Abstract
The ocean is a net source of N2O, a potent greenhouse gas and ozone-depleting agent. However, the removal of N2O via microbial N2O consumption is poorly constrained and rate measurements have been restricted to anoxic waters. Here we expand N2O consumption measurements from anoxic zones to the sharp oxygen gradient above them, and experimentally determine kinetic parameters in both oxic and anoxic seawater for the first time. We find that the substrate affinity, O2 tolerance, and community composition of N2O-consuming microbes in oxic waters differ from those in the underlying anoxic layers. Kinetic parameters determined here are used to model in situ N2O production and consumption rates. Estimated in situ rates differ from measured rates, confirming the necessity to consider kinetics when predicting N2O cycling. Microbes from the oxic layer consume N2O under anoxic conditions at a much faster rate than microbes from anoxic zones. These experimental results are in keeping with model results which indicate that N2O consumption likely takes place above the oxygen deficient zone (ODZ). Thus, the dynamic layer with steep O2 and N2O gradients right above the ODZ is a previously ignored potential gatekeeper of N2O and should be accounted for in the marine N2O budget.
Similar content being viewed by others
Introduction
Nitrous oxide (N2O) is not only a greenhouse gas with about 300 times greater radiative forcing per mole than carbon dioxide, it is also the dominant ozone-depleting agent emitted in the 21st century [1]. The N2O concentration in the atmosphere is increasing [2], and the rate of N2O emission is accelerating [3]. From 2007 to 2016, the ocean contributed 20% of global N2O emissions, and 35% of the natural sources on average [4]. N2O cycling in the ocean thus has the potential to exacerbate climate change, as well as being affected by associated chemical changes, such as ocean acidification [5]. The most intense sources and sinks of N2O in the ocean occur in oxygen minimum zones (OMZs) [6], which are marine regions characterized by a sharp O2 gradient (oxycline) overlying an oxygen deficient zone (ODZ) where O2 concentration is below the detection limit of a switchable trace oxygen (STOX) sensor (10 nM) [7]. There are multiple biological sources of N2O [8,9,10], but there is only one major biological sink [11] (Fig. S1): the reduction of N2O to N2 by N2O-consuming microbes using the nitrous oxide reductase enzyme (N2OR). The possibility of N2O fixation has been suggested [12], but its mechanism is yet to be determined. N2O consumption in oxic waters, including the oxic layer of OMZs, has been ignored because this process was assumed to be part of the complete denitrification pathway (reduction of nitrate to N2 gas) and to be restricted to suboxic/anoxic environments (such as ODZs) [13]. However, the oxic surface layer and the oxycline of OMZs above the ODZ could be of vital importance in regulating N2O emissions if N2O consumption occurs there. N2O concentration in the oxycline or the anoxic ODZ of OMZs can be ≥10-fold higher than atmospheric saturation at the air–sea interface [11]. If not consumed in situ, this excess N2O could diffuse through the oxycline and the surface layer or be upwelled into the surface where it can exchange with the atmosphere.
The functional marker of the operon encoding N2OR, nosZ, has been used as a proxy for the presence of N2O-consuming microbes. Recent detection of nosZ genes and transcripts in oxic seawater [14, 15] implies the potential for N2O consumption there. The presence of genes and transcripts, however, does not guarantee the successful translation or activity of the enzyme. Direct rate measurements are required to determine whether this microbial potential actually results in N2O consumption. Here, the abundance and community composition of N2O-consuming microbes were determined by qPCR and microarray, respectively. N2O-consuming microbes that contain only nosZ (i.e., none of the other genes in the complete denitrification pathway) are of particular interest, because their activity results in net N2O consumption. Based on the analysis of 652 draft or complete microbial genomes with one or more dentification genes, nosZ-only microbes are overrepresented among these isolates from the ocean compared to other ecosystems [16]. N2O consumption rates were also measured under a matrix of controlled N2O and O2 concentrations in ~3000 samples collected from oxic and anoxic depths in the Eastern Tropical North Pacific (ETNP) OMZ, one of the three major oceanic OMZs. O2 tolerance and substrate kinetics of N2O consumption were determined and used to estimate in situ N2O consumption and production rates, which reflect the in situ conditions more accurately than directly using measured rates from incubation experiments without correcting for substrate additions.
Results and discussion
N2O consumption rates and N2O-consuming microbes in the OMZ
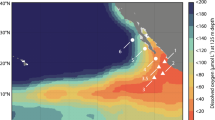
Potential N2O consumption rates (hereafter “measured rates”) were determined in March and April 2018 at three stations in the ETNP OMZ (Fig. 1a). Anoxic incubations were amended with standard additions of (15N)2O tracer with a final concentration of 50 nM at stations PS1 (on the west margin of the OMZ), PS2 (the open ocean station) and PS3 (the coastal station). Measured N2O consumption rates varied from zero to 5.1 nM d−1 at different depths (Fig. 1c, f, i). Measured rates in oxygen deficient waters were on the same order of magnitude (a few nM d−1) as previously measured rates in the ETNP, but lower than rates at one coastal station in that study [6], indicating high variability of N2O cycling in the coastal regions as previously suggested [17]. Notably, significant N2O consumption rates were measured in these anoxic incubations, even in samples collected from the oxycline and the oxygenated surface ocean (in situ [O2] up to 199 µM, Tables S1, S2), where N2O-consuming microbes were present and the nosZ gene was transcribed (Fig. 1d, g, j). In the upper water column, measured N2O consumption rates were highest in the upper oxycline, above the peak of in situ N2O concentrations at each station (Fig. 1b, e, h), and the rate maximum at station PS2 was detected in samples collected from 60 m where in situ O2 was 173.9 µM (Tables S1, S2). The rates of N2O consumption measured in surface waters stripped of oxygen were similar to or larger than rates measured in the ODZ. The larger consumption rates in the oxic layer given the same N2O and O2 concentrations as the ODZ layer might be due to more available dissolved organic matter at the shallower depths compared to the deeper ODZ layer [18] and/or the presence of different microbial communities at these depths.
