Abstract
Microbial skin assemblages, including fungal communities, can influence host resistance to infectious diseases. The diversity-invasibility hypothesis predicts that high-diversity communities are less easily invaded than species-poor communities, and thus diverse microbial communities may prevent pathogens from colonizing a host. To explore the hypothesis that host fungal communities mediate resistance to infection by fungal pathogens, we investigated characteristics of bat skin fungal communities as they relate to susceptibility to the emerging disease white-nose syndrome (WNS). Using a culture-based approach, we compared skin fungal assemblage characteristics of 10 bat species that differ in susceptibility to WNS across 10 eastern U.S. states. The fungal assemblages on WNS-susceptible bat species had significantly lower alpha diversity and abundance compared to WNS-resistant species. Overall fungal assemblage structure did not vary based on WNS-susceptibility, but several yeast species were differentially abundant on WNS-resistant bat species. One yeast species inhibited Pseudogymnoascus destructans (Pd), the causative agent on WNS, in vitro under certain conditions, suggesting a possible role in host protection. Further exploration of interactions between Pd and constituents of skin fungal assemblages may prove useful for predicting susceptibility of bat populations to WNS and for developing effective mitigation strategies.
Similar content being viewed by others
Introduction
A potential microbial invader into an ecosystem must overcome barriers to invasion including abiotic (e.g., pH, temperature, and salinity) and biotic (e.g., competition, antagonism, and predation) resistance [1]. The diversity-invasibility hypothesis predicts that high-diversity communities are less easily invaded than species-poor communities [1,2,3]. For example, reduction of skin microflora on amphibians increases host susceptibility to and mortality from the chytrid fungus Batrachochytrium dendrobatidis [4, 5]. Diversity is not the only characteristic of the microbiome that may influence disease susceptibility. Specific components of microbiome may also inhibit pathogen invasion through production of antimicrobial compounds, stimulation or training of the host immune system, and occupation of adherence sites and metabolic niches [6, 7]. Thus, the presence of certain microorganisms rather than the overall diversity or community structure may be responsible for host resistance.
Although most work on skin microbiomes focus on bacterial communities, the role of skin fungal assemblages in health and disease is an emerging field [8,9,10]. For example, a recent study found cutaneous fungal assemblages may contribute even more to defense against B. dendrobatidis than do bacteria [11]. Diverse skin fungal assemblage patterns are associated with various diseases in humans and can play a role as preventive or therapeutic agents [10]. For instance, the severity of skin disorders such as atopic dermatitis and psoriasis are inversely correlated with the diversity of skin fungal assemblages [9]. Commensal microorganisms in humans, including fungi, play a role in appropriately tuning immune activity to ensure efficient responses to pathogens while limiting responses directed toward host tissues and innocuous agents such as allergens [6, 12].
The role of the fungal skin microbiome in susceptibility to and severity of emerging fungal diseases in wildlife is an active area of investigation, with previous work focusing on amphibian chytridiomycosis and snake fungal disease [11, 13]. However, examination of how the fungal skin microbiome mediates white-nose syndrome (WNS) in bats has not been studied. White-nose syndrome, a cutaneous fungal infection of hibernating bats caused by Pseudogymnoascus destructans (Pd), was introduced to North America from Eurasia [14, 15]. The fungal pathogen damages wing membranes, resulting in physiological disruptions that can lead to death [16]. Since its initial detection in North America in 2006, WNS has killed more than 6.5 million bats [17], making it one of the most devastating wildlife diseases ever documented. Some species of bats have experienced catastrophic population declines due to WNS and are now listed as endangered [18].
Variation in host susceptibility to WNS has been documented within and between bat species. For example, the little brown bat (Myotis lucifugus), northern long-eared bat (My. septentrionalis), and tricolored bat (Perimyotis subflavus) have experienced massive population declines due to WNS, while other species [e.g., big brown bat (Eptesicus fuscus), eastern small-footed bat (My. leibii), and Indiana bat (My. sodalis)] appear to resist infection by Pd (have lower pathogen burdens) and have persisted, even in areas where Pd appears to be established in hibernacula [19,20,21,22]. Still other species of hibernating bats [e.g., Townsend’s big-eared bat (Corynorhinus townsendii) and Rafinesque’s big-eared bat (Co. rafinesquii)] appear to be unaffected by the disease, in that they display no lesions diagnostic for infection despite exposure to Pd [19, 23,24,25]. Variation in host susceptibility has also been documented within species, for example some colonies of My. lucifugus in New York state are persisting after more than a decade of exposure to Pd [26]. In all, 11 bat species have been documented with WNS in North America, and Pd has been detected on an additional six species without clinical signs of WNS [19, 23,24,25]. Multiple host traits have been hypothesized as determining WNS-susceptibility including body size, length of hibernation period, physio-chemistry (e.g., sebaceous lipid composition), immune defenses, hibernation behavior, and skin microbial assemblages [21, 27,28,29,30,31,32]. However, the precise mechanisms of WNS-resistance remain unknown.
We examined skin fungal assemblage characteristics of the wings of 10 bat species across the eastern United States. We hypothesized that culturable constituents of bat skin fungal assemblages differ based on host WNS-susceptibility. Due to complex nature of a host’s microbiome and the various ways in which it can confer resistance to disease, no one analysis is sufficient for testing this hypothesis. Therefore, we tested two sub-hypotheses to address our larger question. For our first sub-hypothesis, we hypothesized that WNS-resistant bat species would have higher fungal diversity and abundance on their wings compared to WNS-susceptible species (consistent with the pre-existing diversity-invasibility hypothesis). For our second sub-hypothesis, we hypothesized that we would detect fungal species on WNS-resistant bats that were rare or absent from susceptible bats, and that these fungal species would directly inhibit Pd growth in vitro.
Methods
Sampling
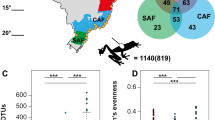
Field sampling was approved by the U.S. Geological Survey National Wildlife Health Center Institutional Animal Care and Use Committee (Protocols #EP140212 and #EP081124-A2). Samples were collected from 25 hibernacula in the eastern USA (10 states) from January to March, 2014‒2017 (Table 1; Fig. 1). Sampling targeted hibernating bats during winter because Pd primarily grows on bats during hibernation.
Each sampling site is represented by a circular chart and the bat species sampled at each site are represented by the colors displayed by its respective chart. Whether or not a species was sampled at a site is displayed as a binary factor, i.e., size of each chart segment is not related to the number of each species sampled at a site. Bat species categorized as white-nose syndrome (WNS)-susceptible are displayed in hues of red, species categorized as WNS-resistant are displayed in hues of blue, and species categorized as WNS-impervious are displayed in hues of yellow. a sampling sites and bat species across the United States. Sampled states are shown in gray. b A finer scale plot of sampling sites within the state of Wisconsin.
