Abstract
The rhizosphere is a hotspot for microbial carbon transformations, and is the entry point for root polysaccharides and polymeric carbohydrates that are important precursors to soil organic matter (SOM). However, the ecological mechanisms that underpin rhizosphere carbohydrate depolymerization are poorly understood. Using Avena fatua, a common annual grass, we analyzed time-resolved metatranscriptomes to compare microbial functions in rhizosphere, detritusphere, and combined rhizosphere–detritusphere habitats. Transcripts were binned using a unique reference database generated from soil isolate genomes, single-cell amplified genomes, metagenomes, and stable isotope probing metagenomes. While soil habitat significantly affected both community composition and overall gene expression, the succession of microbial functions occurred at a faster time scale than compositional changes. Using hierarchical clustering of upregulated decomposition genes, we identified four distinct microbial guilds populated by taxa whose functional succession patterns suggest specialization for substrates provided by fresh growing roots, decaying root detritus, the combination of live and decaying root biomass, or aging root material. Carbohydrate depolymerization genes were consistently upregulated in the rhizosphere, and both taxonomic and functional diversity were highest in the combined rhizosphere–detritusphere, suggesting coexistence of rhizosphere guilds is facilitated by niche differentiation. Metatranscriptome-defined guilds provide a framework to model rhizosphere succession and its consequences for soil carbon cycling.
Similar content being viewed by others
Introduction
The rhizosphere is a critical zone for C transformations in the terrestrial biosphere, since roots are the primary source of soil organic matter (SOM) [1,2,3,4,5] and can significantly alter the rate of soil C turnover [6,7,8]. Plants deposit a significant proportion of their photosynthates into soil as root biomass or exudates [9], and plant-derived polymeric carbohydrates such as cellulose and hemicellulose are the most abundant polysaccharides in soil [10, 11]. These rhizodeposits create a high-resource, high-activity environment, and stimulate a bloom of microbial biomass [12] that undergoes ecological succession as roots grow and senesce [13, 14], selecting for organisms that benefit mineral nutrition [15] and overall plant health [16]. Rhizodeposits also stimulate depolymerization by cellulases [17], chitinases, proteases [18], and hydrocarbon degradation genes [19], and can lead to higher [6, 20] or lower [6] rates of SOM decomposition in the region surrounding both living roots and decaying root detritus. However, the ecological controls of rhizosphere carbohydrate depolymerization are not well understood, which limits our ability to predict the outcome of soil C dynamics [21] and plant-microbe interactions.
Previous studies suggest that rhizosphere community assembly is controlled by abiotic and biotic selective factors [22,23,24,25]. The rhizosphere appears to select for microbial genomes that are enriched in carbohydrate active enzyme (CAZy) genes compared with bulk soil [22, 24, 26, 27]. However, it is unclear if this large genomic potential translates to high carbohydrate degradation activity in the environment. Genomic composition represents the full functional repertoire of a microorganism, which is the “fundamental metabolic niche” that constrains all the potential habitats it could hypothetically occupy [28, 29]. But microbial communities contain functional redundancy that is not necessarily realized or expressed in the ecosystem [30]. To understand “realized” metabolic niches within complex rapidly changing microbial communities [30], it is essential to consider expressed functional measurements—such as transcripts, proteins, or metabolites—that can reflect niche differentiation in real time [31, 32].
Measurements of expressed functions provide a useful way to study community assembly based on shared activities rather than shared phylogeny, and allow us to define microbial guilds—cohorts of organisms defined by similar function that are not dependent on phylogeny [33]. In cases like the rhizosphere and detritusphere, where communities might logically be defined by functional traits rather than taxonomy, guilds defined by gene expression, rather than species, may be the most relevant parameter for understanding patterns of diversity [34], modeling community interactions [35], and identifying the gene transcripts that mediate root-accelerated decomposition. However, the ideal parameters for identifying or operationally defining guilds in microbial communities are unresolved. Microbial guilds have been identified previously by adding single substrates to soil and measuring subsequent increases in taxonomic relative abundance [36], as well as by defining groups based on functional genes [37] and metagenomic gene content [34]. We predict that functional guilds can also be identified using population-resolved gene expression, where guild members turn on and off genes in a coherent spatial or temporal manner in response to the same habitat, resources, or environmental perturbations. Genome-centric analyses now enable us to track transcription in specific taxonomic groups, which may be a more relevant approach than grouping transcripts across disparate classes or phyla [38].
Using comparative metatranscriptomics, we studied microbial degradation of macromolecular plant compounds over time, hypothesizing that gene expression would reflect distinct functional succession patterns in the rhizosphere and detritusphere—consistent with niche differentiation. Niche differentiation is difficult to detect in natural environments [25, 39], and therefore we accentuated the role of taxa functioning in distinct niches over space and time by examining the rhizosphere and detritusphere in isolation and where they coexist (the “combined rhizosphere–detritusphere”). The transcripts were extracted from soil near live and decaying roots in microcosms containing Avena fatua, a common annual grass, growing in its native soil. Aggregated population transcripts measured over the course of 3 weeks were binned using a genome-resolved reference database specific to our experimental soil. We found that carbohydrate depolymerization was executed by a series of microbial guilds, with distinct spatial and temporal response patterns in gene expression. We tested whether these guilds had differing life history traits based on their preferred substrate (rhizosphere or detritusphere), and assessed whether carbohydrate depolymerization expression was controlled by: (a) increasing abundance, (b) upregulating transcription, or (c) synergistically upregulating transcription in response to combined resources (i.e., combined rhizosphere–detritusphere). Our work provides a mechanistic framework for understanding the drivers of rhizosphere succession and identifies gene transcripts mediating decomposition in the rhizosphere.
Methods
Experimental design
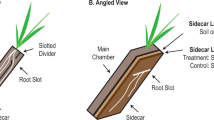
Common wild oat (Avena fatua, Pacific Coast Seed Inc) was grown in two-chamber microcosms with a sidecar region designed to allow access to the rhizosphere (Fig. S1) [12, 18, 40]. The outer wall of the sidecar was clear plastic, allowing us to monitor root growth and rhizosphere age. As previously [13], A. fatua microcosms were packed with soil (1.2 g/cm3) collected beneath a stand of A. barbata at the Hopland Research and Extension Center (Hopland, CA, USA). The soil is a Bearwallow–Hellman loam, pH 5.6, with 2% total C [41]. Plants were grown in the main chamber for 6 weeks before starting the experiment. Six days prior, the divider separating the main chamber and sidecar was replaced with a slotted divider, the sidecar was filled with experimental soil, and microcosms were tilted 40°, allowing roots to grow into the sidecar. The surfaces of half of the sidecars were amended with dried A. fatua root detritus chopped to 1 mm (also called “root litter”); detritus was grown in sand, triple washed, dried, and aged 1 year. Detritus was applied by amending 50 g soil with 0.4 g root detritus and spread on top of 100 g of sidecar soil, and unamended soil was spread on top of the no-detritus controls; living roots were only collected from this surface zone. A 1 µm mesh “bulk soil” bag was placed on top of each sidecar, which was designed to exclude roots and prevent development of a rhizosphere environment but allow moisture equilibration. Bulk soil bags contained 2 g soil, either amended with 0.016 g detritus (bulk + detritus) or unamended (bulk).
Root age was tracked in order to collect rhizosphere soil of defined age. New root growth was marked on the outside of the plastic wall using a waterproof marker 3 days after the microcosms were tilted, and harvests took place 3, 6, 12, or 22 days later (Fig. S1). At each timepoint, we destructively sampled paired rhizosphere and bulk soil for two treatments (with and without detritus) with three biological replicates, collecting 48 total samples (24 rhizosphere, 24 bulk).