a Scale bar indicates depth of water. b, e, h Note breaks in the y-axes. Gray lines indicate O2 concentration, black dots indicate N2O concentration from (Kelly et al. accepted) [37] and red dashed lines indicate the N2O concentration at equilibrium with the atmosphere [38]. c, f, i Black circles with black bars indicate measured N2O consumption rates with standard errors (also shown in Table S2). Error bars are standard errors of linear regression slope calculated from 15 point time-course incubations (three replicates at 5 time points). Blue circles indicate estimated in situ rates, which will be discussed in the “Estimated in situ N2O consumption and production rates” section. d, g, j Filled black circles indicate copy number of nosZ DNA in samples collected from the same casts from which rates were measured, filled gray circles indicate nosZ DNA from other casts at the same stations sampled within the same week, and red circles indicate copy numbers of nosZ RNA from the same casts. None of the RNA error bars overlapped with the zero line. Error bars are standard deviation (n = 3). Error bars are not shown if smaller than the symbol.
N2O-consuming microbes in the oxic surface water and oxycline were at least as abundant (DNA) and transcriptionally active (in terms of nosZ RNA abundance) as those in ODZs (Fig. 1d, g, j). Diverse archetypes of N2O-consuming microbes were detected in the oxic water above the ODZ at three stations in the ETNP, one station in the Arabian Sea, and were previously detected at two stations in the Eastern Tropical South Pacific (ETSP) (Fig. 2a) using a nosZ microarray. The microarray is not quantitative, but it can detect diverse, low abundance microbes from environmental microbial assemblages. Even though the microarray cannot represent every nosZ variant, the probe set (which includes marine, salt marsh, and terrestrial representatives) allowed us to determine that the community composition of the ETSP N2O-consuming microbial assemblages differs from the other two OMZs (Fig. 2a). Within the ETNP, the community composition of nosZ microbes at the RNA level differed between oxic and ODZ waters (Fig. 2b). The detection of N2O consumption in samples from the oxycline and the oxygenated surface seawater of the ETNP OMZ, and the presence of N2O-consuming microbes in all three major OMZs (Fig. 2a), indicate that microbes in the oxic layer above ODZs have the capacity to consume N2O at least under anoxic incubation conditions.
a N2O-consuming microbes from all three major OMZs (ETNP, ETSP (Eastern Tropical South Pacific) and AS (Arabian Sea)). Arabian Sea samples include two samples from the oxic layer above the ODZ and two inside the ODZ at station 1 (19N, 66E) collected on a previous cruise [34], and ETSP samples include four samples from oxic layers and four inside ODZs. ETSP data were obtained from a previous study [15]. b nosZ data from the ETNP only.
O2 tolerance of N2O-consuming microbes
The O2 tolerance of N2O-consuming microbes was determined by incubating seawater under a range of O2 conditions and measuring the O2 concentration corresponding to each rate using Pyroscience optical O2 sensors. N2O consumption rates were highest when O2 was lowest in almost all incubations (Fig. 3). The highest rates were measured in anoxic incubations with samples from the oxic seawater (Fig. 3b, c and Table S3), and surprisingly, these rates were much higher than rates measured in samples collected from anoxic depths at the same station (Fig. 3d–g). These results suggest that N2O-consuming microbes found in the oxic layer have the potential to metabolize N2O more rapidly than anaerobic organisms when conditions become anoxic (Fig. 1d, g, j). At stations PS1 and PS3, the higher potential of N2O consumption also corresponds to higher nosZ gene copy numbers in the oxic layer (Fig. 1d, j). N2O consumption rates in samples from oxic seawater, but not the ODZ, decreased sharply with increasing O2, indicating that N2O-consuming microbes from oxic seawater (Fig. 3a–c) were more sensitive to O2 than those from anoxic seawater (Fig. 3d–g). Although N2O consumption in samples from the oxic layer did not occur at high O2 concentrations, it started rapidly (≤1 day) after transitioning from oxic in situ conditions to anoxic incubation conditions (Fig. S2). The speed of this response might be due to the growth of N2O-consuming microbes, fast enzyme (N2OR) translation, or a response by already translated N2OR in the oxic seawater prior to sampling.
Samples were from (a, b, c) the oxic layer, (d, e) the top of the ODZ, which is the oxic–anoxic interface, and (f, g) anoxic ODZ core at stations (a) PS1, (b, d, f) PS2, and (c, e, g) PS3. Sample depths and in situ O2 concentrations are shown in Table S3. O2 concentrations on the x-axis were measured in incubation bottles using PyroScience optical O2 sensors. Dashed lines are fitted inhibition curves (see “Methods”). No half-inhibition constant (Ki) is significantly different from zero. The gray subplot (h) includes all data from the other plots (station PS1: blue, PS2: red, PS3: black). Error bars for each rate are standard errors of linear regression slope calculated from 15 point time-course incubations (three replicates at 5 time points). Error bars are not shown if smaller than the symbol or when rates are not significantly different from zero.