Categorizing bat species into WNS-susceptibility groups is an area of active debate because disease processes act on a context-dependent continuum [33]. For this study, we classified My. lucifugus, My. septentrionalis, and Per. subflavus as WNS-susceptible because these species have experienced WNS-associated population declines of over 70% in multiple hibernacula [19, 34]. We classified Co. rafinesquii and Virginia big-eared bat (Co. townsendii virginianus) as WNS-impervious as these species have not been documented with Pd infections, despite repeated detection of Pd on their wing skin (indicating exposure) [23, 24]. We classified species as WNS-resistant if they are documented to develop Pd infections, but do not exhibit large-scale mortality as a direct result of WNS. Under this classification, E. fuscus and My. leibii were categorized as WNS-resistant as they have exhibited <50% WNS-associated declines within Pd-infected populations [19]. Similarly, population declines related to WNS have not been documented in southeastern myotis (My. austroriparius) and gray bat (My. grisescens) despite confirmation of WNS in these species [19, 35,36,37]. Myotis sodalis was also classified as WNS-resistant because declines in Pd-infected My. sodalis populations have been variable. At some locations, My. sodalis colonies have experienced >70% declines after the detection of WNS, while at other sites declines have been <50% (or the colony has even increased) post-WNS [19, 34, 38, 39]. Additionally, fungal loads of Pd on the skin of My. sodalis are more consistent with those found on WNS-resistant species [22]. Some individual bats within a resistant species may develop WNS severe enough to cause mortality, while other individuals have low pathogen loads. We emphasize that our classifications are made at the species level and are not necessarily valid for all individuals within a species as we did not track the fate of individual bats in this study.
We swabbed wings by rolling a sterile Pur-Wraps® polyester-tipped swab (Puritan Medical Products Company LLC, Guilford, Maine, USA), pre-moistened with 150 μl sterile nuclease-free water, three times across the ventral plagiopatagium (wing membrane connecting the hindlimb and forelimb). We stored swabs in individual, sterile, 1.5 ml microcentrifuge tubes at 4 °C. Swabs were stored from 1 to 21 days with a mean of 3.9 ± 3 days. We changed nitrile gloves between handling individual bats to prevent cross-contamination.
Fungal culture and identification
We streaked each swab five times, discretely, across three different media: sabouraud dextrose agar with chloramphenicol and gentamicin (SD; BD Diagnostic Systems, Sparks, Maryland, USA), dermatophyte test medium (DTM) with chloramphenicol, cycloheximide (added to inhibit fast-growing saprophytic fungi), and gentamicin (prepared in-house), and modified Leeming and Notman agar (LNA; prepared in-house [40]). Plates were sealed with laboratory film (Bemis Flexible Packaging, Neenah, Wisconsin) and incubated in darkness at 7 °C (to approximate typical conditions in hibernacula) for 2 months. We checked plates weekly and isolated morphologically unique fungal colonies, including Pd, in pure culture. We counted the number of colonies of each morphotype on each plate weekly to determine colony forming units (CFUs) until confluent growth precluded accurate counts. The final CFU count for each morphotype on each bat was calculated by summing the total CFUs of each morphotype on the three media per each individual bat.
We identified pure cultures by analyzing the full-length internal transcribed spacer (ITS) region of the fungal rRNA gene [41]. Sequences were collapsed into representative operational taxonomic units (OTUs) using USEARCH [42] with a 97% similarity threshold [43]. We applied a 99% similarity threshold for the genus Debaromyces because of minimal genetic variation exhibited in the ITS region among Debaromyces species [44]. We assigned taxonomy to sequences in R, using the assigntaxonomy function (DADA2 package) [45] with UNITE [46, 47]. Some sequences were not identified to genus using UNITE, and we compared these to NCBI’s Genbank database using BLAST [48]. We manually generated a community matrix of annotated OTUs and their CFUs for each bat.
Except where noted, we removed Pd from the dataset for statistical analyses to focus on skin fungal assemblage characteristics associated with WNS-susceptibility rather than differences caused by the pathogen. We also excluded fungal OTUs that were only isolated from one bat, as these are likely transient species rather than commensals.
Statistical analyses
We performed analyses in R [49]. We calculated the Shannon diversity index (hereafter, Shannon Index) of fungi on each bat using the diversity function in the vegan package [50]. To test our first sub-hypothesis that skin fungal assemblage diversity is related to WNS-susceptibility group, we constructed a Gaussian zero-inflated model with Shannon Index as the response variable and bat species (9 level factor), site (23 level factor with New York sites excluded), and WNS-susceptibility group (3 level factor) as explanatory variables (package glmmTMB) [51]. We included the number of days swabs were stored, year of collection, month of collection, state (broad spatial scale), and day-of-year swabs were collected in supplemental models to examine which explanatory variables were predictive of variation. We used the function AICtab (package bbmle) [52] to compare model Akaike information criteria (AIC) values. As yeasts are important components of the cutaneous mycobiome of mammals [53, 54], we repeated this analysis using yeasts-only Shannon Index, fungal abundance (CFU), and yeast abundance as the response variable in separate analyses. We determined the optimum model family using AIC for abundance response variables. We determined the best data transformation with the transformTukey function (package rcompanion) [55] for each response variable.
To determine if wing fungal assemblage composition varied among WNS-susceptibility groups, bat species, and sites, we implemented a non-parametric permutational multivariate analysis of variance (PERMANOVA) on abundance based (CFU) Bray-Curtis dissimilarity coefficients using the function ADONIS (vegan [50]). As Bray-Curtis dissimilarity values cannot be calculated for samples that have no composition, we first removed individual bats with no cultured fungi, individuals from which only Pd was cultured, and bats from which the only fungus cultured was the single representative of that OTU within our dataset (n = 4 Co. townsendii virginianus, n = 3 Co. rafinesquii, n = 1 E. fuscus, n = 1 My. leibii, n = 47 My. lucifugus, n = 14 My. septentrionalis, and n = 64 Per. subflavus). We ran PERMANOVA for 1000 iterations, and report R2 values when the variable enters the model last.
To test our second sub-hypothesis, we used DESeq2 (version 1.10.1, alpha = 0.05) to identify OTUs that were differentially abundant among WNS-susceptibility groups, correcting for multiple pairwise comparisons using Benjamini–Hochberg adjustment [56], and including bats with no cultured fungi (DESeq2 allows non-balanced datasets). As differential abundance analysis does not account for within-group consistency, we also performed an indicator species analysis using the multipatt function (indicspecies package) [57]. We report OTUs with an indicator statistic above 0.4 [31].
