Abstract
Ribosomes are essential to cellular life and the genes for their RNA components are the most conserved and transcribed genes in bacteria and archaea. Ribosomal RNA genes are typically organized into a single operon, an arrangement thought to facilitate gene regulation. In reality, some bacteria and archaea do not share this canonical rRNA arrangement—their 16S and 23S rRNA genes are separated across the genome and referred to as “unlinked”. This rearrangement has previously been treated as an anomaly or a byproduct of genome degradation in intracellular bacteria. Here, we leverage complete genome and long-read metagenomic data to show that unlinked 16S and 23S rRNA genes are more common than previously thought. Unlinked rRNA genes occur in many phyla, most significantly within Deinococcus-Thermus, Chloroflexi, and Planctomycetes, and occur in differential frequencies across natural environments. We found that up to 41% of rRNA genes in soil were unlinked, in contrast to the human gut, where all sequenced rRNA genes were linked. The frequency of unlinked rRNA genes may reflect meaningful life history traits, as they tend to be associated with a mix of slow-growing free-living species and intracellular species. We speculate that unlinked rRNA genes may confer selective advantages in some environments, though the specific nature of these advantages remains undetermined and worthy of further investigation. More generally, the prevalence of unlinked rRNA genes in poorly-studied taxa serves as a reminder that paradigms derived from model organisms do not necessarily extend to the broader diversity of bacteria and archaea.
Similar content being viewed by others
Introduction
Ribosomes are the archetypal “essential proteins”, so much so that they are a key criteria in the division between cellular and viral life [1]. In bacteria and archaea, the genes encoding the RNA components of the ribosome are traditionally arranged in a single operon in the order 16S-23S-5S. The rRNA operon is transcribed into a single RNA precursor called the 30S pre-rRNA, which is separated and processed by a number of RNases [2]. This arrangement of rRNA genes within a single operon is thought to allow rapid responses to changing growth conditions—the production of rRNA under a single promoter allows consistent regulation and conservation of stoichiometry between all three essential components [3]. Indeed, the production of rRNA is the rate-limiting step of ribosome synthesis [4], and fast-growing bacteria and archaea accelerate ribosome synthesis by encoding multiple rRNA operons [5].
Some bacteria and archaea have “unlinked” rRNA genes, where the 16S and 23S rRNA genes are separated by large swaths of genomic space (Fig. 1). This unlinked rRNA gene arrangement was first discovered in the thermophilic bacterium Thermus thermophilus [6]. Reports of unlinked rRNA genes soon followed in additional bacteria, including the planctomycete Pirellula marina [7], the aphid endosymbiont Buchnera aphidicola [8], and the intracellular pathogen Rickettsia prowazekii [9]. Though unlinked rRNA genes were first discovered in a free-living environmental bacterium, their ubiquity among the order Rickettsiales has led to suggestions that unlinked rRNA genes are a result of the genome degradation typical of obligate intracellular lifestyles [10,11,12].
With this study we sought to determine the frequency of unlinked rRNA genes across bacteria and archaea and whether this unique genomic feature is largely confined to those bacteria and archaea with an obligate intracellular lifestyle. We examined the rRNA genes of over 10,000 publicly available complete bacterial and archaeal genomes to identify which taxa have unlinked rRNA genes and to determine if there are any genomic characteristics shared across taxa with this feature. As complete genomes are not typically available for the broader diversity of bacteria and archaea found in environmental samples [13], we also characterized rRNA gene arrangements using long-read metagenomic datasets obtained from a range of environmental samples, which together encompassed over 17 million sequences (≥1000 bp). With these long-read metagenomic datasets, we were able to determine whether unlinked rRNA genes are common in environmental populations and how the distributions of unlinked rRNA genes differ across prokaryotic lineages and across distinct microbial habitats.