While our rate measurements suggest that N2OR is not active under oxic conditions, molecular data obtained here (Fig. 1d, g, j) and previously [15] show that both nosZ RNA and N2OR can be synthesized under oxic conditions. This phenomenon is seen in other environments as well, for example, an obligate aerobe from soil requires O2 to initiate nosZ expression, and can use the N2OR enzyme to consume N2O to survive temporary anoxia [19]. Another microbial culture continually makes N2OR and stores the enzymes inside their cells under oxic conditions, which was proposed as a “bet-hedging” strategy by this facultative anaerobe to allow for a rapid transition into anoxic environments [20]. Regardless of the mechanism of the rapid response, our results indicate that microbes from oxic seawater have the genetic potential to consume N2O, that the consumption was not limited by organic matter supply (in situ limitation by organic matter would prevent the observed increase in rate with increasing N2O concentration, Fig. 4a–c) and that they could consume N2O under anoxic conditions. These anoxic conditions could occur at a small scale in otherwise oxic water; for example they could be associated with phytoplankton colonies [14] and other particles [21, 22], especially in the productive and dynamic oxyclines of the OMZ with strong O2 and N2O gradients at shallow depths.
Samples were from (a, b, c) the oxic layer, (d, e) the top of the ODZ which is the oxic–anoxic interface, and (f, g) anoxic ODZ core at stations (a) PS1, (b, d, f) PS2, and (c, e, g) PS3. The gray subplot (h) includes all data from the other plots (station PS1: blue, PS2: red, PS3: black). Open circles indicate incubations without O2, and dashed lines are Michaelis–Menten curves fitted to these open circles. Stars indicate incubations with O2 additions (final O2 concentrations measured in incubation bottles, in situ O2 and sample depths shown in Table S3), and solid lines are Michaelis–Menten curves fitted to these stars. The blue line in (a) is curve fitted to all except the 4th datapoint, and the red line in (a) is curve fitted to the first four datapoints. Although the increase in rate with increasing N2O concentration was significant in six sets of kinetic experiments, only three curves resulted in significant Km values (Km corresponding to the blue dashed line in a, the dashed line in b, and the solid line in e). Error bars for each rate are standard errors of linear regression slope calculated from 15 point time-course incubations (three replicates at 5 time points). Error bars are not shown if smaller than the symbol.
Substrate affinity of N2O-consuming microbes
In incubations without O2, measured Km values (half-saturation constants of the Michaelis–Menten curve) (Fig. 4) for N2O consumption were in excess of in situ N2O concentrations at every station and depth (Fig. 1b, e, h), indicating that the in situ N2O concentrations were too low to saturate the N2O consumption rate. Notably, the potential maximum rate of N2O consumption in the oxic layer, upon removal of O2, was much higher than that in the ODZ at the same station (Fig. 4), and the substrate affinities of N2O-consuming microbes were distinct between oxic and anoxic depths (Fig. 4). Consistently, the community composition of N2O-consuming microbes at the RNA level was also different between oxic layers and anoxic ODZs of the ETNP (Fig. 2b). These results indicate that different kinetics parameters should be applied to the estimate of the anoxic N2O sink and the newly discovered potential N2O sink in the oxic layer. The difference in substrate affinities is likely due to diverse N2O-consuming microbes occupying different niches, because their affinities for N2O can vary by two orders of magnitude as shown in pure cultures [23]. This difference might be obscured when microbes in different layers are mixed up by physical processes such as upwelling and eddies.
Significant Km for N2O consumption could be determined for the oxic layer at station PS2 and the oxic–anoxic interface at station PS3 (Fig. 4b, e). The lack of Michaelis–Menten kinetics in samples from anoxic ODZs (Fig. 4f, g) implies that factors (e.g., organic matter) other than the added substrate were limiting N2O consumption. Km for the oxic layer at station PS3 was likely larger than that of station PS2, because the rate was not saturated even at the maximum N2O concentration (Fig. 4c). As for station PS1, the decreasing rate at the 4th datapoint suggests a mixture of N2O-consuming microbes with different substrate affinities (Fig. 4a). Although not significant, Km was 110 (±230) nM when only including the first four datapoints. When excluding the 4th datapoint, Km was 1354 (±653) nM for the oxic layer at station PS1. Larger Km values in samples from the oxic layer indicate that microbes there have lower affinity for N2O.
Notably, nosZ archetypes closely related to A. dehalogenans were among the top five most abundant archetypes at almost all examined depths from the ETNP (Tables S4, S5), ETSP [15], and Arabian Sea (Table S6), implying the importance of A. dehalogenans-like N2O-consuming microbes in both oxic layers and ODZs of all major OMZs. Furthermore, the Km (1.3 μM) of Anaeromyxobacter dehalogenans determined in cultures [23] was within the range of the Km values determined in this study (2.8 μM in the oxic layer and 0.3 μM at the top of the ODZ, Fig. 4). Different overall community composition, but similar affiliation of the most abundant archetypes, implies that the difference in community composition between the oxic and ODZ assemblages in the ETNP (Fig. 2b) and the difference between the ETSP and the other two OMZs (Fig. 2a) results from a diversity of low abundance microbes rather than a few abundant clades. The microarray probes cannot identify exact species, but can differentiate among archetypes representing unknown microbes, such as the A. dehalogenans-like N2O-consuming types that were detected in most of these samples. The vital role of A. dehalogenans-like microbes and other microbes in the same clade has been demonstrated in soils [24, 25]. Because A. dehalogenans possesses nosZ but no other denitrification genes [26], A. dehalogenans-like microbes may decouple N2O consumption from its production, resulting in a net N2O sink at depths where they dominate N2O-consuming assemblages.