To determine if the abundance of Pd on a bat, as determined by our culture-dependent results, affected skin fungal assemblages, we constructed zero-inflated models using either fungal abundance (truncated negative binomial) or alpha diversity (represented by Shannon Index; Gaussian distribution) as the response variable, Pd abundance (CFU counts; explanatory variable), and bat species and site (random effects). The models with Shannon Index did not converge, so we determined the best data transformation with the transformTukey function (package rcompanion) [55] for both the response (Shannon Index, lambda = 0.4) and explanatory variables (Pd abundance, lambda = 0.225). To examine whether Pd abundance influenced the fungal community composition of each bat (beta diversity), a PERMANOVA was run as described above. These analyses were only run with bat species on which Pd was cultured from at least one individual bat, but included all individuals sampled within those species (My. lucifugus, My. septentrionalis, My. sodalis, and Per. subflavus).
Mycobiomes on WNS-resistant vs. susceptible populations of Myotis lucifugus
We sampled colonies of My. lucifugus from two sites in New York during winter 2014–2015 as described above. These bat colonies have persisted despite the ongoing presence of Pd since 2006 [26]. We compared samples from these resistant bat colonies to samples collected from My. lucifugus colonies farther west (Wisconsin and Kentucky) that were naïve to WNS at the time of sampling and thus considered susceptible. We used zero-inflated models with either Shannon Index (Gaussian), fungal abundance (truncated negative binomial), or yeast abundance (truncated negative binomial) as the response variable, and WNS-susceptibility group (resistant vs. susceptible) as the sole explanatory variable with site (7-level factor) as a random effect. We also performed a differential abundance analysis as described above.
Inhibition assays
We screened five yeast OTUs that were differentially enriched on WNS-resistant or impervious bats for Pd-antagonism by spore-germination and growth-inhibition assays. The production of toxins by fungi varies with environmental conditions [58]. Therefore, different types of media, pH, and salt conditions were tested, including SD (pH 5.6), brain heart infusion (BHI, pH 7.4), BHI with 10% sheep blood (pH 7.4), yeast morphology (YM) medium with the pH adjusted to either 4.5, 5.0, or 7.0 with 0.1 M citrate-phosphate buffer, and YM supplemented with 6% (w/v) NaCl at pH 5.0. We supplemented media at pH 4.5 with increased agar (2% w/v) to ensure solidification. To determine whether Pd would grow on medium with increased NaCl, we supplemented YM at pH 5.0 with NaCl at half percent intervals, inoculated with pure cultures of Pd, and incubated at 7 °C for 2 months. Pseudogymnoascus destructans grew on YM at both pH 4.5 and 5.0 (no NaCl supplementation). We saw visible Pd growth on YM pH 5.0 supplemented with 0.5% to 2.0% NaCl after 2 weeks incubation, 2.5% to 4.0% NaCl after 1 month, and 4.5% to 6.0% NaCl after 2 months. All yeast strains used in the assays also grew under these conditions.
For inhibition assays, we harvested Pd conidia (ATCC MYA-4855) from 3-month old cultures as described by Lorch et al. [14] and enumerated conidia using a hemocytometer. We spread 150 µl of conidial suspension containing two million conidia onto agar medium. We placed six pre-sterilized Whatman #1 filter paper discs equidistant from one another on each plate. We harvested yeasts from 6-day old cultures grown on SD at 7 °C by scraping cells off agar surfaces with sterile loops and suspending them in sterile phosphate-buffered saline (PBST) containing 0.5% Tween 20. We enumerated yeast cells as described above and 8 µl suspensions each containing 500,000 cells were pipetted directly onto individual filter-paper discs. We tested each yeast-strain in triplicate, incubating all plates in the dark at 7 °C. Negative controls were discs treated with PBST only and positive controls were discs containing voriconazole (30 µg; Sensi-disc; Becton, Dickinson, & Co., Franklin Lakes, New Jersey). We checked plates daily for the first week to assess inhibition of Pd germination near the discs, and then weekly for 2 months or until Pd growth had covered the entire agar surface. We measured zones of inhibition around the discs to the nearest mm after 2 weeks of incubation.
Results
The skin mycobiome differs between susceptible and resistant bat species
We processed 398 swabs from 10 bat species sampled across 10 states (Table 1). Fungi were cultured from 86.9% (346) of swabs, representing 137 fungal morphotypes of 80 genera. We found that 20.9% of OTUs were cultured from a single individual bat (Table S1). The most commonly cultured fungi were Pd (from 32.4% of bats), Debaryomyces hansenii (29.1%), Cutaneotrichosporon moniliiforme (17.3%), Malassezia vespertilionis (14.6%; newly described during this study [40]), two additional unassigned Debaryomyces species [hereafter referred to as Debaryomyces sp. 1 (14.3%) and Debaryomyces sp. 3 (12.3%)], and Cladosporium delicatulum (8.5%). Yeasts (including dimorphic fungi) comprised 32.1% of detected OTUs.
Yeasts, when present, often had high CFU counts compared to filamentous fungi. This is shown in Fig. 2 as patterns among bat species and sites for overall fungal abundance (A) are virtually identical to yeast abundance (C). In sites where multiple bat species were sampled, such as AL-1, AL-2, AL-3, KY-1, MO-1, and WI-7 (Figs. 1 and 2), WNS-resistant species, such as My. grisescens and E. fuscus, always had higher fungal and yeast abundance than WNS-susceptible species. WNS-impervious species had low fungal and yeast abundance at all sites (Fig. 2a, c). Although filamentous fungi were generally present in low abundance, their diversity was higher than yeast diversity on some bats. This is reflected in Fig. 2 where bat species with low fungal abundance (A), such as My. leibii and Corynorhinus spp., had a higher mean Shannon Index (B) compared with bat species with high fungal abundance, such as My. austroriparius. Conversely, some bat species had high abundance of one or two OTUs, but low overall diversity as reflected by the Shannon Index (e.g., My. austroriparius). WNS-susceptible species had higher diversity of filamentous fungi (Fig. 2b) compared to yeast diversity (Fig. 2d). Fungal abundance was similar among the three bat species in the WNS-susceptible group and between the two impervious Corynorhinus spp. (Fig. S1A, B).
Mean counts of fungal abundance and diversity per bat, expressed as colony forming units (CFU), and mean Shannon Index for all fungi (a, b) and yeast only (c, d) on various species of bats at different sites. Pseudogymnoascus destructans has been excluded. AR Arkansas, WV West Virginia, IA Iowa, PA Pennsylvania, WI Wisconsin, AL Alabama, KY Kentucky, NY New York, MO Missouri, OK Oklahoma. Corynorhinus spp. = Co. rafinesquii and Co. townsendii virginianus; WNS-resistant = Eptesicus fuscus, Myotis austroriparius, My. grisescens, My. leibii, My. sodalis; WNS-susceptible = My. lucifugus, My. septentrionalis, and Perimyotis subflavus.