Methods
Analyses of complete genomes
We downloaded all bacterial and archaeal genomes in the RefSeq genome database [14] classified with the assembly level “Complete Genome” from NCBI in January 2019 (12539 genomes). We removed genomes from consideration that had rRNA genes that were split across the genome start and end (96 genomes), >20 reported rRNA genes (2 genomes), or an unequal number of 16S and 23S rRNA genes (219 genomes). This left us with a set of 12222 genomes. We used gene ranges associated with each open reading frame (ORF) to pair the 16S and 23S rRNA genes that were closest to each other in each genome. We then checked for gene directionality (sense/antisense) and calculated the distance between each pair, taking directionality into account (see Supplementary Figure S1 for more detail and a visual representation). rRNA pairs were classified as “unlinked” if the distance between each gene was >1500 bp, “linked” if the distance was ≤1500 bp. We separated genomes that had a 16S or 23S rRNA gene that started or ended within 1500 bp of the beginning or end of its genome and classified these 226 genomes independently to account for the circular nature of bacterial and archaeal genomes. For this subset of genomes, we iteratively adjusted the start and end position of those ‘edge-case’ rRNA genes with respect to genome size and selected the smallest distance between the 16S and 23S rRNA genes as the true distance, using the same formula presented in Supplementary Fig. 1. Each genome was classified as “unlinked”, “linked”, or “mixed” depending on the status of their rRNA genes with “mixed” genomes having multiple rRNA copies with a combination of linked and unlinked rRNA genes. We reassigned taxonomy to each genome using the SILVA 132 SSU database (clustered at 99%) to maintain a consistent taxonomy between our two datasets. All analyses were done in R version 3.5.1 [15]. Information on all genomes included in these analyses (including classification of rRNA genes) is available in Supplementary Dataset S1.
Long-read metagenomic analyses
To investigate the prevalence of unlinked rRNA genes among those bacteria and archaea found in environmental samples (including many taxa for which genomes are not yet available), we analyzed long-read metagenomic datasets generated from soil, sediment, activated sludge, anaerobic digesters, and human gut samples. These metagenomic datasets were generated using either the Oxford Nanopore MinION/PromethION (six samples) or the Illumina synthetic long-read sequencing technology (also known as Moleculo, first described in [16], nine samples). The Moleculo sequences originated from four previously published studies covering: the human gut [17], prairie soil [18], sediment [19], and grassland soils (MG-RAST project mgp14596, [20]. The Nanopore sequences originated from four unpublished studies spanning a diverse range of environment types: anaerobic digesters, activated sludge, sediment, and lawn soil. For these samples, DNA was extracted using DNeasy PowerSoil Kits (Qiagen, DE) and libraries were prepared for sequencing using the LSK108 kit (Oxford Nanopore Technologies, UK) following the manufacturers protocol. The libraries were sequenced on either the MinION or the PromethION sequencing platforms (Oxford Nanopore Technologies, UK). Base calling was conducted using the Albacore v. 2.1.10 for the lawn soil sample (VCsoil) and Albacore v. 2.3.1 for all other samples (Oxford Nanopore Technologies, UK). Across these 15 samples, we compiled 16,870,533 Nanopore sequences and 846,437 Moleculo sequences with a minimum read length of 1000 bp.
We trimmed the first 250 bp of each Nanopore sequence to remove low quality regions, but performed no other quality filtering as not all samples included information on sequence quality (some sequences were fasta format). Instead, we relied on our downstream filtering steps to remove sequences of poor quality. Metaxa2 version 2.1 [21] was run on all sequences with default settings to search for SSU (16S rRNA) and LSU (23S rRNA) gene fragments. Taxonomy was assigned to the partial rRNA sequences using the RDP classifier [22] and the SILVA 132 SSU and LSU databases (both clustered at 99% sequence identity, [23]). If a sequence contained both 16S and 23S rRNA genes we used the taxonomy with the highest resolution (i.e. if the 16S was annotated to family level while the 23S was genus level, we used the 23S taxonomy for both rRNAs). Details on each sample, including number of reads and median read lengths, are available in Supplementary Table S1.
We next used a number of criteria to filter the reads included in downstream analyses and to identify taxa with unlinked rRNA genes. We only included those reads in our final dataset that met the following criteria:
-
(1)
Contained a 16S rRNA gene (to avoid potentially double counting organisms with unlinked 16S and 23S rRNA genes),
-
(2)
Included the last two domains of the 16S rRNA gene (V8|V9) (Metaxa2 uses multiple Hidden Markov Model (HMM) profiles targeting conserved regions of rRNA genes, each of these regions is referred to as a domain),
-
(3)
The length of the 16S rRNA gene was ≤4000 bp and the length of the 23S rRNA gene (if present) was ≤6800 bp. These thresholds were chosen to remove erroneously long rRNA genes while accommodating insertions within rRNA genes such as those that occur in Candidate Phyla Radiation (CPR) taxa [24], Nostoc, Salmonella, and others [25],
-
(4)
Could be classified to at least the phylum level of taxonomic resolution.
Of the subset of reads that met these criteria (112–878 per Moleculo sample, 3817–28056 per Nanopore sample, see Supplementary Table S1 for details), we classified reads as containing unlinked rRNA genes if there was >1500 bp between the 16S and 23S rRNA genes, or if there was no 23S domain found 1500 bp after the end of the 16S rRNA. We note that, unlike the NCBI gene ranges, Metaxa2 takes strand information into account and translates start and stop locations into sense orientation for SSU and LSU. For our final analyses, we removed reads that could not be classified as linked or unlinked rRNA genes (for instance a sequence with only 300 bp after the 3′ end of the 16S rRNA gene). All analyses were done in R version 3.5.1 [15]. Information on all long-read sequences included in these analyses (including classification of rRNA genes) is available in Supplementary Dataset S2.