In addition to anoxic incubations, we also examined N2O consumption kinetics in oxic incubations. Consistent with the low O2 tolerance of N2O-consuming microbes, especially in the oxic layers (Fig. 3), the kinetics of N2O consumption could not be determined under most incubations with O2 additions because N2O consumption rates were not detected (Fig. 4). Only samples from the oxic–anoxic interface at station PS3 showed Michaelis–Menten kinetics in the oxic incubation (Fig. 4e), likely because the O2 addition in this incubation (4.5 μM, Table S3) was less than all the other oxic incubations (8.1–342.0 μM, Table S3) and N2O-consuming microbes in ODZs had higher tolerance to increasing O2 concentration than those from the oxic layer (Fig. 3e). The O2 concentration (≥4.5 μM) allowing the occurrence of N2O consumption (N2O→N2) here was much higher than the previously determined threshold (0.2–0.3 µM) for denitrification (NO2−→ N2O or N2) [27], which might be due to differential O2 sensitivities of microbes possessing different parts of the denitrification pathway. Variable oxygen sensitivities have also been observed in O2 thresholds for N2O production from NO2− and NO3−, the latter showing a higher tolerance to O2 [10, 28]. In addition, unlike the presence of nosZ in the oxic layers, the group of denitrifiers represented by nitrite reductase genes (nirK and nirS) were very rare in the oxic layers [29]. These observations suggest that denitrification can be carried out in a modular fashion by independent organisms possessing different segments of the pathway [16], rather than one process of coupled reactions that occur without exchange of intermediates. Modular denitrification may have implications for the interpretation of classical isotope pairing experiments.
Estimated in situ N2O consumption and production rates
Rates of biogeochemical processes inferred from incubation experiments can be biased away from in situ values due to the dependence of rates on substrate concentrations and other environmental factors (e.g., O2 concentration), which often differ between in situ and incubation conditions. However, the new quantitative information on N2O consumption kinetics and effects of environmental factors like O2 derived here can be used to estimate in situ rates. Using these kinetic parameters most in situ N2O consumption rates were inferred to be zero in oxic layers (Fig. 1c, f, i) based on the high sensitivity of those microbes to O2 (Fig. 3a–c). The exception was 90 m at station PS2 where the in situ O2 concentration (4.4 µM, Table S1) was likely to be low enough to allow N2O consumption based on similar kinetics in anoxic incubations and incubations at PS3 with 4.5 µM O2 (Fig. 4e). In situ N2O consumption rates in anoxic ODZs were simulated by the Michaelis–Menten equation using the Km value determined here (Fig. 4e) and the in situ N2O concentrations (Fig. 1b, e, h). Measured N2O consumption rates had maxima in the upper oxycline above the N2O concentration peaks at all three stations, but the maxima in estimated in situ rates at these stations occurred at or below the oxic–anoxic interface, and the highest rate at station PS2 (6.3 nM d−1) was at 850 m, the lower edge of the ODZ (Fig. 1c, f, i). The secondary peak of N2O consumption inside the ODZ at station PS2 was greatly reduced after corrections were made using in situ N2O concentrations due to low in situ N2O concentrations (Fig. 1e). The peak inside the ODZ at station PS1, however, was larger after correction because the in situ N2O concentration (~80 nM, Fig. 1b) was higher than that in incubations (50 nM). The persistently high N2O concentration in the ODZ core at station PS1 reflects the slow N2O removal by denitrification at the margin of the OMZ. The lack of a SNM, a typical feature for anoxic ODZs, at station PS1 (Fig. S3) is also consistent with its position at the oceanic edge of the OMZ. The difference between measured rates and kinetics-corrected in situ rates indicates the need for more information on the kinetics of N2O consumption under different environmental conditions and in different OMZ regions.
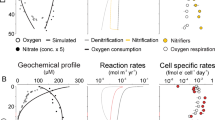
In situ N2O production rates were modeled from estimated in situ N2O consumption rates, N2O concentrations, advection, and diffusion using a 1-D steady-state framework (Fig. 5), which reflects a weak lateral advection and upwelling scenario as in a previous study [6]. Production and consumption rates were mostly balanced but were decoupled at the sharp N2O concentration gradient at station PS3. The decoupling of the production and consumption was due to the strong N2O fluxes from physical processes (i.e., advection and diffusion) in the sharp N2O gradient coinciding with the sharp O2 gradient. This decoupling was not reported previously because measurements of N2O consumption were all below the bottom of the upper oxycline [6]. Notably, the modeled production rates in the oxic layer at stations PS1 and PS3 (Fig. 5) with 199.0 and 89.9 µM in situ O2 were negative considering advection, diffusion and zero estimated consumption rates (Table S1). Since the production rate cannot be negative, this analysis suggests that N2O consumption occurs at least sometimes at these depths to balance the N2O flux from physical processes (assuming steady state). Consistent with the model results, N2O consumption rates were detected when oxygen is above the 4.5 µM threshold especially at station PS3 (Fig. 3c, e, j). The significant rates under oxic conditions might imply more micro-anoxic sites (e.g., particulate organic matter) at the coastal station. Although we chose 4.5 µM as a conservative oxygen threshold here, different thresholds for N2O consumption need to be determined for different environmental conditions in future studies. Particles and other microsites might disintegrate during sampling and purging, so the in situ N2O consumption rates in the oxic layer were potentially underestimated by the incubation experiments.