Fungal diversity differs among bat species
The best model explaining differences in skin fungal diversity (Shannon Index as dependent variable) among individuals included bat species and site (Table S2). Some bat species, such as Per. subflavus and Co. townsendii virginianus, had significantly lower Shannon Index compared to other bat species such as E. fuscus and My. grisescens (Table S3). When bat species and site were included as random variables and WNS-susceptibility group was the sole explanatory variable, WNS-impervious (estimate = 0.1703, std error = 0.1094, p < 0.001) and WNS-susceptible (estimate = 0.1244, std error = 0.0782, p < 0.001) species had significantly lower Shannon Indices than WNS-resistant species (estimate = 0.5988, std error = 0.0600, p < 0.001) in the conditional model. None of the factors explained variance in the zero-inflation model (estimate = −23.77, std error = 13427.55, p = 0.999; i.e., there was no pattern among groups, bat species, or sites for which individuals would have a Shannon Index of zero).
Yeast-only diversity differs among bat species
The best model with yeast-only Shannon Indices as the dependent variable included bat species and month of collection (Table S2). This model indicated that some bat species, such as E. fuscus, My. grisescens, and My. austroriparius, had significantly higher yeast Shannon Indices than other bat species such as Per. subflavus (Table S3). When bat species and site were included as random variables and WNS-susceptibility group was the sole explanatory variable, WNS-impervious (estimate = 0.0938, std error = 0.0918, p < 0.001) and WNS-susceptible (estimate = 0, std error = 0.0681, p < 0.001) species had significantly lower Shannon Indices than WNS-resistant species (estimate = 0.4883, std error = 0.0509, p < 0.001) in the conditional model. None of the factors explained variance in the zero-inflation model (estimate = −25.97, std error = 45955.42, p = 1).
Fungal abundance differs among bat species
The best models with fungal abundance (CFU counts; response variable) all included bat species (Table S2). The best model (bat species and day-of-year swabs were collected) indicated that My. grisescens, My. austroriparius, and My. sodalis had significantly higher fungal abundance than other bat species (Table S3). When bat species and site were included as random variables and WNS-susceptibility group was the sole explanatory variable, WNS-impervious (estimate = 1.4299, std error = 0.4333, p = 0.0065) and WNS-susceptible (estimate = 1.0361, std error = 0.3632, p < 0.001) species had lower fungal abundance compared to WNS-resistant species (estimate = 2.6087, std error = 0.2395, p < 0.001) in the conditional model.
Yeast-only abundance differs among bat species
The best models with yeast abundance (response variable) all included bat species (Table S2). The best model (bat species and day-of-year swabs were collected) indicated that My. grisescens, My. austroriparius, and My. sodalis had significantly higher yeast abundance than other bat species (Table S3). When bat species and site were included as random variables and WNS-susceptibility group was the sole explanatory variable, WNS-susceptible (estimate = 21.25, std error = 74.84, p = 0.0013) and WNS-impervious (estimate = 23.25, std error = 86.32, p = 0.0057) species had significantly lower fungal abundance compared to WNS-resistant species (estimate = 262.09, std error = 48.54, p < 0.001).
Fungal composition differs among bat species
Skin fungal assemblage composition was significantly associated with both bat species (pseudo-F5,207 = 2.833 R2 = 0.0386, p = 0.001) and site of collection (pseudo-F18,207 = 2.725, R2 = 0.1335, p = 0.001). White-nose syndrome susceptibility group, month of collection, year of collection, number of days swabs were stored, state, and day of year swabs were collected were not significant predictors of skin fungal assemblage composition (these variables were removed because they did not improve the model). Six OTUs (all yeast) were differentially abundant among WNS-susceptibility groups and identified as indicator species (Fig. 3).
For each pairwise comparison, the WNS-susceptibility group listed first was the baseline and the second was the comparison. Log2change values above zero indicate that the OTUs were more abundant in the comparison group compared to the baseline. OTUs that were significant indicator species (indicator statistic > 0.4, p < 0.05) for each WNS-susceptibility group are shown with blue boxes. Debaryomyces sp. 3 is an indicator species for both WNS-resistant and WNS-impervious species of bats. All other fungal indicator species are for WNS-resistant species of bats alone. P values were corrected for multiple hypothesis testing in both tests.
Relationship between Pd and the skin mycobiome
Pd was only isolated from four bat species: from 5.1% of My. septentrionalis (n = 39), 53.6% of My. sodalis (n = 28), 49.4% of Per. subflauvs (n = 79), 91.8% of My. lucifugus sampled in New York (n = 61), and from 32.6% of My. lucifugus sampled elsewhere (n = 43; Table S4). When present, Pd was abundant (high CFU counts) on My. lucifugus, but had low abundance on the only WNS-resistant species from which Pd was cultured during this study, My. sodalis (Table S4). Neither Shannon Index nor fungal abundance significantly varied with Pd abundance in either the conditional model (estimate = 0.2692, std error = 0.0194, p = 0.495 for Shannon Index; estimate = −0.0178 on log scale, std error = 0.0172, p = 0.302) or zero-inflation model (estimate = −25.17, std error = 31587.79, p = 0.999 for Shannon Index; estimate = 0.0006, std error = 0.0012, p = 0.583 for fungal abundance). The composition of bat skin fungal assemblages was predicted by both bat species (pseudo-F1,127 = 4.8693, R2 = 0.0287, p = 0.001) and site (pseudo-F13,127 = 2.2349, R2 = 0.1713, p = 0.001), but not Pd abundance (pseudo-F1,127 = 1.4216, R2 = 0.0084, p = 0.086).
Mycobiomes of WNS-resistant versus WNS-susceptible populations of Myotis lucifugus
Resistant My. lucifugus in New York had lower fungal abundance (estimate = −16.934 on log scale, std error = 0.6056, p < 0.001), lower yeast abundance (estimate = −14.8541, std error = 0.7492, p < 0.001), and more individuals with zero yeast colonies (estimate = 0.5521, std error = 0.2036, p = 0.0067) compared to susceptible My. lucifugus in Wisconsin and Kentucky. However, resistant My. lucifugus colonies did not have more individuals with zero fungal colonies (estimate = −0.1929, std error = 0.197, p = 0.328) or different Shannon Indices (estimate = 0.0484, std error = 0.08554, p = 0.1627) than susceptible My. lucifugus. Malassezia vespertilionis was identified as differentially abundant on susceptible as compared to resistant My. lucifugus (log2Change = −5.897, p < 0.001).
Skin mycobiome constituents from WNS-resistant bats inhibit Pd under certain conditions
Of the yeast strains tested, two isolates of Cu. moniliiforme inhibited Pd in vitro under certain conditions (Table 2). On YM at pH 5.0 with 6% NaCl medium, Pd grew slowly with contorted hyphae (germination of spores was limited and first seen after 4 weeks, but inhibition results could only be assessed 7 weeks post-inoculation), indicating Pd was likely stressed. The diameter of inhibition rings decreased over time and completely disappeared on all plates, including positive controls, after 4 weeks of incubation. The only exception was the positive control YM pH 5 with 6% NaCl, on which very limited germination of Pd was first observed after 10 weeks of incubation.
