BP, Semel MA, Gillespie TR, et al. Local habitat, not phylogenetic relatedness, predicts gut microbiome structure within frugivorous and folivorous lemur lineages. Biol Lett. 2019;15:20190028.
Greene LK, McKenney EA, O’Connell TM, Drea CM. The critical role of dietary foliage in maintaining the gut microbiome and metabolome of folivorous sifakas. Sci Rep. 2018;8:14482.
Dickinson S, Berner PO. Ambatovy project: Mining in a challenging biodiversity setting in Madagascar. In: Goodman SM, Mass V, editors. Biodiversity, exploration, and conservation of the natural habitats associated with the Ambatovy project. Malagasy nature. 2010;3 p. 2–13.
Junge RE, Williams CV, Rakotondrainibe H, Mahefarisoa KL, Rajaonarivelo T, Faulkner C, et al. Baseline health and nutrition evoluation of two sympatric nocturnal lemur species (Avahi laniger) and (Lepilemur mustelinus) residing near an active mine site at Ambatovy, Madagascar. J Zoo Wildl Med. 2017;48:794–803.
Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith A, et al. Reproducible, interactive, scalable and extensive microbiome data science using QIIME 2. Nat Biotechnol. 2019. https://doi.org/10.1038/s41587-019-0209-9.
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucl Acids Res. 2013;41:D590–D596.
Skaug H, Fournier D, Bolker B, Magnusson A, Nielsen A. Generalized Linear Mixed Models using AD Model Builder. 2016; R package version 0.8.3.3.
R Core Team. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2017. https://www.R-project.org/.
RStudio Team. RStudio: integrated development for R. Boston, MA: RStudio, Inc.; 2016. http://www.rstudio.com/.
Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, et al. Vegan community ecology package: ordination methods, diversity analysis and other functions for community and vegetation ecologists. 2016. https://cran.r-project.org/web/packages/vegan/index.html.
Franzosa EA, McIver LJ, Rahnavard G, Thompson LR, Schirmer M, Weingart G, et al. Species-level functional profiling of metagenomes and metatranscriptomes. Nat Methods 2018;15:962–8.
Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60.
Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 1995;57:289–300.
Ezenwa VO, Gerardo NM, Inouye DW, Medina M, Xavier JB. Animal behavior and the microbiome. Science 2012;338:198–9.
Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes 2012;3:289–306.
Daniel H, Gholami AM, Berry D, Desmarchelier C, Hahne H, Loh G, et al. High-fat diet alters gut microbiota physiology in mice. ISME J. 2014;8:295–308.
Schwarz WH. The cellulosome and cellulose degradation by anaerobic bacteria. Appl Microbiol Biotechnol. 2001;56:634–49.
Biddle A, Stewart L, Blanchard J, Leschine S. Untangling the genetic basis of fibrolytic specialization by Lachnospiraceae and Ruminococcaceae in diverse gut communities. Diversity 2013;5:627–40.
den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud D-J, Bakker BM. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res. 2013;54:2325–40.
Dudley R. Ethanol, fruit ripening, and the historical origins of human alcoholism in primate frugivory. Integr Comp Biol. 2004;44:315–23.
Forbes SL, Perrault KA. Decomposition odour profiling in the air and soil surrounding vertebrate carrion. PLoS ONE 2014;9:e95107.
Shin NR, Whon TW, Bae JW. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015;33:496–503.
Lee PC, Lee SY, Chang HN. Succinic acid production by Anaerobiospirillium succiniciproducens ATCC 29305 growing on galactose, galactose/glucose, and galactose/lactose. J Microbiol Biotechnol. 2008;18:1792–6.
Förster AH, Gescher J. Metabolic engineering of Escherichia coli for production of mixed-acid fermentation end products. Front Bioeng Biotechnol. 2014;2:16.
Ganzhorn JU, Wright PC. Temporal patterns in primate leaf eating: the possible role of leaf chemistry. Folia Primatol. 1994;63:203–8.
Chesson A, Stewart CS, Wallace RJ. Influence of plant phenolic acids on growth and cellulolytic activity of rumen bacteria. Appl Environ Microbiol. 1982;44:597–603.
Pei K, Ou J, Huang J, Ou S. p-Coumaric acid and its conjugates: dietary sources, pharmacokinetic properties and biological activities. J Sci Food Agric. 2016;96:2952–62.
Filannino P, Di Cagno R, Gobbetti M. Metabolic and functional paths of lactic acid bacteria in plant foods: get out of the labyrinth. Curr Opin Biotechnol. 2018;48:64–72.
Burlingame R, Chapman PJ. Catabolism of phenylpropionic acid and its 3-hydroxy derivative by Escherichia coli. J Bacteriol. 1983;155:113–21.
Kobayashi Y, Shinkai T, Koike S. Ecological and physiological characterization shows that Fibrobacter succinogens is important in rumen fiber digestion- review. Folia Microbiol. 2008;53:195–200.