Implications for the oceanic N2O budget
The annual N2O emission rate of the ocean is 3–5 Tg-N yr−1 based on recent estimates [4, 30]. Developing an understanding of what controls the major biological sink of N2O is vitally important to better constrain estimation of the highly uncertain [17] marine N2O budget. Using direct rate measurements, we demonstrated the ability of microbial assemblages in the oxic layer above N2O production hotspots to consume N2O under anoxic conditions and quantified the rate dependence on N2O and O2 concentrations. The potential N2O consumption rate in oxic seawaters was at least two orders of magnitude faster than that in ODZs under favorable conditions (i.e., low O2 and high N2O concentrations). Even though N2O is unlikely to be consumed when surface seawater is saturated with O2, N2O consumption started rapidly after switching to anoxic conditions. N2O consumption also occurred in the oxycline where O2 concentration was low and N2O concentration was high. High N2O concentrations in the oxycline are attributed to production from both denitrification and nitrification, with dentification as the main source [10]. Assuming anoxia occurs transiently in the layer above the ODZ, the consumption rate (0.7–10.3 nM d−1) estimated from measured rates using in situ N2O concentrations (Table S1) are on the same order of magnitude as N2O production rates measured in the ETNP OMZ [10]. These rates suggest a potential gross N2O sink in the layer above the ODZ of all OMZs of 0.02–0.32 Tg-N yr−1 assuming a 10 cm transiently anoxic depth interval of this layer with a volume of 3.04 × 1011 m3 [31]. Although anoxia is unlikely to occur in such a large area at once, this potential gross sink of N2O is considerable relative to an annual oceanic N2O emission of 3–5 Tg-N yr−1 [4, 30].
Anoxic conditions could occur in microsites such as particles [21] and phytoplankton aggregates [14] in the oxic layer. nosZ transcripts were found to be highly enriched in particle-associated fractions compared to free-living fractions in the ETNP OMZ [22]. Additionally, eddies, upwelling and other dynamic mixing events could lead to the shoaling of low oxygen, high N2O seawater, and N2O-consuming microbes [32]. O2 concentrations in the oxycline above the ODZ of the ETNP varied from nearly 100 µM to below detection within days or weeks based on both Argo float data (Fig. S4) and conductivity–temperature–depth (CTD) Seabird data (Fig. S5). Thus, microbes in the surface layer or subsurface microbes being brought to the surface layer could consume a portion of N2O before it escapes into the atmosphere when surface anoxia occurs.
The detection of viable N2O-consuming microbes in the upper oxyclines of all three OMZs implies a potential role for unconventional nosZ-containing microbes in regulating the N2O budget. The presence of these microbes in all three OMZs and their potential N2O sink raises the necessity of quantifying this potential in the other two OMZs. The substrate kinetics and biological information obtained in this study provide previously lacking parameters for the characterization of N2O consumption in marine N2O models. Applying the new O2 threshold obtained here for N2O consumption (4.5 µM) to a mechanistic 1-D biogeochemistry model [6] produces N2O peaks and O2 profiles similar to our measurements at the open ocean station when the O2 threshold for N2O production via denitrification was also increased (to 20 µM [10, 28]) (Fig. S6). The O2 thresholds for N2O production and consumption along the redox gradient of the ocean need further investigation, but the high O2 tolerance of N2O production from denitrification in the modified model is consistent with previous experimental results showing the persistence of N2O production from NO3− at 7 and 23 µM O2 in the ETNP [10] and ETSP [28], respectively. Further kinetics and molecular experiments are required to investigate the co-occurrence of microbes with different substrate kinetics in the same sample (Fig. 4a) and the spatial variation of microbial communities for both N2O consumption and production. The OMZs are not only the most intense N2O cycling regions [6, 10], but also contribute to a large seasonal variation to the global N2O emissions [30]. Thus, the findings here will not only improve estimates of these N2O sinks, but also will improve estimates of N2O sources, two crucial variables to constrain in a changing ocean.
Methods
Sampling, incubations, and rate measurements
The sampling sites are within the ETNP OMZ, one of the three major OMZs in the world. We sampled at three stations (OMZ margin station PS1, open ocean OMZ station PS2, and coastal OMZ station PS3; Fig. 1a) in March and April 2018 on board R/V Sally Ride (Cruise ID: SR 1805). The three sampling stations represent a transect from offshore to onshore, along a gradient from low to high productivity, and from the oceanic edge of the OMZ to the intense ODZ of the coastal station. Station PS1 is on the margin of the ETNP OMZ, so oxygen intrusion events likely occur below the upper oxycline at this station. Station PS1 does not have a secondary nitrite maximum (SNM), while stations PS2 and PS3 both have SNM in their ODZs (Fig. S3). PS1 and PS2 are open ocean stations, and PS3 is a coastal station. Station PS3 has a very shallow oxycline close to the surface of the ocean. This shallow oxyline is probably due to the intense heterotrophic respiration fueled by the primary production (implied by chlorophyll concentrations of 3 µg/L at 10 m) at the surface.