Discussion
We hypothesized that the skin microbiomes of bats would vary based on WNS-susceptibility group. More specifically, we tested the sub-hypothesis that WNS-resistant bat species would have more diverse and abundant skin fungal assemblages as compared to susceptible bat species. Although resistant species did have higher Shannon Indices and fungal abundance compared to susceptible species, bat species that are impervious to Pd infection did not. Njus [59] also found that bat species with low WNS-associated mortality (Co. townsendii and E. fuscus) were disproportionally colonized by yeasts, particularly Debaryomyces spp., compared to species with high WNS-associated mortality (My. lucifugus). The lack of culturable fungi on WNS-susceptible and impervious species was supported by the absence of non-Pd fungi observed on My. lucifugus, Per. subflavus, and Co. rafinesquii wings using SEM (Appendix 1; Fig. S2). We also found that susceptible My. lucifugus colonies had higher fungal abundance compared to resistant colonies. Therefore, mechanisms other than skin fungal assemblages contribute to infection outcomes in WNS-impervious Corynorhinus spp. and persisting colonies of WNS-resistant My. lucifugus. Disease resistance is often multifactorial, and combinations of host genetic, physiological, and behavioral characteristics also likely influence WNS susceptibility [60].
Distinguishing transient fungi from commensals is a common challenge for skin microbial assemblage studies [61, 62]. Many genera we isolated (e.g., Penicillium, Cladosporium, non-Pd Pseudogymnoascus, and Mortierella) are commonly isolated from cave environments and considered saprotrophs that are unlikely to colonize bat skin [63,64,65,66]. In contrast, the high abundance of certain yeast taxa (e.g., Debaryomyces, Cutaneotrichosporon, Leucosporidium, and Ma. vespertilionis), combined with our SEM observations of yeasts budding on the skin of resistant bat species, indicate they are commensals. Several of these yeasts may represent novel taxa (Appendix 1; Fig. S3; Tables S5 and S6) that could be adapted to living on bat skin, and further work is needed to characterize these bat-associated strains. Many yeasts we isolated have been documented from cold regions (e.g., Antarctica and glacial habitats), saline and acidic environments [67,68,69], human skin fungal assemblages [53, 70, 71], and bats in tropical habitats [72,73,74,75,76,77,78]. These patterns suggest that these yeasts are commensals and thus might affect disease susceptibility.
In this study, we employed a culture-based approach to examine the mycobiome of bats, which has several limitations. Specifically, taxa that are rare or unculturable may be overlooked. However, when we compared our culture results to those based on next generation sequencing (NGS) from a subset of samples (Appendix 1; Tables S7 and S8), we found that our culture-based methods typically detected the fungi most likely to be skin commensals on bats. Further, the use of NGS presents a different set of drawbacks. For example, although NGS can determine relative abundance of various taxa, calculating absolute abundance of particular organisms is more difficult. Samples that do not yield satisfactory sequence data are routinely excluded from further analyses, meaning that negative datapoints are often discarded rather than given equal weight in analyses. Through the use of zero-inflated models, we were able to retain samples with many non-detections in our dataset. This demonstrated that WNS-resistant species of bats were significantly more likely to have culturable fungi than WNS-susceptible or WNS-impervious species of bats. Finally, our culture-based analysis yielded isolates of skin commensals for downstream experiments such as Pd inhibition assays and further genetic characterization of potentially novel fungal taxa.
Geographic location (site) is an important explanatory variable for the overall diversity and composition of skin fungal assemblages. This pattern was previously found for bat-skin fungal assemblages [66, 79], bat-skin bacterial assemblages [31, 80,81,82], and amphibian skin bacterial assemblages [83, 84]. Microbes that constitute an organism’s skin microbial assemblages are primarily drawn from local environments [84], and microbial diversity is influenced by numerous abiotic factors [85]. Factors that influence cave fungal assemblages include quantity of organic material and water, cave chemistry, temperature, cave size and depth, and number and diversity of animals [86]. Although we did not characterize environmental fungal assemblages, likely each location we sampled possessed a unique fungal assemblage. Each site was only sampled once and sites within a state were all sampled by the same collection team (which could affect results). Therefore, factors such as day-of-year and month samples were collected, and number of days swabs were stored are proxies for the variable site. Overall, bats in southern states (Alabama, Kentucky, Missouri) had higher fungal abundance than bats in northern states, indicating that temperature or other environmental factors associated with the region could be an important determinant of fungal abundance. However, because many bat species with relatively high fungal abundance (My. austroriparius, My. sodalis, and My. grisescens) were exclusively sampled in southern states (due primarily to restricted geographic ranges for those bat species), it is difficult to decouple the effects of location and certain environmental parameters from species.
The diversity of fungal species occupying the skin is not the only way in which the microbiome may modulate disease resistance. Instead, resistance may be related to the presence of a single key species that acts as an antagonist to a pathogen. For our second sub-hypothesis, we hypothesized WNS-resistant species would harbor fungal species that were comparatively rare on WNS-susceptible species, and that these fungi would inhibit Pd in vitro. Each WNS-susceptibility group contained at least one fungal species that was enriched within it, all of which were yeasts. The WNS-resistant group had multiple differentially enriched yeasts, one of which, Cu. moniliiforme, reduced Pd growth in vitro, indicating direct antagonistic interactions of this yeast with Pd may be a mechanism of WNS-resistance by slowing disease progression. Inhibition was limited to specific pH and salinity conditions, and thus other yeasts might inhibit Pd under untested conditions. The production of antifungal compounds by fungi varies with pH, salinity, temperature, and nitrogen source included in media [58, 87]. Studies of physical conditions on the surface of bat skin are necessary to determine the biological relevance of our inhibition assays. Other mechanisms by commensal yeasts may also mediate resistance to Pd infection, such as through competition or facilitating immunostimulation in response to infection.
Microbes that inhibit fungi have been proposed for use in WNS control efforts [88, 89], but their effectiveness is difficult to gauge without understanding underlying host factors that influence microbiomes and favor potentially protective microorganisms. We found fungal taxa that were abundant on resistant bats were rare on susceptible bats sampled at the same time from the same locations. This may indicate that potentially protective microbes are not easily established on WNS-susceptible hosts, which may have different skin chemistries or different microclimate preferences within hibernacula compared to WNS-resistant bats. Therefore, applications of these yeasts on bats may not confer resistance if they cannot colonize, or if environmental conditions are not conducive for yeasts to inhibit Pd. Furthermore, although we demonstrated an association between components of skin fungal assemblages and resistance to WNS, it does not prove that commensal fungi directly protect bats from Pd. Skin chemistry, physiological, or behavioral traits that promote yeast commensals on some bats may also result in suboptimal conditions for Pd growth. Nonetheless, the potential of certain yeasts to protect bats from WNS warrants further investigation, as does the ability to predict WNS-susceptibility of bat populations using skin fungal assemblage characteristics.