Sample collection
Rhizosphere soil <2 mm from the root was excised with a scalpel. Root sections and adhering soil were placed immediately in ice-cold Lifeguard Soil Preservation Reagent (MoBio), vortexed for 2 min on medium speed, and pelleted according to the Lifeguard protocol. Roots were removed using flame-sterilized tweezers and supernatant removed. Pelleted soils were frozen on dry ice and stored at −80°C. Bulk soils were processed identically. Approximately 1 g of soil was collected per sample. Rhizosphere soil and root endophyte microbial cells were also prepared for single cell sorting to create single amplified genomes (SAGs). Briefly, roots with attached rhizosphere soil were washed in cell release buffer (0.5% tween, 2.24 mM Na pyrophosphate in PBS) to create a rhizosphere cell suspension; roots were later sterilized and then macerated (see Supplementary Methods for more detail).
The remaining sidecar soil was collected for edaphic characterization. Soil pH was measured as per [42] with a Corning 340 pH meter. Gravimetric moisture content was determined by measuring water loss from 10 g fresh soil after 2 days at 105 °C. Total carbon was measured on a subset of the samples to calculate C addition due to the root material using an elemental analyzer IRMS (PDZ Europa, Limited, Crewe, UK).
DNA/RNA extraction
DNA and RNA were co-extracted from 0.5 g of frozen soil using a phenol–chloroform extraction protocol [43, 44]. DNA and RNA were separated using the Qiagen AllPrep kit. RNA was treated with TURBO DNase (Thermo Fisher Scientific) following the manufacturer’s protocol and concentrated by ethanol precipitation. DNA and RNA were quantified using the Qubit DNA BR Assay Kit and Qubit RNA BR Assay Kit (Thermo Fisher Scientific), respectively. RNA integrity was visualized using an Experion Electrophoresis System (Bio-Rad).
Sequencing library preparation
Metatranscriptomes, iTags (16S, ITS), and SAGs were sequenced at the Joint Genome Institute (JGI); see Supplementary Methods for full details. Briefly, for metatranscriptomic libraries, ribosomal RNA was depleted using the Ribo-Zero rRNA Removal Kit (Epicentre) for plants and bacteria and reverse transcribed into cDNA. cDNA was sequenced (2 × 150 bp) on an Illumina HiSeq2000 sequencer using a TruSeq SBS sequencing kit (v3). For complimentary 16S and ITS community analysis using iTag sequencing, paired DNA and RNA were amplified from the same nucleic acid extract prepared for metatranscriptomics. iTag libraries targeted the bacterial 16S V4 region (primers 515F, 805R) [45, 46] and the fungal ITS2 region (primers ITS9, ITS4) [47, 48] using barcoded reverse primers [45]. Amplicons were sequenced (2 × 300 bp) on an Illumina MiSeq platform using a MiSeq Reagent Kit (v3 600 cycle). SAGs that successfully amplified 16S rRNA genes were sequenced using the Illumina NextSeq platform [49].
Sequence processing
Metatranscriptomic raw reads were quality-trimmed (Q20) using fastqTrimmer, and artifacts were removed using DUK [50]. Contaminating ribosomal RNA and transfer RNA were identified and removed with bowtie2 [51] by mapping reads against SILVA [52], greengenes [53], IMG rRNA [54], GtRNAdb [55], and tRNADB-CE [56] databases. In total we sequenced 408 Gbp of RNA, and after in silico contaminant filtering, we obtained an average of 43 million paired-end metatranscriptomic reads per library (see Table S1 for repository IDs and sequencing statistics). We did not detect any bias toward rhizosphere or bulk soil in either sequencing library size (Fig. S2) or gene diversity (Fig. S3).
Amplicons were analyzed on JGI’s iTag analysis pipeline (iTagger) [45], which created OTUs at the 97 and 95% identity level for bacterial 16S and fungal ITS, respectively. Contaminants were removed using DUK, merged with FLASH [57], and dereplicated. Dereplicated sequences were sorted by decreasing abundance, clustered with USEARCH [58], and assigned taxonomy using the RDP classifier [59].
SAG sequences were processed using the BBTools suite [60]. Sequences were filtered using BBDuk and mapped against masked contaminant references (human, cat, dog) using BBMap and BBMask. Normalization was performed with BBNorm and error correction with tadpole [60]. Sequences were assembled using SPAdes (version v3.7.1) [61]; 200 bp was trimmed from contig ends; contigs were discarded if length was <2 kbp or read coverage was <2. Cross contaminated contigs were removed using CrossBlock [60]. Automated SAG decontamination was performed with ProDeGe (version 2.3) [62] and assemblies were discarded if the total size was <200 kbp. Details about the final draft assemblies are listed in Table S2.
Soil-specific reference database
Transcripts were mapped against a genome database specific to our Hopland CA experimental soil, composed of metagenome-assembled genomes (MAGs) (NCBI PRJNA517182), MAGs from a stable isotope probing (SIP) rhizosphere density gradient [63] (http://ggkbase.berkeley.edu/), isolate genomes [64], and SAGs (this study; Table S1). Reference genomes were dereplicated using whole pairwise genome alignments at 98% nucleotide identity [65]; we selected the highest quality representative based on completeness of single copy genes [66]. To ensure we did not include multiple fragmented genomes from the same organism, genomes >70% complete were clustered into groups that overlapped by at least 50%; genomes <70% complete were clustered in a second round using a 30% overlap. The highest quality representative was selected for each cluster (score = # single copy genes − 2 × multiple single copy genes; the genome with the highest N50 was selected to break a tie). This resulted in 282 total genomes for our custom reference database (see Table S3 for quality information), composed of 197 rhizosphere SIP-MAGs (64%), 22 soil MAGs (16%), 39 isolate genomes (12%), and 24 SAGs (8%). This database includes most of the major groups that significantly responded to the rhizosphere by 16S community analysis (Table S4), but is not a complete genomic inventory of our soil.
Gene annotation and counts
Gene prediction was performed on all genome bins using Prodigal in metagenome mode [67]. Protein sequences were annotated using dbCAN2 [68] (accessed April 2017), KEGG [69], and ggKbase (http://ggkbase.berkeley.edu/). Recent work has shown that creating consensus annotations from multiple annotation sources can improve annotation confidence [70]. We used three annotations to generate a consensus annotation (CAZy, KEGG, ggKbase). Proteins with CAZyme functional domains were manually curated to generate a consensus annotation: CAZymes without KEGG or ggKbase annotations were ignored, and if the KEGG and ggKbase annotations disagreed, KEGG was selected. Genes containing signal peptide signatures for extracellular protein transport were annotated using SignalP 4.1 [71].
Trimmed and filtered reads were mapped against our soil-specific database using BBSplit [60]. Reads that mapped ambiguously to multiple reference genomes were discarded to prevent double counts. Transcripts were binned into “aggregated population transcriptomes” using a relaxed similarity cutoff (80% min identity) and should not be interpreted as genome or species transcriptomes, as these are aggregated transcriptomes for closely related taxa that could include the genus or family. Gene counts were determined using featureCounts (R package: Rsubread).
Data analysis
Statistical analyses
Metatranscriptomic and amplicon sequencing data were normalized using DESeq2 to account for differences in sequencing effort [72], except for Shannon diversity analysis, where reads were rarified and diversity indices calculated using QIIME 1.9.1 [73]. Presence/absence in our treatments was statistically defined in comparison to unamended bulk soil communities, as our operational definition of rhizosphere likely contains some amount of bulk soil. At each timepoint, significant differential expression relative to bulk soil was determined using DESeq2 (p < 0.05), which adjusts p values for multiple comparisons. Ordination and graph visualization were conducted in R [74]. Data were ordinated using nonmetric multidimensional scaling (R package: vegan), and significantly different clusters were determined using adonis [75]. Correlations between environmental data and ordination data were tested using envfit (R package: vegan).