Kaakoush NO. Insights into the role of Erysipelotrichaceae in the human host. Front Cell Infect Microbiol. 2015;5:84.
Wu GD, Chen J, Hoffmann C, Bittinger K, Chen Y-Y, Keilbaugh SA, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011;334:105–8.
Jia Y, Wilkins D, Lu H, Cai M, Lee PKH. Long-term enrichment on cellulose or xylan causes functional and taxonomic convergence of microbial communities from anaerobic digesters. Appl Environ Microbiol. 2016;82:1519–29.
Maruo T, Sakamoto M, Ito C, Toda T, Benno Y. Adlercreutzia equolifaciens gen. nov., sp. nov., an equol-producing bacterium isolated from human faeces, and emended description of the genus Eggerthella. Int J Syst Evol Microbiol. 2008;58:1221–7.
Muthyala RS, Ju YH, Williams LD, Doerge DR, Katzenellenbogen BS, Helferich WG, et al. Equol, a natural estrogenic metabolite from soy isoflavones: convenient preparation and resolution of R- and S-equols and their differing binding and biological activity through estrogen receptors alpha and beta. Bioorg Med Chem. 2004;12:1559–67.
Lu M-F, Xiao Z-T, Zhang H-Y. Where do health benefits of flavonoids come from? Insights from flavonoid targets and their evolutionary history. Biochem Biophys Res Commun. 2013;434:701–4.
Madagascar Catalogue. Catalogue of the vascular plants of Madagascar. Missouri Botanical Garden, St. Louis, USA & Antananarivo, Madagascar. 2019; http://www.tropicos.org/Project/Madagascar.
Gurib-Fakim A. Medicinal plants: traditions of yesterday and drugs of tomorrow. Mol Asp Med. 2006;27:1–93.
Kohl KD, Stengel A, Dearing MD. Inoculation of tannin-degrading bacteria into novel hosts increases performance on tannin-rich diets. Environ Microbiol. 2016;18:1720–9.
IUCN. The IUCN Red List of Threatened Species. Version 2019-1. http://www.iucnredlist.org.
Estrada A, Garber PA, Rylands AB, Roos C, Fernandez-Duque E, Di Fiore A, et al. Impending extinction crisis of the world’s primates: why primates matter. Sci Adv. 2017;3:e1600946.
Thomas CD, Cameron A, Green RE, Bakkenes M, Beaumont LJ, Collingham YC, et al. Extinction risk from climate change. Nature 2004;427:145–8.
Colles A, Liow LH, Prinzing A. Are specialists at risk under environmental change? Neoecological, paleoecological and phylogenetic approaches. Ecol Lett. 2009;12:849–63.
Jernvall J, Wright PC. Diversity components of impending primate extinctions. Proc Natl Acad Sci. 1998;95:11279–83.
Acknowledgements
We thank the Ambatovy Biocamp Agents for locating and tracking the lemurs. Vanessa Mass and Josia Razafindramanana facilitated logistics on site. Conversations with Marina Blanco, Erin McKenney, and Sally Bornbusch contributed to many of the ideas presented herein. Argonne National Laboratory and the New York Genome Center provided amplicon and metagenomic sequences, respectively. André Corvelo and Amrita Kar performed bioinformatic analyses of metagenomic data. The David H. Murdock Research Institute provided NMR spectral data. Funding was provided by two Margot Marsh Biodiversity Foundation awards (to LKG and CMD), a Duke University International Travel Award (to LKG), Duke University research funds (to CMD), the Duke Lemur Center (to CVW), and by Ambatovy Minerals, S.A., Madagascar. This is Duke Lemur Center publication number 1450.
Author information
Authors and Affiliations
Contributions
LKG, CVW, and CMD conceived the study. All authors contributed to study design. CVW, REJ, KLM, TR, and HR performed field work, with assistance from LKG and CMD. LKG and TMO performed sample and data analyses. LKG and CMD wrote the manuscript, and all authors contributed to final preparation.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Greene, L.K., Williams, C., Junge, R.E. et al. A role for gut microbiota in host niche differentiation. ISME J 14, 1675–1687 (2020). https://doi.org/10.1038/s41396-020-0640-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-020-0640-4
This article is cited by
-
Effects of age, seasonality, and reproductive status on the gut microbiome of Southern White Rhinoceros (Ceratotherium simum simum) at the North Carolina zoo
Animal Microbiome (2023)
-
Bifidobacteria define gut microbiome profiles of golden lion tamarin (Leontopithecus rosalia) and marmoset (Callithrix sp.) metagenomic shotgun pools
Scientific Reports (2023)
-
Gut Site and Gut Morphology Predict Microbiome Structure and Function in Ecologically Diverse Lemurs
Microbial Ecology (2023)
-
Host-Specific Differences in Gut Microbiota Between Cricetulus barabensis and Phodopus sungorus
Current Microbiology (2023)
-
Lemur Gut Microeukaryotic Community Variation Is Not Associated with Host Phylogeny, Diet, or Habitat
Microbial Ecology (2023)