References
Mallon CA, Van Elsas JD, Salles JF. Microbial invasions: the process, patterns, and mechanisms. Trends Microbiol. 2015;23:719–29.
Case TJ. Invasion resistance arises in strongly interacting species-rich model competition communities. Proc Natl Acad Sci. 1990;87:9610–4.
Kennedy TA, Naeem S, Howe KM, Knops JM, Tilman D, Reich P. Biodiversity as a barrier to ecological invasion. Nature. 2002;417:636–8.
Harris RN, Brucker R, Walke J, Becker M, Schwantes C. Skin microbes on frogs prevent morbidity and mortality caused by a lethal skin fungus. ISME J. 2009;3:818–24.
Becker MH, Harris RN. Cutaneous bacteria of the redback salamander prevent morbidity associated with a lethal disease. PLoS ONE. 2010;5:e10957.
Chen EY, Fischbach MA, Belkaid Y. Skin microbiota-host interactions. Nature. 2018;553:427–36.
Harder J, Schröder JM, Gläser R. The skin surface as antimicrobial barrier: present concepts and future outlooks. Exp Dermatol. 2013;22:1–5.
Huffnagle GB, Noverr MC. The emerging world of the fungal microbiome. Trends Microbiol. 2013;21:334–41.
Kong HH, Morris A. The emerging importance and challenges of the human mycobiome. Virulence. 2017;8:310–2.
Cui L, Morris A, Ghedin E. The human mycobiome in health and disease. Genome Med. 2013;5:63.
Kearns PJ, Fischer S, Fernández-Beaskoetxea S, Gabor CR, Bosch J, Bowen JL, et al. Fight fungi with fungi: antifungal properties of the amphibian mycobiome. Front Microbiol. 2017;8:2494.
Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–73.
Allender MC, Baker S, Britton M, Kent AD. Snake fungal disease alters skin bacterial and fungal diversity in an endangered rattlesnake. Sci Rep. 2018;8:1–9.
Lorch JM, Meteyer CU, Behr MJ, Boyles JG, Cryan PM, Hicks AC, et al. Experimental infection of bats with Geomyces destructans causes white-nose syndrome. Nature. 2011;480:376–8.
Frick W, Puechmaille S, Willis C. White-nose syndrome in bats. In: Voigt C, Kingston T, editors. Bats in the Anthropocene: conservation of bats in a changing world. Heidelberg, New York: Springer International Publishing; 2016. p. 245–62.
Cryan PM, Meteyer CU, Blehert DS, Lorch JM, Reeder DM, Turner GG, et al. Electrolyte depletion in white-nose syndrome bats. J Wildl Dis. 2013;49:398–402.
US Fish and Wildlife Service. North American bat death toll exceeds 5.5 million from white-nose syndrome. 2012. http://static.whitenosesyndrome.org/sites/default/%0Afiles/files/wns_mortality_2012_nr_final_0.pdf.
Solari S. Myotis lucifugus. IUCN Red List of Threatened Species. Version 2020-2. 2018;e.T14176A22056344. https://www.iucnredlist.org.
Turner G, Reeder D, Coleman J. A five-year assessment of mortality and geographic spread of white-nose syndrome in North American bats and a look to the future. Bat Res N. 2011;52:13–27.
Langwig KE, Frick WF, Bried JT, Hicks AC, Kunz TH, Marm, et al. Sociality, density-dependence and microclimates determine the persistence of populations suffering from a novel fungal disease, white-nose syndrome. Ecol Lett. 2012;15:1050–7.
Frank CL, Michalski A, McDonough AA, Rahimian M, Rudd RJ, Herzog C. The resistance of a North American bat species (Eptesicus fuscus) to white-nose syndrome (WNS). PLoS ONE. 2014;9:e113958.
Frick W, Cheng T, Langwig K, Hoyt J, Janicki A, Parise K, et al. Pathogen dynamics during invasion and establishment of white-nose syndrome explain mechanisms of host persistence. Ecology. 2017;98:624–31.
Bernard RF, Willcox EV, Parise KL, Foster JT, McCracken GF. White-nose syndrome fungus, Pseudogymnoascus destructans, on bats captured emerging from caves during winter in the southeastern United States. BMC Zool. 2017;2:12.
Bernard RF, Foster JT, Willcox EV, Parise KL, McCracken GF. Molecular detection of the causative agent of white-nose syndrome on Rafinesque’s Big-eared Bats (Corynorhinus rafinesquii) and two species of migratory bats in the Southeastern USA. J Wildl Dis. 2015;51:519–22.
White-Nose Syndrome Response Team. Bats affected by WNS. 2018. https://www.whitenosesyndrome.org/static-page/bats-affected-by-wns.
Reichard JD, Fuller NW, Bennett AB, Darling SR, Moore MS, Langwig KE, et al. Interannual survival of Myotis lucifugus (Chiroptera: Vespertilionidae) near the epicenter of white-nose syndrome. Northeast Nat. 2014;21:N56–9.
Warnecke L, Turner JM, Bollinger TK, Lorch JM, Misra V, Cryan PM, et al. Inoculation of bats with European Geomyces destructans supports the novel pathogen hypothesis for the origin of white-nose syndrome. Proc Natl Acad Sci. 2012;109:6999–7003.
Frank CL, Ingala MR, Ravenelle RE, Dougherty-Howard K, Wicks SO, Herzog C, et al. The effects of cutaneous fatty acids on the growth of Pseudogymnoascus destructans, the etiological agent of white-nose syndrome (WNS). PLoS ONE. 2016;11:e0153535.
Field KA, Johnson JS, Lilley TM, Reeder SM, Rogers EJ, Behr MJ, et al. The white-nose syndrome transcriptome: activation of anti-fungal host responses in wing tissue of hibernating Little Brown Myotis. PLoS Pathog. 2015;11:e1005168.
Hayman DTS, Cryan PM, Fricker PD, Dannemiller NG. Long-term video surveillance and automated analyses reveal arousal patterns in groups of hibernating bats. Methods Ecol Evol. 2017;8:1813–21.
Lemieux-Labonté V, Simard A, Willis CKR, Lapointe F-J. Enrichment of beneficial bacteria in the skin microbiota of bats persisting with white-nose syndrome. Microbiome. 2017;5:115.
Moore MS, Field KA, Behr MJ, Turner GG, Furze ME, Stern DWF, et al. Energy conserving thermoregulatory patterns and lower disease severity in a bat resistant to the impacts of white-nose syndrome. J Comp Physiol B Biochem Syst Environ Physiol. 2018;188:163–76.