Carbohydrate depolymerization CAZymes (d-CAZy)
We selected aggregated population transcriptomes with 4+ upregulated carbohydrate depolymerization genes for further analysis, which identified 24 of the 282 populations. Target substrates for d-CAZy were initially classified based on Berlemont and Martiny [76] and then refined into the following putative substrate categories using the consensus annotation described above: cellulose, xylan, xyloglucan, pectin, plant polysaccharides, microbial cell walls, starch and glycogen, xylose and cellulose oligosaccharides, oligosaccharides, and mono- and disaccharides (see Table S5 for gene names and references). Area-proportional Venn diagrams visualized the subset of the community that significantly upregulated d-CAZy transcripts relative to the total d-CAZy genomic potential (R package: venneuler).
Guild assignment
We defined guilds based on d-CAZy expression over time and across treatments. Average d-CAZy differential gene expression (log2-fold change) relative to bulk soil was visualized using heatmaps (R package: pheatmap). One-dimensional hierarchical clustering was used to assign heatmap groups, which were classified as guilds based on resource preference and timing of peak gene expression (“Rhizosphere,” “Detritusphere,” “Aging roots,” “Low Response”). Rhizosphere and Detritusphere guilds responded early in response to their eponymous substrates, while the Aging Root guild responded late as the rhizosphere aged; the low response guild had a significant but low responses to the treatments. Maximum gene expression log2-fold change relative to bulk soil (per enzyme, per treatment) was plotted by reference genome using ggplot2. Phylogeny is presented in accordance with current taxonomic nomenclature (e.g., Actinobacteria now “Actinobacteriota”) [77].
d-CAZy transcription strategies
Decomposition transcription strategies were assessed in two ways: by evaluating gene expression levels relative to abundance, and by comparing expression in the rhizosphere or detritusphere to the combined rhizosphere–detritusphere. To distinguish changes in transcription due to growth versus gene upregulation, we conducted additional analyses using gyrase A and B transcription as a proxy for relative abundance. Gyrases are housekeeping genes that often have stable expression patterns over a variety of treatment conditions, and are commonly used in qPCR gene expression studies to normalize the data to account for changes in abundance [78]. To determine if taxa increased in abundance, we compared gyrase gene expression in the treatment to bulk soil expression using DESeq2; taxa with significantly higher gyrase expression were assigned to the “Grower” strategy. To determine if d-CAZy gene expression was upregulated above population size, we divided d-CAZy fold change by gyrase fold change to normalize for abundance; if d-CAZy expression was >3× higher than gyrase expression, the taxa were assigned to the “Upregulator” strategy. Taxa that had the highest gene expression when combined resources were available, with 3× higher gene expression in the combined rhizosphere–detritusphere compared with the rhizosphere or detritusphere alone, were assigned to the “Synergist” strategy.
Results
Microcosm soil properties
Root detritus additions increased soil carbon from 2.0% ± 0.1 to 2.8% ± 0.1. Gravimetric soil moisture at the time of harvest averaged 0.34 ± 0.067 g water g−1 dry soil, with the exception of the final timepoint, when microcosms had an average of 0.11 ± 0.013 g water g−1 dry soil due to plant transpiration (Fig. S4a). The addition of root detritus significantly increased bulk soil pH by 0.14 pH units at the first timepoint, and this difference decreased over time (Fig. S4b).
Rapid community and functional assembly in the rhizosphere and detritusphere
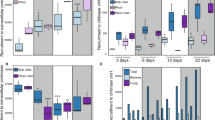
Living roots and detritus rapidly altered bacterial community structure and functional assembly. All treatments diverged from bulk soil within 3 days, as seen by clear groupings in NMDS ordination space for both 16S cDNA and mRNA transcripts (Fig. 1a, c, respectively). For 16S cDNA, the rhizosphere and detritusphere significantly shaped community composition (Fig. 1) (see Table S6 for PERMANOVA F tables). Changes due to time explained 19% of the community variability, indicating that some taxonomic succession occurred within the treatments (Fig. S5a). In contrast, fungal community composition, measured by ITS cDNA, was indistinguishable between rhizosphere and bulk soil (p > 0.1), and was instead significantly altered by both the detritus amendment and time (Fig. 1b) (Table S6).
Microbial community composition was measured by (a) bacterial 16S cDNA amplicons and (b) fungal ITS cDNA amplicons. Expressed functional composition was measured by (c) total mRNA transcripts. Symbols represent four experimental habitats: rhizosphere (filled symbols), bulk soil (hollow symbols); each with added root detritus (red), or without added root detritus (blue). Ellipses represent the standard error of the weighted average of the centroid scores (calculated by ordiellipse). n = 3 for each habitat and timepoint. To view data grouped by timepoint, see Fig. S5.
Time was the dominant factor structuring bacterial gene expression; transcripts from the final (22-day) timepoint for all treatments clearly separated from earlier timepoints (Fig. 1c) (Table S6). This shift was correlated with both soil moisture (envfit: r2 = 0.87, p < 0.001) (Fig. S5a) and time (envfit: r2 = 0.57, p < 0.001) (Fig. S5b). When only the first three timepoints are considered (days 3, 6, 12), the rhizosphere and detritus treatments were the dominant factors structuring community gene expression (Table S6).
Early colonists of the rhizosphere rapidly increased in relative abundance within 3 days (16S cDNA, Table S4), and included Proteobacteria (Burkholderiaceae) and Verrucomicrobiota (Opitutaceae). Early colonists of the detritusphere included Fibrobacterota, Verrucomicrobiota (Chthoniobacteraceae, Opitutaceae), Armatimonadota, Bacteroidota, and Proteobacteria. In contrast, relatively few Actinobacteriota and Acidobacteriota significantly responded to either the rhizosphere or detritusphere on this time scale by 16S cDNA (3–22 days). Taxa that decreased in relative abundance are listed in Table S7.
Root detritus increased taxonomic and functional diversity
Adding root detritus increased the taxonomic and functional Shannon diversity of both rhizosphere and bulk soil, and the combined rhizosphere–detritusphere had the highest overall taxonomic diversity by the final timepoint (Tukey HSD analysis, Fig. 2a). Taxonomic Shannon diversity was calculated based on 16S rRNA gene sequences. Adding root detritus to bulk soil significantly increased KEGG functional diversity, and appeared to have a similar effect on rhizosphere soil, although the trend was not significant at p < 0.05 (Tukey HSD analysis, Fig. 2b). Of our four treatments, rhizosphere soils (with and without root detritus) had the highest expressed functional diversity after 22 days of root growth.
Average α-diversity (Shannon diversity) for (a) 16S rRNA genes, and (b) KEGG functional genes derived from community mRNA transcripts. Error bars reflect one standard error. In order to make our results more comparable to a prior study of bacterial succession in the Avena rhizosphere, Shannon taxonomic diversity was calculated based on 16S rRNA genes [13]. The different letters represent significant differences measured by Tukey HSD analysis at the final timepoint. (c) The cumulative number of significant differentially upregulated decomposition CAZy (d-CAZy) genes relative to bulk soil, measured by DESeq (p < 0.05). Treatments are: rhizosphere + detritus (filled red circle, solid line), rhizosphere (hollow red circle, dashed line), bulk soil + detritus (filled blue square, solid line), and untreated bulk soil (hollow blue square, dashed line).
Roots stimulated expression of carbohydrate depolymerization transcripts
We curated a set of CAZyme genes relevant for plant and microbial carbohydrate depolymerization (d-CAZy, Table S5) and assessed expression of carbohydrate depolymerization transcripts relative to the bulk soil treatment. Overall, rhizosphere communities had the most significantly upregulated d-CAZy genes (Fig. 2c). The combined rhizosphere–detritusphere had the largest number of significantly upregulated d-CAZy genes, with the exception of the final timepoint, when unamended rhizosphere soil had the largest number of upregulated genes. This result was generally consistent across the four major CAZyme classes (auxiliary activity, carbohydrate esterases, glycoside hydrolases, polysaccharide lyases) (Fig. S6). In the bulk soil, root detritus additions initially stimulated a large pulse of d-CAZy activity, but this dropped dramatically over time; by the final timepoint, only 10–20% of the genes were distinguishable from bulk soil expression levels.