Twelve 30 L Niskin bottles on a rosette with a CTD profiler were used to collect seawater from different depths while recording temperature, pressure, salinity, chlorophyll, and in situ O2 concentration with both Seabird (Sea-Bird SBE 9, Sea-Bird Electronics, Bellevue, NA) and STOX sensors [7]. Samples for measurements of in situ N2O concentration were collected from Niskin bottles into 160 mL bottles after overflowing three times and preserved with saturated HgCl2. N2O concentrations were measured on an isotope ratio mass spectrometer based on the major ion (m/z = 44) peak area [33]. Particulate material for microbial DNA and RNA analysis was collected by filtering up to 4 L seawater from Niskin bottles through Sterivex filters (0.22 µm). Filters were flash frozen in liquid N2 on board, and then preserved at −80 °C until DNA and RNA extraction in the lab. Seawater samples for determining N2O consumption rate were collected into 320 mL ground glass-stoppered glass bottles after overflowing three times to minimize O2 contamination. Seawater (8 mL) was then aliquoted into 12 mL exetainers inside a N2 flushed glove bag to leave a 4 mL headspace for purging. After sealing in the glove bag, exetainers were purged with helium for 5 min to reach anoxia for depth profiles shown in Fig. 1 and kinetics determined under anoxic conditions shown in Fig. 4. (15N)2O tracer (Cambridge Isotope Laboratories, purity ≥98%) was added as gas into each exetainer using a helium-flushed gas-tight glass syringe to reach a final concentration of 50 nM N2O (standard additions shown in Figs. 1, 3) or varying from 50 to 1978 nM N2O for kinetics experiments in Fig. 4. A 50 nM N2O addition was made to ensure the produced N2 was detectable. A set of 15 exetainers incubated in a time series (triplicates for each time point, 5 time points in total including three time zero bottles as abiotic controls) was used to determine a single rate. Incubations were sampled approximately every 12 h for 2 days and were terminated by adding 0.05 mL of 50% (w/v) ZnCl2 following previous procedures [6]. The amount and isotopic composition of N2 in each exetainer was measured on a mass spectrometer (Europa Scientific 20-20, Crewe, UK), and the rate of N2O consumption was calculated from the linear regression of the excess of 30N2 over the incubation time following the previous method [34].
N2O consumption rates were measured at 10 depths at each station, including oxic surface water, upper and lower oxycline, top of the anoxic ODZ (oxic–anoxic interface), and core of the ODZ. The position of the upper oxycline varies among the three stations (station PS2: 59–96 m; station PS3: 48–84 m; station PS3: 0–35 m). The lower oxycline starts at around 700 m for station PS1, 850 m for station PS2, and 860 m for station PS3. N2O kinetics and O2 tolerance experiments were performed in the oxic layer, at the top of the ODZ (oxic–anoxic interface), and in the core of the ODZ. Ambient O2 concentrations at both the top and the core of the ODZ were below detection limit of the Seabird sensor (Table S3). The kinetics of N2O consumption were determined by measuring rates with varying added (15N)2O concentrations (50, 99, 198, 371, 865, and 1978 nM). Different O2 concentrations in incubations investigating the O2 tolerance of N2O consumption in Figs. 3, 4 were achieved prior to initiation of the experiment by varying flow rates of O2 and helium gases using a custom-assembled gas flow manifold with two gas flow meters on board. The O2 concentration for each set of exetainers was monitored by direct measurement using optical oxygen sensors with a detection limit of 0.06 µM (PyroScience GmbH, Aachen, Germany), and is shown on the x-axis in Fig. 3.
Kinetics models and in situ rate estimation
Half-saturation constant (Km) and the maximum rate (Vm) were determined by fitting N2O consumption rate and N2O concentration data to the Michaelis–Menten equation (Eq. (1)). Km is the N2O concentration at which the rate (V) equals half of Vm. Fitting was performed by the curve fitting tool in Matlab. The 95% confidence interval was used to determine whether a parameter is significantly different from zero. The half-inhibition constant (Ki), analogous to Km, is the O2 concentration that causes half of the potential maximum inhibition (Im). Ki was calculated from fitting an inhibition curve (Eq. (2)). The unit of V or Vm is nM d−1, the unit of [N2O] or Km is nM, the unit of [O2] or Ki is µM and Im is unitless.
The in situ N2O consumption rate was estimated from in situ O2 concentration, in situ N2O concentration, and calculated Km and Vm. First, Km in Fig. 4, measured rates (V) and measured [N2O] were used to calculate Vm for each depth based on Eq. (1). Km and Vm determined in samples from the oxic layer were used to calculate in situ rates in oxic seawaters, and those determined in samples from the ODZ were used in ODZ rate estimation. Then, in situ rates were set to zero if in situ O2 concentrations were above a threshold level of 4.5 µM. This threshold was chosen because it was the highest oxygen concentration at which N2O consumption was dependent upon N2O concentration i.e., N2O consumption showed similar kinetics in response to N2O concentration under undetectable O2 concentration and at 4.5 µM O2 (Fig. 4e), but not at higher O2 concentrations. This is a conservative threshold because we did not determine the absolute highest O2 level that allowed N2O consumption.
In situ N2O production rate was estimated by subtracting the advection and diffusion of N2O from in situ consumption rates assuming a steady state (Eq. (3)). v (1 × 10−7 m s−1) is advection coefficient and D (2 × 10−5 m2 s–1) is the diffusivity coefficient. Coefficients and the steady-state 1-D model follow (Babbin et al.) [6].
A mechanistic 1-D biogeochemistry model [6] was updated based on the experimental results in this study to assess the effect of different O2 thresholds on N2O predictions (Fig. S6). Briefly, the model was built upon the balance of physical and biological processes at steady state. Physical processes in the model include advection, diffusion, and gravitational sinking. Biological processes include aerobic respiration (i.e., remineralization fueled by O2), nitrification, and denitrification. The production of N2O from both nitrification and denitrification, and the consumption of N2O from denitrification are considered. The O2 threshold for N2O consumption is 0.3 µM in the original model and 4.5 µM in the updated model. The O2 threshold for N2O production via denitrification is 1 µM in the original model and 20 µM in the updated model. The only differences between the updated model and the original model are the O2 threshold values. Model code, detailed evaluation of the model and all the other parameters of the model are available in the previous study [6].