Davy CM, Donaldson ME, Willis CKR, Saville BJ, McGuire LP, Mayberry H, et al. Environmentally persistent pathogens present unique challenges for studies of host-pathogen interactions: Reply to Field (2018). Ecol Evol. 2018;8:5238–41.
Thogmartin WE, King RA, McKann PC, Szymanski JA, Pruitt L. Population-level impact of white-nose syndrome on the endangered Indiana bat. J Mammal. 2012;93:1086–98.
Powers KE, Reynolds RJ, Orndorff W, Hyzy BA, Hobson CS, Ford WM. Monitoring the status of Gray Bats (Myotis grisescens) in Virginia, 2009–2014, and potential impacts of white-nose syndrome. Southeast Nat. 2016;15:127–37.
Bernard RF, McCracken GF. Winter behavior of bats and the progression of white-nose syndrome in the southeastern United States. Ecol Evol. 2017;7:1487–96.
US Geological Survey. Alabama survey finds southeastern bat with white-nose syndrome. 2017. https://www.usgs.gov/news/alabama-survey-finds-first-southeastern-bat-white-nose-syndrome.
Powers KE, Reynolds RJ, Orndorff W, Ford WM, Hobson CS. Post-white-nose syndrome trends in Virginias cave bats. 2008-2013 J Ecol Nat Environ. 2015;7:113–23.
Ingersoll TE, Sewall BJ, Amelon SK. Improved analysis of long-term monitoring data demonstrates marked regional declines of bat populations in the Eastern United States. PLoS ONE. 2013;8:e65907.
Lorch J, Palmer J, Vanderwolf K, Schmidt K, Verant M, Weller T, et al. Malassezia vespertilionis sp. nov.: a new cold-tolerant species of yeast isolated from bats. Persoonia. 2018;41:56–70. https://doi.org/10.3767/persoonia.2018.41.04.
Lorch JM, Minnis AM, Meteyer CU, Redell JA, White JP, Kaarakka HM, et al. The fungus Trichophyton redellii sp. nov. causes skin infections that resemble white-nose syndrome of hibernating bats. J Wildl Dis. 2015;51:36–47.
Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–1.
O’Brien HE, Parrent JL, Jackson JA, Moncalvo JM, Vilgalys R. Fungal community analysis by large-scale sequencing of environmental samples. Appl Environ Microbiol. 2005;71:5544–50.
Martorell P, Fernández-Espinar MT, Querol A. Sequence-based identification of species belonging to the genus Debaryomyces. FEMS Yeast Res. 2005;5:1157–65.
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–3.
Kõljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AFS, Bahram M, et al. Towards a unified paradigm for sequence-based identification of fungi. Mol Ecol. 2013;22:5271–7.
Community U. UNITE general FASTA release. 2017. p. Version 01.12.2017. https://doi.org/10.15156/BIO/587475.
Altschul S, Gish W, Miller W, Myers E, Lipman D. Basic local alignment search tool. J Mol Biol. 1990;215:403–10.
Team RC. R: a language and environment for statistical computing. R Foundation for Statistical Computing. 2017. https://www.r-project.org/.
Oksanen J, Blanchet F, Friendly M, Kindt R, Legendre P, McGlinn D, et al. Vegan: community ecology package. R package version 2.5-2. 2018. https://cran.r-project.org/package=vegan.
Brooks ME, Kristensen K, van Benthem KJ, Magnusson A, Berg CW, Nielsen A, et al. Modeling zero-inflated count data with glmmTMB. 2017;132753. https://www.biorxiv.org/content/early/2017/05/01/132753.
Bolker B, Team RDC. bbmle: tools for general maximum likelihood estimation. R Packag version 1020. 2017. https://cran.r-project.org/package=bbmle.
Byrd A, Belkaid Y, Segre J. The human skin microbiome. Nat Rev Microbiol. 2018;16:143–55.
Mason IS, Mason KV, Lloyd DH. A review of the biology of canine skin with respect to the commensals Staphylococcus intermedius, Demodex canis and Malassezia pachydermatis. Vet Dermatol. 1996;7:119–32.
Mangiafico S. rcompanion: functions to support extension education program evaluation. R Packag version 2010. 2019. https://cran.r-project.org/package=rcompanion.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. https://doi.org/10.1186/s13059-014-0550-8.
De Cáceres M, Legendre P. Associations between species and groups of sites: indices and statistical inference. Ecology. 2009;90:3566–74.
Marquina D, Barroso J, Santos A, Peinado JM. Production and characteristics of Debaryomyces hansenii killer toxin. Microbiol Res. 2001;156:387–91.
Njus K. Molecular techniques for the identification of commensal fungal populations on cave roosting bats. University of Akron; 2014.
Medzhitov R, Schneider D, Soares M. Disease tolerance as a defense strategy. Science. 2012;335:936–41.
Kramer R, Sauer-Heilborn A, Welte T, Guzman CA, Abraham WR, Höfle MG. Cohort study of airway mycobiome in adult cystic fibrosis patients: differences in community structure between fungi and bacteria reveal predominance of transient fungal elements. J Clin Microbiol. 2015;53:2900–7.
Nash AK, Auchtung TA, Wong MC, Smith DP, Gesell JR, Ross MC, et al. The gut mycobiome of the Human Microbiome Project healthy cohort. Microbiome. 2017;5:153.
Johnson LJAN, Miller AN, McCleery RA, McClanahan R, Kath JA, Lueschow S, et al. Psychrophilic and psychrotolerant fungi on bats and the presence of Geomyces spp. on bat wings prior to the arrival of white nose syndrom. Appl Environ Microbiol. 2013;79:5465–71.
Lorch JM, Lindner DL, Gargas A, Muller LK, Minnis AM, Blehert DS. A culture-based survey of fungi in soil from bat hibernacula in the eastern United States and its implications for detection of Geomyces destructans, the causal agent of bat white-nose syndrome. Mycologia. 2013;105:237–52.
Vanderwolf KJ, McAlpine DF, Malloch D, Forbes GJ. Ectomycota associated with hibernating bats in Eastern Canadian Caves prior to the emergence of white-nose syndrome. Northeast Nat. 2013;20:115–30.
Vanderwolf KJ, Malloch D, McAlpine DF. Fungi on white-nose infected bats (Myotis spp.) in Eastern Canada show no decline in diversity associated with Pseudogymnoascus destructans (Ascomycota: Pseudeurotiaceae). Int J Speleol. 2016;45:43–50.
Middelhoven WJ, Koorevaar M, Schuur GW. Degradation of benzene compounds by yeasts in acidic soils. Plant Soil. 1992;145:37–43.