Realized niches in rhizosphere and detritusphere
Using our reference database, we identified aggregated populations with statistically significant gene expression and compared it with genomic content in an effort to identify the “realized” metabolic niches within our bacterial community (Fig. 3). While many taxa had the genomic capacity for carbohydrate depolymerization, only a small fraction significantly upregulated these genes relative to bulk soil by DESeq2 (p < 0.05). Populations upregulating cellulases and xylanases relative to bulk soil across the three treatments represented 12–16 and 11–21% of the total genomic potential across all the reference genomes, respectively (Fig. 3). The relative expression patterns for gyrase A and B housekeeping genes indicate that the general population dynamics followed similar patterns by treatment as was observed for functional genes.
Area-proportional Venn diagrams indicate the number of functionally active taxa by soil habitat relative to the total metagenomic capacity for 282 assembled soil genomes. The outer circle (brown) indicates the number of unique genomes in the reference database with the genomic potential for the specified class of genes; inner circles reflect the number of taxa that differentially upregulated each class of genes relative to bulk soil for each treatment: rhizosphere (green), rhizosphere + detritus (pink), and bulk soil + detritus (blue). Overlapping regions represent shared niche space, with the number of genomes shared between different treatments. Genome classes analyzed include: gyrase A, B (housekeeping gene), oligosaccharide hydrolases (e.g., glycosidases, xylanases), cellulases, and xylanases (see Table S5 for full gene list). The bottom panel lists the number of active genomes by treatment; the percentage of active genomes relative to total genomic potential is denoted in parentheses.
Guilds defined by temporal and habitat gene expression
We assessed gene transcription patterns over time and across habitats to define ecological guilds. Most of the d-CAZy gene expression was attributed to 24 of our 282 reference genomes; these taxa had at least four d-CAZy genes significantly upregulated relative to bulk soil. Most of these populations (15 of 24) were derived from our rhizosphere SIP-metagenome database (Fig. 4), and had distinct rhizosphere versus detritusphere transcriptional preferences, as shown by one-dimensional hierarchical clustering (Fig. 4). We averaged d-CAZy differential expression per aggregated population to show broad differential expression patterns (full heatmaps available in Fig. S7). Using statistical upregulation of depolymerization genes relative to bulk soil (p < 0.05) as a proxy for resource preference and guild membership, we defined “Rhizosphere,” “Detritusphere,” and “Aging Root” guilds, and a “Low Response” group where there was no discernable habitat preference.
Red indicates log2-fold gene upregulation in the treatment, blue indicates gene upregulation in bulk soil. Reference genome taxonomy is listed for the aggregated population transcriptomes (rows), as is the source of the genome (in parentheses): rhizosphere SIP-metagenome (SIP-MG), soil metagenome (MG), cultured isolate genome (I), single amplified genome (SAG). Time (days) is indicated by the columns. Metatranscriptomic guild assignment was accomplished through one-dimensional hierarchical clustering and is denoted by the left gray bars and numbers: high d-CAZy gene expression when living roots were present were assigned to the “Rhizosphere” guild; high d-CAZy expression when added detritus was present formed the “Detritusphere” guild; high d-CAZy expression when living roots were present, but where expression peaked at the last timepoint, formed the “Aging Root” guild. Stars indicate d-CAZy transcription strategy: blue stars indicate taxa that significantly increased abundance (“Growers”, Fig. S8); purple stars indicate taxa where per capita gene expression was 3× > abundance, where gyrase gene expression serves as a proxy for abundance (“Upregulators”, Fig. S9); red stars indicate taxa where gene expression in the combined rhizosphere–detritusphere was 3× > than either rhizosphere or detritusphere alone (“Synergist”, Fig. S10).
Carbohydrate depolymerization guilds undergo functional succession
Transcriptionally defined guilds captured a functional succession in carbohydrate depolymerization, for both polysaccharides and also oligosaccharide breakdown products. The Rhizosphere and Detritusphere guilds had high d-CAZy expression within the first 6 days, then between 12 and 22 days an additional Aging Root guild emerged (Fig. 4).
The Rhizosphere guild contained Proteobacteria (order Burkholderiales) and a Verrucomicrobiota taxa from the Opitutaceae (Fig. 4, Group 1). Cellulases (endoglucanases), xylanases, and xyloglucanases were most highly expressed at 3 days, as were enzymes for potential breakdown products like cellulose- and xylan-oligosaccharide hydrolases (beta-glucosidases and beta-xylosidases, respectively) (Fig. S7a–d). One Burkholderiaceae taxon did not follow this pattern, and instead had high d-CAZy expression at the final timepoint. Overall, xyloglucan hydrolases were characteristic of rhizosphere taxa, and observed only once in the detritusphere (bulk + detritus) (Fig. 5).
Ovals and their size indicate maximum differential expression relative to bulk soil (log2-fold change) over the time course for the treatments: rhizosphere (blue ovals), rhizosphere + detritus (brown ovals), and bulk + detritus (yellow ovals). Genes are grouped by the enzyme’s putative target substrate: plant polysaccharides (cellulose, xylan, xyloglucan, pectin, other plant polysaccharides), microbial cell walls, starch and glycogen, xylan- and/or cellulose oligosaccharides, other oligosaccharides, and mono- and disaccharides. Phylum abbreviations: γ-Proteobacteria (γ-Proteo), Verrucomicrobiota (Verruco), α-Proteobacteria (α-Proteo), Actinobacteriota (Actino).
The Detritusphere guild was phylogenetically diverse, including taxa from Proteobacteria, Fibrobacterota, Bacteroidota, and Actinobacteriota phyla (Fig. 4, Group 2). With the exception of Rhizobiaceae taxa, members of the Detritusphere guild typically upregulated cellulases and xylanase (or both) soon after detritus was added (3, 6 days), and cellulose- or xylan-oligosaccharide hydrolases for potential breakdown products (Fig. 5).
In the Aging Root guild, Actinobacteriota taxa from the Streptomycetaceae and Catenulisporales had high d-CAZy gene expression at the final timepoint (22 days) in the rhizosphere, and early gene expression in the detritus-amended treatments (Fig. 4, Group 3). The Aging Root guild had almost no upregulated genes for starch, glycogen, cell wall, and disaccharide decomposition (Fig. 5).
Guild-based assessment of d-CAZy transcription strategies
We used metatranscriptomic expression patterns to determine the prevalence of three d-CAZy transcription strategies within the microbial guilds (Fig. 4): (a) increased abundance, (b) upregulated gene expression (above per capita abundance), or (c) synergistic gene upregulation when combined resources were available (i.e., combined rhizosphere–detritusphere). We interpreted significant gyrase upregulation relative to bulk soil as an increase in relative abundance (DESeq2, Fig. S8).
Decomposition strategies varied by guild membership, and were not mutually exclusive. All members of the Rhizosphere and Detritusphere guilds were “Growers” and increased in abundance (Figs. 4 and S8). Rhizosphere guild taxa were also “Upregulators” (threefold higher gene expression per capita) (Figs. 4 and S9). In contrast, enzyme expression in the Detritusphere guild tracked abundance with no “Upregulators”. Over half of the Aging Root guild did not change in abundance relative to bulk soil. This guild, composed entirely of Actinobacteriota taxa, had “Upregulators” with relative abundances statistically indistinguishable from bulk soil.