DNA and RNA extraction, quantitative PCR (qPCR) assays, and nosZ microarray
These experiments were performed as previously described [15]. Briefly, DNA and RNA were extracted from Sterivex filters including four filters collected from a previous cruise in the Arabian Sea OMZ [34]. Each DNA or RNA copy number value corresponds to one Sterivex. RNA was reverse transcribed into cDNA. qPCR was used to estimate the abundance of total and transcribed nosZ assemblages using the nosZ1F (5′ -WCSYTGTTCMTCGACAGCCAG-3′) and nosZ1R (5′-ATGTCGATCARCTGVKCRTTYTC-3′) primer set [35]. The qPCR products were purified from agarose gels and then used as targets for microarray experiments following a previous protocol [36]. The detection limit of qPCR is 18.1 copies mL−1. The microarray contains 114 nosZ archetype probes and the sequences of all probes are published in the supplementary dataset in a previous study [15]. The fluorescence ratio of each archetype on the microarray is defined as the ratio of Cy3 to Cy5 fluorescence. Normalized fluorescence ratio (FRn) was calculated by dividing the fluorescence ratio of each archetype by the maximum fluorescence ratio on the same microarray. FRn is used as a proxy for the relative abundance of each nosZ archetype. FRn was used to determine the top five most abundant archetypes in each sample. Detrended correspondence analysis was performed on FRn to analyze the community composition of N2O-consuming assemblages using the vegan package in R (version 3.6.0). Identification of nosZ sequences is limited by the probe selection on the array and the larger database of nosZ sequences now available might help identify the oxic nosZ more precisely. Nonetheless, the limited database represented on the array sufficed to detect significant differences among samples and to identify phylogenetic affinities of OMZ nosZ genes.
References
Ravishankara AR, Daniel JS, Portmann RW. Nitrous oxide (N2O): the dominant ozone-depleting substance emitted in the 21st century. Science. 2009;326:123–5.
Park S, Croteau P, Boering KA, Etheridge DM, Ferretti D, Fraser PJ, et al. Trends and seasonal cycles in the isotopic composition of nitrous oxide since 1940. Nat Geosci. 2012;5:261–5.
Thompson RL, Lassaletta L, Patra PK, Wilson C, Wells KC, Gressent A, et al. Acceleration of global N2O emissions seen from two decades of atmospheric inversion. Nat Clim Change. 2019;9:993–8.
Tian H, Xu R, Canadell JG, Thompson RL, Winiwarter W, Suntharalingam P, et al. A comprehensive quantification of global nitrous oxide sources and sinks. Nature. 2020;586:248–56.
Breider F, Yoshikawa C, Makabe A, Toyoda S, Wakita M, Matsui Y, et al. Response of N2O production rate to ocean acidification in the western North Pacific. Nat Clim Change. 2019;9:954–8.
Babbin AR, Bianchi D, Jayakumar A, Ward BB. Rapid nitrous oxide cycling in the suboxic ocean. Science. 2015;348:1127–9.
Revsbech NP, Larsen LH, Gundersen J, Dalsgaard T, Ulloa O, Thamdrup B. Determination of ultra-low oxygen concentrations in oxygen minimum zones by the STOX sensor. Limnol Oceanogr Methods. 2009;7:371–81.
Löscher CR, Kock A, Könneke M, Laroche J, Bange HW, Schmitz RA. Production of oceanic nitrous oxide by ammonia-oxidizing archaea. Biogeosciences. 2012;9:2419–29.
Trimmer M, Chronopoulou P-M, Maanoja ST, Upstill-Goddard RC, Kitidis V, Purdy KJ. Nitrous oxide as a function of oxygen and archaeal gene abundance in the North Pacific. Nat Commun. 2016;7:13451.
Ji Q, Buitenhuis E, Suntharalingam P, Sarmiento JL, Ward BB. Global nitrous oxide production determined by oxygen sensitivity of nitrification and denitrification. Glob Biogeochem Cycles. 2018;32:1790–802.
Bourbonnais A, Letscher RT, Bange HW, Échevin V, Larkum J, Mohn J, et al. N2O production and consumption from stable isotopic and concentration data in the Peruvian coastal upwelling system. Glob Biogeochem Cycles. 2017;31:678–98.
Farías L, Faúndez J, Fernández C, Cornejo M, Sanhueza S, Carrasco C. Biological N2O fixation in the eastern South Pacific Ocean and marine cyanobacterial cultures. PLoS ONE. 2013;8:e63956.
Bonin P, Gilewicz M, Bertrand JC. Effects of oxygen on each step of denitrification on Pseudomonas nautica. Can J Microbiol. 1989;35:1061–4.
Coates CJ, Wyman M. A denitrifying community associated with a major, marine nitrogen fixer. Environ Microbiol. 2017;19:4978–92.
Sun X, Jayakumar A, Ward BB. Community composition of nitrous oxide consuming bacteria in the oxygen minimum zone of the eastern Tropical South Pacific. Front Microbiol. 2017;8:1–11.
Graf DRH, Jones CM, Hallin S. Intergenomic comparisons highlight modularity of the denitrification pathway and underpin the importance of community structure for N2O emissions. PLoS ONE. 2014;9:e114118.
Arévalo-Martínez DL, Kock A, Löscher CR, Schmitz RA, Bange HW. Massive nitrous oxide emissions from the tropical South Pacific Ocean. Nat Geosci. 2015;8:530–3.
Cantarero SI, Henríquez-Castillo C, Dildar N, Vargas CA, von Dassow P, Cornejo-D’Ottone M, et al. Size-fractionated contribution of microbial biomass to suspended organic matter in the eastern Tropical South Pacific oxygen minimum zone. Front Mar Sci. 2020;7:1–20.
Park D, Kim H, Yoon S. Nitrous oxide reduction by an obligate aerobic bacterium Gemmatimonas aurantiaca strain T-27. Appl Environ Microbiol. 2017;83:00502–17.