Buzzini P, Branda E, Goretti M, Turchetti B. Psychrophilic yeasts from worldwide glacial habitats: diversity, adaptation strategies and biotechnological potential. FEMS Microbiol Ecol. 2012;82:217–41.
Mokhtarnejad L, Arzanlou M, Babai-Ahari A, Di Mauro S, Onofri A, Buzzini P, et al. Characterization of basidiomycetous yeasts in hypersaline soils of the Urmia Lake National Park, Iran. Extremophiles. 2016;20:915–28.
Pfaller MA, Diekema DJ, Rinaldi MG, Barnes R, Hu B, Veselov AV, et al. Results from the ARTEMIS DISK global antifungal surveillance study: a 6.5-year analysis of susceptibilities of Candida and other yeast species to fluconazole and voriconazole by standardized disk diffusion testing. J Clin Microbiol. 2005;43:5848–59.
Jo JH, Kennedy EA, Kong HH. Topographical and physiological differences of the skin mycobiome in health and disease. Virulence. 2017;8:324–33.
Mok WY, Luizao RCC, Barreto Da Silva MS. Isolation of fungi from bats of the Amazon Basin. Appl Environ Microbiol. 1982;44:570–5.
Gandra RF, Gambale W, De Cássia Garcia Simão R, Da Silva Ruiz L, Durigon EL, De Camargo LMA, et al. Malassezia spp. in acoustic meatus of bats (Molossus molossus) of the Amazon region, Brazil. Mycopathologia. 2008;165:21–6.
Belisle M, Mendenhall CD, Oviedo Brenes F, Fukami T. Temporal variation in fungal communities associated with tropical hummingbirds and nectarivorous bats. Fungal Ecol. 2014;12:44–51.
Brilhante RSN, Maia-Júnior JE, Oliveira JS, Guedes GMM, Silva AL, Moura FBP, et al. Yeasts from the microbiota of bats: a focus on the identification and antimicrobial susceptibility of cryptic species of Candida. J Med Microbiol. 2016;65:1225–8.
Oyeka C. Isolation of Candida species from bats in Nigeria. Mycoses. 1994;37:353–5.
Grose E, Marinkelle C, Striegel C. The use of tissue cultures in the identification of Cryptococcus neoformans isolated from Colombian bats. Sabouraudia. 1968;6:127–32.
Grose E, Marinkelle C. Species of Sporotrichum, Trichophyton, and Microsporum from Columbian bats. Trop Geogr Med. 1966;18:260–3.
Vanderwolf KJ, Malloch D, McAlpine DF. Fungi associated with over-wintering tricolored bats, Perimyotis subflavus, in a white-nose syndrome region of eastern Canada. J Cave Karst Stud. 2015;77:145–51.
Avena CV, Parfrey LW, Leff JW, Archer HM, Frick WF, Langwig KE, et al. Deconstructing the bat skin microbiome: Influences of the host and the environment. Front Microbiol. 2016;7:1753.
Winter AS, Hathaway JJM, Kimble JC, Buecher DC, Valdez EW, Porras-Alfaro A, et al. Skin and fur bacterial diversity and community structure on American southwestern bats: effects of habitat, geography and bat traits. PeerJ. 2017;5:e3944.
Lemieux-Labonté V, Tromas N, Shapiro BJ, Lapointe F-J. Environment and host species shape the skin microbiome of captive neotropical bats. PeerJ. 2016;4:e2430.
Walke JB, Becker MH, Loftus SC, House LL, Cormier G, Jensen RV, et al. Amphibian skin may select for rare environmental microbes. ISME J. 2014;8:2207–17.
Loudon AH, Woodhams DC, Parfrey LW, Archer H, Knight R, McKenzie V, et al. Microbial community dynamics and effect of environmental microbial reservoirs on red-backed salamanders (Plethodon cinereus). ISME J. 2014;8:830–40.
Fierer N, Jackson RB. The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA. 2006;103:626–31.
Vanderwolf KJ, Malloch D, Mcalpine DF, Forbes GJ. A world review of fungi, yeasts, and slime molds in caves. Int J Speleol. 2013;42:77–96.
Gasparetti C, Buzzini P, Cramarossa MR, Turchetti B, Pagnoni UM, Forti L. Application of the response surface methodology (RSM) for optimizing the production of volatile organic compounds (VOCs) by Trichosporon moniliiforme. Enzym Micro Technol. 2006;39:1341–6.
Hoyt J, Cheng T, Langwig K, Hee M, Frick W, Kilpatrick A. Bacteria isolated from bats inhibit the growth of Pseudogymnoascus destructans, the causative agent of white-nose syndrome. PLoS ONE. 2015;10:e0121329. https://doi.org/10.1371/journal.pone.0121329.
Cheng TL, Mayberry H, McGuire LP, Hoyt JR, Langwig KE, Nguyen H, et al. Efficacy of a probiotic bacterium to treat bats affected by the disease white-nose syndrome. J Appl Ecol. 2017;54:701–8.
Vanderwolf KJ, Campbell LJ, Lorch JM. Skin mycobiomes of eastern North American bats. USGS Data Release 2020. https://www.sciencebase.gov/catalog/item/5cdef4c2e4b038687e296912.
Acknowledgements
We thank the many field biologists who coordinated sampling efforts and collected samples, including Anne Ballmann, Shelly Colatskie, Katie O’Connor, Tony Elliott, Kyle George, Lindsey Heffernan, Carl Herzog, Catherine Hoose, Daryl Howell, Jennifer Redell, Blake Sasse, Michael Scafini, Nick Sharp, Kirk Silas, Craig Stihler, Greg Turner, Paul White, Rebecca Williams, and Michelle Verant. We are most grateful to Rick Lankau for facilitating the culture-independent analysis, Doerte Doepfer and James Paterson for providing statistical advice, and Katie Schmidt and Daniel Taylor for providing technical assistance with laboratory work. We thank the University of Wisconsin Biotechnology Center DNA Sequencing Facility and Newcomb Imaging Center for providing next-generation sequencing and electron microscopy services, respectively. This work was funded by the U.S. Fish and Wildlife Service, the U.S. Geological Survey, and a Natural Sciences and Engineering Research Council of Canada postgraduate scholarship to KJV. Portions of this work were conducted by KJV and LJC while serving as volunteers at the U.S. Geological Survey. The use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. Government. Metadata associated with this publication is available in machine-readable format at https://doi.org/10.5066/P9Y54WW4 [90]. Scripts used in the analysis of this project are available from the GitHub repository URL https://github.com/zoolew/Bat-Wing-Mycobiomes.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing financial interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Vanderwolf, K.J., Campbell, L.J., Goldberg, T.L. et al. Skin fungal assemblages of bats vary based on susceptibility to white-nose syndrome. ISME J 15, 909–920 (2021). https://doi.org/10.1038/s41396-020-00821-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-020-00821-w