The combined rhizosphere–detritusphere stimulated three “Synergist” taxa from the Opitutaceae (Verrucomicrobiota), Burkholderiales, and Fibrobacterota (Figs. 4 and S10). The Opitutaceae ribosomal protein S3 (RP-S3) was 93% similar to an obligate anaerobe isolated from rice paddy soil, Opitutus terrae, by blastx [79]. The Burkholderiales and Fibrobacterota reference genomes were most closely related to uncultivated MAGs (RP-S3 85 and 73% similar, respectively). Both the Optitutaceae and Fibrobacterota reference genomes contained putative CBB3 cytochrome oxidases, a microaerophilic version of cytochrome oxidase; these genes were actively expressed but not upregulated relative to bulk soil. At the early timepoints, Opitutaceae taxa upregulated enzymes for xylan degradation (arabinoxylan arabinofuranosidase) and xylan breakdown products (xylan 1,4-beta-xylosidase, alpha-D-xyloside xylohydrolase) (Figs. 5 and S7d). Fibrobacterota taxa upregulated endoglucanases, endo-1,4-beta-xylanases, and enzymes targeting their potential breakdown products (beta-xylosidase, cellobiose phosphorylase) (Fig. S7h). At later timepoints, the Burkholderiales upregulated putative lignocellulosic enzymes such as endoglucanase, tannase and feruloyl esterase, and rhamnogalacturonan lyase (Fig. S7e).
Discussion
Rapid community and functional assembly in the nascent rhizosphere and detritusphere
The soil microbial community surrounding roots undergoes a compositional succession corresponding to the phenological stages of plant growth [13, 14]. However, little is known about microbial gene expression during rhizosphere succession, and the temporal relationship between functional succession versus community changes. We used genome-centric, comparative metatranscriptomics to identify taxa mediating root-enhanced decomposition using carbohydrate gene transcripts. Since many soil taxa are noncultivable by conventional methods, this approach offers insights into the physiologies of populations only known by sequencing [38]. Our results, among the first using a genome-centric metatranscriptome approach in soil [80], illustrate that microbial taxa have specialized functions and life strategies based on spatiotemporal differences in root habitats.
We found community and functional assembly proceeded at different rates—while taxonomic composition underwent minor successional changes over 3 weeks, expressed functional composition distinctly shifted between 12 and 22 days. mRNA has a short half-life relative to DNA, and is a sensitive indicator about ongoing ecological processes and near-real-time conditions experienced by cells [81]. Our previous work indicates that rhizosphere community composition continues to shift from 3 weeks until senescence [13], but the faster changes we observed for transcript inventories suggest microbes experienced changes in rhizodeposits, environmental conditions (e.g., moisture, pH, O2), or other signals on the scale of days [77]. The relative speed of functional shifts suggests that expressed functional succession occurs at a faster time scale than compositional changes, and possibly presages the rhizosphere microbial community succession that occurs over longer time scales. This illustrates a benefit of using time-resolved metatranscriptomics to assess activity of specific microbial taxa and the processes that lead to community assembly, since organisms transcriptionally respond to stimuli on a shorter time scale than evinced by replication.
SIP-metagenomes produced the most useful genomes for soil metatranscriptomics
The proportion of transcripts mapping to our reference genomes illustrates the comparative benefits of the four sequence products in our custom database: single-cell amplified genomes, isolate genomes, deeply sequenced bulk soil metagenomes, and rhizosphere SIP metagenomes. Genomes derived from rhizosphere SIP-metagenomes proved to be the most relevant for transcript mapping, and the source of most of the aggregated populations with 4+ upregulated carbohydrate depolymerization genes (15 of 24 populations). In a previous study [63], these taxa also showed high 13C-incorporation by rhizosphere 13C-SIP (unpublished data), where the plants were continuously-labeled with 13CO2 for 6 weeks. Our results suggest that SIP-metagenome datasets are a highly fruitful genomic resource for environmental metatranscriptomics and other omics analyses.
Metatranscriptomic guilds provide a framework to understand rhizosphere succession
By assigning expressed carbohydrate depolymerization genes to taxa derived from our custom genome database, we stepped beyond gene-centric studies that have shown rhizosphere gene expression with plant development [12] or environmental changes [19] and identified carbohydrate depolymerization guilds based on shared spatiotemporal gene expression. We identified four guilds based on spatiotemporal CAZy expression patterns (Rhizosphere, Detritusphere, Aging Root, and Low Response). In macroecology, the guild concept is a common way to group populations as functional ecological units, based on their resource utilization traits or life history strategies [29, 33]. In microbial ecology, advances in next-generation sequencing have allowed researchers to taxonomically profile microbial communities in a high throughput manner [82], but taxonomy and function may not correspond [30, 34, 83]. Over the past 2 decades, the guild categorization framework has gained traction in microbial ecology [30, 34, 36, 37], and may be particularly useful for phylogenetically ubiquitous microbial functions—for example, SOM decomposition [36, 83,84,85], stress, or nitrogen mineralization—where guilds are based on shared life history traits rather than phylogeny.
Within each guild, many taxa engaged multiple catabolic pathways for carbohydrate degradation, including potential degradation by-products such as cellulose- and xylose-oligosaccharides. For example, taxa from the Rhizosphere and Detritusphere guilds not only expressed enzymes for cellulose and xylan degradation, but also their breakdown products. Recent work suggests that facilitative processes such as cross-feeding in large networks can act to stabilize coexisting competitors for resources [86,87,88]. Genome-resolved metagenomic analyses indicate the importance of metabolic byproduct handoff in linking together interacting members of microbial communities [80, 87, 88]. The breadth of carbohydrate degradation pathways that Rhizosphere and Detritusphere guilds engage in may be a potential explanation for the stable, positive and repeatable interaction networks in the rhizosphere that we observed in a previous study [89]. We hypothesize that complex cross-feedings networks promote coexistence within highly interconnected rhizosphere communities.
Niche differentiation promotes coexistence of rhizosphere and detritusphere guilds
By combining taxonomy and function with genome-resolved metatranscriptomics, we examined both the “fundamental” and “realized” metabolic niches [30, 38] of bacteria in our experiment. The niche differentiation concept asserts that organisms coexist by subdividing available resources, such as food or space [29, 36]. As the number of niches increases in a system, so should the number of coexisting species [90]. When detritus was added to the rhizosphere, most taxa demonstrated spatial and temporal coexistence rather than synergistic consumption of resources (only three observed “Synergists”). These results are reflected by the higher functional alpha diversity in the combined rhizosphere–detritusphere; we saw approximately additive increases in functional diversity when a new resource (root detritus) was added to the system. Our work suggests that spatial and temporal niche differentiation promotes microbial coexistence in the rhizosphere and detritusphere.
Guild-based assessment of d-CAZy transcription strategies
We further assessed if our guilds had differing d-CAZy transcription strategies and evaluated if our increases in gene expression (a) tracked increases in abundance (“Growers”), (b) were upregulated per capita (“Upregulators”), or (c) synergistically upregulated when both root exudates and detritus were available (“Synergists”). These strategies were not mutually exclusive, and their prevalence varied according to guild membership. All organisms in the Rhizosphere guild were both “Growers” and “Upregulators,” while the Detritusphere guild were primarily “Growers”. Multiple studies have shown that the input of organic-C substrates can increase or decrease the rates of C degradation of surrounding SOM, which is a phenomenon known as priming [6, 7, 91]. Due to the large number of significantly upregulated decomposition transcripts in the rhizosphere, both with and without detritus amendments, rhizosphere soil has a high potential for increased rates of decomposition, as was previously observed in this plant-soil system [8]. This is consistent with the expectations for positive rhizosphere priming, where fresh organic matter provided by the rhizosphere stimulates the production of enzymes that can degrade SOM [20, 92, 93].