Lycus P, Soriano-Laguna MJ, Kjos M, Richardson DJ, Gates AJ, Milligan DA, et al. A bet-hedging strategy for denitrifying bacteria curtails their release of N2O. Proc Natl Acad Sci USA. 2018;115:11820–5.
Bianchi D, Weber TS, Kiko R, Deutsch C. Global niche of marine anaerobic metabolisms expanded by particle microenvironments. Nat Geosci. 2018;11:263–8.
Ganesh S, Bristow LA, Larsen M, Sarode N, Thamdrup B, Stewart FJ. Size-fraction partitioning of community gene transcription and nitrogen metabolism in a marine oxygen minimum zone. ISME J. 2015;9:2682–96.
Yoon S, Nissen S, Park D, Sanford RA, Löffler E. Nitrous oxide reduction kinetics distinguish bacteria harboring clade I NosZ from those harboring clade II NosZ. Appl Environ Microbiol. 2016;82:3793–800.
Jones CM, Spor A, Brennan FP, Breuil MC, Bru D, Lemanceau P, et al. Recently identified microbial guild mediates soil N2O sink capacity. Nat Clim Change. 2014;4:801–5.
Jones CM, Graf DR, Bru D, Philippot L, Hallin S. The unaccounted yet abundant nitrous oxide-reducing microbial community: a potential nitrous oxide sink. ISME J. 2013;7:417–26.
Sanford RA, Wagner DD, Wu Q, Chee-Sanford JC, Thomas SH, Cruz-Garcia C, et al. Unexpected nondenitrifier nitrous oxide reductase gene diversity and abundance in soils. Proc Natl Acad Sci USA. 2012;109:19709–14.
Dalsgaard T, Stewart FJ, Thamdrup B, Brabandere LDE, Revsbech P, Ulloa O, et al. Oxygen at nanomolar levels reversibly suppresses process rates and gene expression in anammox and denitrification in the oxygen. MBio. 2014;5:1–14.
Frey C, Bange HW, Achterberg EP, Jayakumar A, Löscher CR, Arévalo-Martínez DL, et al. Regulation of nitrous oxide production in low-oxygen waters off the coast of Peru. Biogeosciences. 2020;17:2263–87.
Jayakumar A, Peng X, Ward BB. Community composition of bacteria involved in fixed nitrogen loss in the water column of two major oxygen minimum zones in the ocean. Aquat Microb Ecol. 2013;70:245–59.
Yang S, Chang BX, Warner MJ, Weber TS, Bourbonnais AM, Santoro AE, et al. Global reconstruction reduces the uncertainty of oceanic nitrous oxide emissions and reveals a vigorous seasonal cycle. Proc Natl Acad Sci USA. 2020;117:11954–60.
Paulmier A, Ruiz-Pino D. Oxygen minimum zones (OMZs) in the modern ocean. Prog Oceanogr. 2009;80:113–28.
Grundle DS, Löscher CR, Krahmann G, Altabet MA, Bange HW, Karstensen J, et al. Low oxygen eddies in the eastern tropical North Atlantic: implications for N2O cycling. Sci Rep. 2017;7:1–10.
Mcilvin MR, Casciotti KL. Fully automated system for stable isotopic analyses of dissolved nitrous oxide at natural abundance levels. Limnol Oceanogr Methods. 2010;8:54–66.
Bulow SE, Rich JJ, Naik HS, Pratihary AK, Ward BB. Denitrification exceeds anammox as a nitrogen loss pathway in the Arabian Sea oxygen minimum zone. Deep Res Part I Oceanogr Res Pap. 2010;57:384–93.
Henry S, Bru D, Stres B, Hallet S, Philippot L, Burgundy I, et al. Quantitative detection of the nosZ gene, encoding nitrous oxide reductase, and comparison of the abundances of 16S rRNA, narG, nirK, and nosZ genes in soils. Appl Environ Microbiol. 2006;72:5181–9.
Ward BB, Bouskill NJ. The utility of functional gene arrays for assessing community composition, relative abundance, and distribution of ammonia-oxidizing bacteria and archaea. Methods Enzymol. Academic Press. 2011;496:373–96.
Kelly CL, Travis NM, Baya PA, Casciotti KL. Quantifying nitrous oxide cycling regimes in the Eastern Tropical North Pacific Ocean with isotopomer analysis. Glob Biogeochem Cycles. Accepted.
Weiss RF, Price BA. Nitrous oxide solubility in water and seawater. Mar Chem. 1980;8:347–59.
Acknowledgements
We thank all the crew and scientists on R/V Sally Ride for assistance. This research was supported by an NSF grant to BBW and KLC. We thank Sergey Oleynik for maintaining the mass spectrometer.
Author information
Authors and Affiliations
Contributions
XS and BBW conceptualized the research. XS, BBW, and JCT sampled and performed incubation experiments on board. CLK and KLC sampled and measured N2O concentrations. AJ collected microbial samples, extracted DNA, and performed qPCR and microarray experiments. XS and EW assisted with DNA extraction and qPCR experiments. XS, AJ, and BBW analyzed microbial data. XS measured N2O consumption rates, analyzed the data, and modeled the rates. XS and BBW wrote the paper with inputs from all authors.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sun, X., Jayakumar, A., Tracey, J.C. et al. Microbial N2O consumption in and above marine N2O production hotspots. ISME J 15, 1434–1444 (2021). https://doi.org/10.1038/s41396-020-00861-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-020-00861-2
This article is cited by
-
Partitioning of the denitrification pathway and other nitrite metabolisms within global oxygen deficient zones
ISME Communications (2023)
-
Direct biological fixation provides a freshwater sink for N2O
Nature Communications (2023)
-
The marine nitrogen cycle: new developments and global change
Nature Reviews Microbiology (2022)