In keeping with the rhizosphere priming hypothesis, we not only expected that the rhizosphere would stimulate d-CAZy expression, but we also expected that amending the rhizosphere with detritus would stimulate the greatest gene expression, and these “Synergists” would be prevalent when combined resources were present. Interestingly, we only observed three “Synergist” taxa, and two of these were putative microaerophiles. This suggests that these taxa may also be partitioning their niches based on changes in the edaphic environment, such as oxygen or pH, rather than simply by consuming combined resources. The Verrucomicrobiota MAG is distantly related to Opitutaceae isolates derived from oxygen-limited rice patties and insect guts [94,95,96,97]. Fibrobacterota include cellulose degrading bacteria found in mammal rumens [98], termite guts [99], anaerobic cellulose reactors [100], and rice paddy soil [101]. Both MAGs contain cytochrome oxidases with high oxygen affinity (CBB3), which is associated with organisms living in microaerophilic environments [102]. High amounts of heterotrophic respiration can create microaerophilic zones in otherwise aerobic environments, such as the rhizosphere [103, 104]. The combined oxygen demand from both the rhizosphere and detritusphere may have been sufficiently high to create microaerophilic niche for root detritus decomposition, thus providing a possible mechanism for the observed synergistic response. Both of these taxa are rhizosphere inhabitants found in our other studies [13, 23] suggesting that this synergistic decomposition in the combined rhizosphere–detritusphere may be functionally significant in semiarid grasslands.
During the functional succession of guilds, one guild emerged during the latter half of the experiment as the rhizosphere aged and potentially started to become more like a detritusphere habitat. Interestingly, more than half of the Aging Root guild had relative abundances that were indistinguishable from bulk soil based on gyrase housekeeping gene expression, but in some cases were categorized as “Upregulators”. Similarly, by 16S relative abundance analysis, few actinobacterial taxa were distinguishable from bulk soil in response to the treatments. This suggests that these taxa were actively utilizing carbohydrates and not appreciably changing their abundances over the time scale we measured. A recent SIP study on forest soils found that Actinobacteriota only accumulated 13C after 21 days [105]. Since SIP requires replication to increase the isotopic enrichment of DNA, the authors hypothesized that this could be due to slow growth. Our results support this hypothesis, and indicate that populations with minimal growth can still be active and functionally relevant in the community [106]. We also note that some actinobacterial taxa in the Detritusphere guild had significant d-CAZy transcription as early as 3 days. Thus, metatranscriptomics provides a way to assess functional relevance that is independent of changes in taxonomic relative abundance.
Niche differentiation in soil based on habitat and substrate preferences (root exudates, detrital organic matter, and the combination of the two) may have broader system-level implications. For example, SOM produced by these distinct communities may persist at different rates—stable SOM has been linked to the biochemical quality of root inputs [1, 3]. In addition, the ecophysiological properties of taxa in each guild and the chemical composition of their necromass likely affect SOM properties, nutrient availability, and plant performance [107, 108]. We also expect that the physical association of detritusphere and rhizosphere organisms with minerals will differ, as the detritusphere may promote transient attachment to organic matter while the rhizosphere can promote growth directly on minerals without requiring contact with the root [109]. Experimental tests and representation of these processes in ecosystem biogeochemical models is currently lacking, and they represent important targets to improve predictions of soil-plant-microbial feedbacks.
Conclusions
Niche differentiation is central to theories of coexistence [29, 36, 90]. Recent advances in metagenomic sequencing have allowed us to define the fundamental metabolic niches of representatives from poorly known phyla with limited phenotypic data [63, 110]. Using genome-centric metatranscriptomics to define the realized metabolic niches for soil taxa, we found that carbohydrate depolymerization guilds rapidly emerged during rhizosphere community assembly. Using these guilds, we determined the prevalence of three d-CAZy transcriptional strategies in the rhizosphere, and found that rhizosphere organisms upregulate decomposition transcripts in addition to increasing in abundance. Further, these taxa used both primary and breakdown products; these results support recent observations that metabolic handoffs link together interacting members of microbial communities [86,87,88]. Guild dynamics of carbohydrate depolymerization during rhizosphere succession provides a key step toward developing microbially constrained models to predict the fate of soil carbon.
References
Rasse DP, Rumpel C, Dignac M. Is soil carbon mostly root carbon? Mechanisms for a specific stabilisation. Plant Soil. 2005;269:341–56.
Pett-Ridge J, Firestone MK. Using stable isotopes to explore root-microbe-mineral interactions in soil. Rhizosphere. 2017;3:244–53.
Sokol NW, Bradford MA. Microbial formation of stable soil carbon is more efficient from belowground than aboveground input. Nat Geosci. 2019;12:46–53.
Jackson RB, Lajtha K, Crow SE, Hugelius G, Kramer MG, Piñeiro G. The ecology of soil carbon: pools, vulnerabilities, and biotic and abiotic controls. Annu Rev Ecol Evol Syst. 2017;48:419–45.
Austin EE, Wickings K, McDaniel MD, Robertson GP, Grandy AS. Cover crop root contributions to soil carbon in a no‐till corn bioenergy cropping system. Glob Change Biol Bioenergy. 2017;9:1252–63.
Cheng W, Parton WJ, Gonzalez Meler MA, Phillips R, Asao S, McNickle GG, et al. Synthesis and modeling perspectives of rhizosphere priming. N. Phytol. 2014;201:31–44.
Huo C, Luo Y, Cheng W. Rhizosphere priming effect: a meta-analysis. Fungal Genet Biol. 2017;111:78–84.
Bird JA, Herman DJ, Firestone MK. Rhizosphere priming of soil organic matter by bacterial groups in a grassland soil. Soil Biol Biochem. 2011;43:718–25.
Pausch J, Kuzyakov Y. Carbon input by roots into the soil: quantification of rhizodeposition from root to ecosystem scale. Glob Change Biol. 2018;24:1–12.
Eichorst SA, Kuske CR. Identification of cellulose-responsive bacterial and fungal communities in geographically and edaphically different soils by using stable isotope probing. Appl Environ Microbiol. 2012;78:2316–27.
Berlemont R, Martiny AC. Phylogenetic distribution of potential cellulases in bacteria. Appl Environ Microbiol. 2013;79:1545–54.
DeAngelis KM, Brodie EL, DeSantis T, Andersen G, Lindow S, Firestone MK. Selective progressive response of soil microbial community to wild oat. ISME J. 2009;3:168–78.
Shi SJ, Nuccio E, Herman DJ, Rijkers R, Estera K, Li JB, et al. Successional trajectories of rhizosphere bacterial communities over consecutive seasons. Mbio. 2015;6:e00746–15.
Chaparro JM, Badri DV, Vivanco JM. Rhizosphere microbiome assemblage is affected by plant development. ISME J. 2014;8:790–803.
van der Heijden MGA, Bardgett RD, van Straalen NM. The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol Lett. 2008;11:296–310.
Chaparro JM, Sheflin AM, Manter DK, Vivanco JM. Manipulating the soil microbiome to increase soil health and plant fertility. Biol Fertil Soils. 2012;48:489–99.
Spohn M, Kuzyakov Y. Spatial and temporal dynamics of hotspots of enzyme activity in soil as affected by living and dead roots-a soil zymography analysis. Plant Soil. 2014;379:67–77.
DeAngelis KM, Lindow S, Firestone MK. Bacterial quorum sensing and nitrogen cycling in rhizosphere soil. FEMS Micobiol Ecol. 2008;66:197–207.
Yergeau E, Sanschagrin S, Maynard C, St-Arnaud M, Greer CW. Microbial expression profiles in the rhizosphere of willows depend on soil contamination. ISME J. 2014;8:344–58.
Kuzyakov Y, Blagodatskaya E. Microbial hotspots and hot moments in soil: Concept & review. Soil Biol Biochem. 2015;83:184–99.
Wang K, Peng C, Zhu Q, Zhou X, Wang M, Zhang K, et al. Modeling global soil carbon and soil microbial carbon by integrating microbial processes into the ecosystem process model TRIPLEX‐GHG. J Adv Model Earth Syst. 2017;9:2368–84.
Mendes LW, Kuramae EE, Navarrete AA, van Veen JA, Tsai SM. Taxonomical and functional microbial community selection in soybean rhizosphere. ISME J. 2014;8:1577–87.
Nuccio EE, Anderson-Furgeson J, Estera K, Pett-Ridge J, de Valpine P, Brodie EL, et al. Climate and edaphic controllers influence rhizosphere community assembly for a wild annual grass. Ecology. 2016;97:1307–18.
Ofek-Lalzar M, Sela N, Goldman-Voronov M, Green SJ, Hadar Y, Minz D. Niche and host-associated functional signatures of the root surface microbiome. Nat Commun. 2014;5:4950.
Nemergut DR, Schmidt SK, Fukami T, O'Neill SP, Bilinski TM. et al. Patterns and processes of microbial community assembly. Microbiol Mol Biol Rev. 2013;77:342–56.
Levy A, Salas Gonzalez I, Mittelviefhaus M, Clingenpeel S, Herrera Paredes S, Miao J, et al. Genomic features of bacterial adaptation to plants. Nat Genet. 2018;50:138–50.
Yan Y, Kuramae EE, de Hollander M, Klinkhamer PGL, van Veen JA. Functional traits dominate the diversity-related selection of bacterial communities in the rhizosphere. ISME J. 2017;11:56–66.
Hutchinson GE. Concluding remarks. Cold Spring Harb Symp Quant Biol. 1957;22:415–27.
Silvertown J. Plant coexistence and the niche. Trends Ecol Evol. 2004;19:605–11.
Louca S, Polz MF, Mazel F, Albright MBN, Huber JA, O’Connor MI, et al. Function and functional redundancy in microbial systems. Nat Ecol Evol. 2018;2:936–43.
Tannock GW, Wilson CM, Loach D, Cook GM, Eason J, O'Toole PW, et al. Resource partitioning in relation to cohabitation of Lactobacillus species in the mouse forestomach. ISME J. 2012;6:927–38.
Plichta DR, Juncker AS, Bertalan M, Rettedal E, Gautier L, Varela E, et al. Transcriptional interactions suggest niche segregation among microorganisms in the human gut. Nat Microbiol. 2016;1:16152.
Root RB. The niche exploitation pattern of the blue‐gray gnatcatcher. Ecol Monogr. 1967;37:317–50.
Burke C, Steinberg P, Rusch D, Kjelleberg S, Thomas T. Bacterial community assembly based on functional genes rather than species. Proc Natl Acad Sci USA. 2011;108:14288–93.
McGill B, Enquist B, Weiher E, Westoby M. Rebuilding community ecology from functional traits. Trends Ecol Evol. 2006;21:178–85.
Bhatnagar JM, Peay KG, Treseder KK. Litter chemistry influences decomposition through activity of specific microbial functional guilds. Ecol Monogr. 2018;88:429–44.
Taroncher-Oldenburg G, Griner EM, Francis CA, Ward BB. Oligonucleotide microarray for the study of functional gene diversity in the nitrogen cycle in the environment. Appl Environ Microbiol. 2003;69:1159–71.
Prosser JI. Dispersing misconceptions and identifying opportunities for the use of ‘omics’ in soil microbial ecology. Nat Rev Microbiol. 2015;13:439–46.
Bardgett RD. Causes and consequences of biological diversity in soil. Zoology. 2002;105:367–74.
Jaeger CH, Lindow SE, Miller W, Clark E, Firestone MK. Mapping of sugar and amino acid availability in soil around roots with bacterial sensors of sucrose and tryptophan. Appl Environ Microbiol. 1999;65:2685–90.
Placella SA, Brodie EL, Firestone MK. Rainfall-induced carbon dioxide pulses result from sequential resuscitation of phylogenetically clustered microbial groups. Proc Natl Acad Sci USA. 2012;109:10931–6.
Fierer N, Jackson RB. The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA. 2006;103:626–31.
Griffiths RI, Whiteley AS, O’Donnell AG, Bailey M. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl Environ Microbiol. 2000;66:5488–91.
Barnard RL, Osborne CA, Firestone MK. Responses of soil bacterial and fungal communities to extreme desiccation and rewetting. ISME J. 2013;7:2229–41.
Tremblay J, Singh K, Fern A, Kirton ES, He S, Woyke T, et al. Primer and platform effects on 16S rRNA tag sequencing. Front Microbiol. 2015;6:771.
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA. 2011;108:4516–22.
White TJ, Bruns T, Lee S, Taylor J Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M, Gelfland D, Sninsky J, White T, editors. PCR protocols: a guide to methods and applications. San Diego, CA: Academic Press; 1990. p. 315–22.
Ihrmark K, Bödeker ITM, Cruz-Martínez K, Friberg H, Kubartova A, Schenck J, et al. New primers to amplify the fungal ITS2 region – evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol Ecol. 2012;82:666–77.
Rinke C, Lee J, Nath N, Goudeau D, Thompson B, Poulton N, et al. Obtaining genomes from uncultivated environmental microorganisms using FACS–based single-cell genomics. Nat Protoc. 2014;9:1038–48.
Li M, Copeland A, Han J. DUK - a fast and efficient kmer based sequence matching tool. LBNL-4516E-Poster. 2011. duk.sourceforge.net.
Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9.
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2012;41:D590–D6.
McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–8.
Chen IMA, Chu K, Palaniappan K, Pillay M, Ratner A, Huang J, et al. IMG/M v.5.0: an integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res. 2019;47:D666–77.
Chan PP, Lowe TM. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009;37:D93–7.
Abe T, Ikemura T, Sugahara J, Kanai A, Ohara Y, Uehara H, et al. tRNADB-CE 2011: tRNA gene database curated manually by experts. Nucleic Acids Res. 2011;39:D210–3.
Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–63.
Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–1.
Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–7.
Bushnell B. BBTools software package. sourceforge.net/projects/bbmap/.
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77.
Tennessen K, Andersen E, Clingenpeel S, Rinke C, Lundberg DS, Han J, et al. ProDeGe: a computational protocol for fully automated decontamination of genomes. ISME J. 2016;10:269–72.
Starr EP, Shi S, Blazewicz SJ, Probst AJ, Herman DJ, Firestone MK, et al. Stable isotope informed genome-resolved metagenomics reveals that Saccharibacteria utilize microbially-processed plant-derived carbon. Microbiome. 2018;6:122.
Zhalnina K, Louie KB, Hao Z, Mansoori N, da Rocha UN, Shi S, et al. Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat Microbiol. 2018;3:470–80.
Probst AJ, Ladd B, Jarett JK, Geller-McGrath DE, Sieber CMK, Emerson JB, et al. Differential depth distribution of microbial function and putative symbionts through sediment-hosted aquifers in the deep terrestrial subsurface. Nat Microbiol. 2018;3:328–36.
Raes J, Korbel JO, Lercher MJ, von Mering C, Bork P. Prediction of effective genome size in metagenomic samples. Genome Biol. 2007;8:R10.
Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinforma. 2010;11:119.
Yin Y, Mao X, Yang J, Chen X, Mao F, Xu Y. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012;40:W445–51.
Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30.
Griesemer M, Kimbrel JA, Zhou CE, Navid A, D'haeseleer P. Combining multiple functional annotation tools increases coverage of metabolic annotation. BMC Genom. 2018;19:948.
Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–6.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550.
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6.
Team RC. R: a language and environment for statistical computing. R Foundation for Statistical Computing; 2017. http://www.R-project.org/.
Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, et al. Vegan: community ecology package. R package version 2.0-6 2013. http://CRAN.R-project.org/package=vegan.
Berlemont R, Martiny AC. Genomic potential for polysaccharide deconstruction in bacteria. Appl Environ Microbiol. 2015;81:1513–9.
Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, Chaumeil P-A, et al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol. 2018;36:996.
Rocha DJP, Santos CS, Pacheco LGC. Bacterial reference genes for gene expression studies by RT-qPCR: survey and analysis. Antonie Van Leeuwenhoek. 2015;108:685–93.
Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008;36:W5–9.
Woodcroft BJ, Singleton CM, Boyd JA, Evans PN, Emerson JB, Zayed AAF, et al. Genome-centric view of carbon processing in thawing permafrost. Nature. 2018;560:49–54.
Moran MA, Satinsky B, Gifford SM, Luo H, Rivers A, Chan L-K, et al. Sizing up metatranscriptomics. ISME J. 2013;7:237–43.
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–4.
Schimel JP, Schaeffer SM. Microbial control over carbon cycling in soil. Front Microbiol. 2012;3:1–11.
Martiny JBH, Jones SE, Lennon JT, Martiny AC. Microbiomes in light of traits: a phylogenetic perspective. Science. 2015;350:aac9323–1–8.
Moorhead DL, Sinsabaugh RL. A theoretical model of litter decay and microbial interaction. Ecol Monogr. 2006;76:151–74.
Goldford JE, Lu N, Bajić D, Estrela S, Tikhonov M, Sanchez-Gorostiaga A, et al. Emergent simplicity in microbial community assembly. Science. 2018;361:469–74.
Anantharaman K, Brown CT, Hug LA, Sharon I, Castelle CJ, Probst AJ, et al. Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system. Nat Commun. 2016;7:13219.
Diamond S, Andeer P, Li Z, Crits-Christoph A, Burstein D, Anantharaman K, et al. Processing of grassland soil C-N compounds into soluble and volatile molecules is depth stratified and mediated by genomically novel bacteria and archaea. 2018; 445817. https://doi.org/10.1101/445817.
Shi SJ, Nuccio EE, Shi ZJ, He ZL, Zhou JZ, Firestone MK. The interconnected rhizosphere: high network complexity dominates rhizosphere assemblages. Ecol Lett. 2016;19:926–36.
Macarthur R, Levins R. The limiting similarity, convergence, and divergence of coexisting species. Am Nat. 1967;101:377–85.
Kuzyakov Y, Friedel J, Stahr K. Review of mechanisms and quantification of priming effects. Soil Biol Biochem. 2000;32:1485–98.
Fontaine S, Mariotti A, Abbadie L. The priming effect of organic matter: a question of microbial competition? Soil Biol Biochem. 2003;35:837–43.
Pascault N, Ranjard L, Kaisermann A, Bachar D, Christen R, Terrat S, et al. Stimulation of different functional groups of bacteria by various plant residues as a driver of soil priming effect. Ecosystems. 2013;16:810–22.
van Passel MWJ, Kant R, Palva A, Copeland A, Lucas S, Lapidus A, et al. Genome sequence of the Verrucomicrobium Opitutus terrae PB90-1, an abundant inhabitant of rice paddy soil ecosystems. J Bacteriol. 2011;193:2367–8.
Wertz JT, Kim E, Breznak JA, Schmidt TM, Rodrigues JLM. Genomic and physiological characterization of the Verrucomicrobia isolate Diplosphaera colitermitum gen. nov., sp. nov., reveals microaerophily and nitrogen fixation genes. Appl Environ Microbiol. 2012;78:1544–55.
Lin JY, Russell JA, Sanders JG, Wertz JT. Cephaloticoccus gen. nov., a new genus of ‘Verrucomicrobia’ containing two novel species isolated from Cephalotes ant guts. Int J Syst Evol Microbiol. 2016;66:3034–40.
Hünninghaus M, Dibbern D, Kramer S, Koller R, Pausch J, Schloter-Hai B, et al. Disentangling carbon flow across microbial kingdoms in the rhizosphere of maize. Soil Biol Biochem. 2019;134:122–30.
Montgomery L, Flesher B, Stahl D. Transfer of Bacteroides succinogenes (Hungate) to Fibrobacter gen. nov. as Fibrobacter succinogenes comb. nov. and description of Fibrobacter intestinalis sp. nov. Int J Syst Bacteriol. 1988;38:430–5.
Ivanova AA, Wegner CE, Kim Y, Liesack W, Dedysh SN. Identification of microbial populations driving biopolymer degradation in acidic peatlands by metatranscriptomic analysis. Mol Ecol. 2016;25:4818–35.
Rahman NA, Parks DH, Vanwonterghem I, Morrison M, Tyson GW, Hugenholtz P. A phylogenomic analysis of the bacterial phylum Fibrobacteres. Front Microbiol. 2016;6:1469.
Edwards J, Johnson C, Santos-Medellín C, Lurie E, Podishetty NK, Bhatnagar S, et al. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc Natl Acad Sci USA. 2015;112:E911–20.
Pitcher RS, Brittain T, Watmugh NJ. Cytochrome cbb3 oxidase and bacterial microaerobic metabolism. Biochem Soc Trans. 2002;30:653–8.
Højberg O, Sørensen J. Microgradients of microbial oxygen consumption in a barley rhizosphere model system. Appl Environ Microbiol. 1993;59:431–7.
Keiluweit M, Bougoure JJ, Nico PS, Pett-Ridge J, Weber PK, Kleber M. Mineral protection of soil carbon counteracted by root exudates. Nat Clim Change. 2015;5:588–95.
López-Mondéjar R, Brabcová V, Štursová M, Davidová A, Jansa J, Cajthaml T, et al. Decomposer food web in a deciduous forest shows high share of generalist microorganisms and importance of microbial biomass recycling. ISME J. 2018;12:1768–78.
Li J, Mau R, Dijkstra P, Koch B, Schwartz E, Purcell A, et al. Genomic traits predict microbial growth in culture but fail in soils, except during resource pulses. ISME J. 2019;13:2162–72.
Pett-Ridge J, Shi S, Estera-Molina K, Nuccio EE, Yuan M, Rijkers R, et al. Rhizosphere carbon turnover from cradle to grave: the role of microbe-plant interactions. In: Gupta V, Sharma AK, editors. Rhizosphere biology: interactions with plants. Springer Nature Press; 2019. In press.
Kallenbach CM, Frey SD, Grandy AS. Direct evidence for microbial-derived soil organic matter formation and its ecophysiological controls. Nat Commun. 2016;7:13630.
Whitman T, Neurath R, Perera A, Chu-Jacoby I, Ning D, Zhou J, et al. Microbial community assembly differs across minerals in a rhizosphere microcosm. Environ Microbiol. 2018;20:4444–60.
Hug LA, Baker BJ, Anantharaman K, Brown CT, Probst AJ, Castelle CJ, et al. A new view of the tree of life. Nat Microbiol. 2016;1:16048.
Acknowledgements
This research was supported by the U.S. Department of Energy Office of Science, Office of Biological and Environmental Research Genomic Science program under awards SCW1589 and SCW1039 to JPR, DE-SC0010570 and DOE-SC0016247 to MKF, and DOE-SC10010566 to JFB. Sequencing was conducted as part of Community Science Program awards 1487 to JPR and 1472 to MKF. Work conducted at Lawrence Livermore National Laboratory was supported under the auspices of the U.S. DOE under Contract DE-AC52-07NA27344. Work conducted at Lawrence Berkeley National Laboratory and the U.S. DOE JGI, a DOE Office of Science User Facility, was supported under Contract DE-AC02-05CH11231. This research used resources of the National Energy Research Scientific Computing Center, a U.S. DOE Office of Science User Facility operated under Contract No. DE-AC02-05CH11231. We thank Shengjing Shi, Katerina Estera, and Jia Tian for sample processing assistance and Alex Probst for bioinformatics assistance, and the reviewers for their insightful comments that improved this manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Nuccio, E.E., Starr, E., Karaoz, U. et al. Niche differentiation is spatially and temporally regulated in the rhizosphere. ISME J 14, 999–1014 (2020). https://doi.org/10.1038/s41396-019-0582-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-019-0582-x
This article is cited by
-
Biogeographic patterns and drivers of soil viromes
Nature Ecology & Evolution (2024)
-
Predictions of rhizosphere microbiome dynamics with a genome-informed and trait-based energy budget model
Nature Microbiology (2024)
-
Soil microbial ecology through the lens of metatranscriptomics
Soil Ecology Letters (2024)
-
Trait biases in microbial reference genomes
Scientific Data (2023)
-
Viral but not bacterial community successional patterns reflect extreme turnover shortly after rewetting dry soils
Nature Ecology & Evolution (2023)