Phylogenetic tree combining long-read and NCBI datasets
A phylogenetic tree was created from full-length 16S rRNA gene sequences by combining both the NCBI complete genomes and representatives of the long-read metagenomic datasets. For the NCBI genome sequences, we selected one 16S rRNA gene sequence per unique species. For the long-read datasets, we first matched the partial 16S rRNA genes recovered by metaxa2 [21] to full-length 16S rRNA gene sequences in the SILVA 132 SSU database [23] using the usearch10 version 10.0.240 command usearch_global (settings: -id 0.95 -strand both -maxaccepts 0 -maxrejects 0; [26]). The full-length SILVA 16S rRNA genes sequences that matched to the long-read sequences ≥95% percent identity and ≥500 bp alignment length were used as representatives of their long-read sequence match. We used 95% percent identity as our cutoff as we found unlinked rRNA gene status to generally be conserved within genera (see below and Supplemental Figure S2). The NCBI and SILVA sequences were then aligned with PyNAST version 0.1 [27] and the phylogenetic tree was constructed using FastTree version 2.1.10 SSE3 [28], and plotted with iTOL [29].
Genomic attributes associated with unlinked rRNA genes
All tests for genomic attributes were done with a subset of our complete genome dataset—we reduced the dataset to include only one representative genome per unique species and operon status. For example, if a species had 24 genomes with linked rRNA genes and three genomes with unlinked rRNA genes, we retained two genomes total, one linked and one unlinked. Species with heterogeneous rRNA gene status accounted for only 0.71% of species and we found that the presence of unlinked rRNA genes was strongly conserved at the species and genus level (Supplementary Figure S2).
With this set of reduced genomes (3967 genomes in total), we first calculated Pagel’s lambda [30] to determine whether there was a phylogenetic signal associated with unlinked rRNA genes using the phylosig function of the phytools package version 0.6.60 [31]. The results of this test indicated that there was a strong phylogenetic signal (lambda = 0.96, p < 0.0001), so we controlled for phylogeny in all of our subsequent tests by using a Phylogenetic Generalized Linear Model for continuous variables (with the function phyloglm in the phylolm package version 2.6 [32]).
To determine if taxa with unlinked rRNA genes have a lower predicted growth rate, we calculated the codon usage proxy ΔENC′ [33, 34], which provides an estimate of minimum generation times [35]. We calculated ΔENC′ with the program ENCprime [33] with default options, on both the concatenated ORF sequences and concatenated ribosomal protein sequences for each genome following Vieira-Silva and Rocha (2009). To determine if RNaseIII was present in each genome, we used HMMER version 3.1b2 [36] to search for three RNaseIII pfams (bacterial PF00636, PF14622, and archaeal PF11469) in the translated protein files of each genome. We used the gathering thresholds (GA) associated with each of these pfams to set all cutoffs and reduce the likelihood of false positives (–cut_ga).
Results
Unlinked rRNA genes occur frequently in complete genomes
We used a set of 12222 “complete” bacterial and archaeal genomes extracted from NCBI in January 2019 to determine how frequently unlinked 16S and 23S rRNA genes occur. We analyzed the distribution of distances between the closest edges of the closest pairs of 16S and 23S rRNA genes (known as the Internally Transcribed Spacer—ITS) in each genome and found that the vast majority of 16S and 23S rRNA gene pairs (98.7%) had an ITS ≤1500 bp with an average ITS length of 418.7 bp (±169.7 bp, Fig. 2a). However, pairs with ITS lengths >1500 bp showed a scattered distribution of distances, with an average ITS length of 410374 bp (±521792 bp). Hence, for this classification scheme we called rRNA genes “unlinked” if the ITS was greater than 1500 bp in length. This 1500 bp threshold is in some ways conservative, as the distance between genes in an operon is usually quite low—peaking between 20 and 30 bp in most genomes [37]. In addition, tRNA are the most common genes found in the space between the 16S and 23S rRNA genes, and range from only 75–90 bp in length [38].
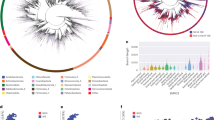
Unlinked rRNA genes can be found in 30 phyla. a The distribution of ITS lengths in complete genomes from NCBI. 1.3% of NCBI rRNA genes have an ITS region >1500 bp in length. The majority of unlinked rRNA genes have an ITS of >6000 bp (682/778) with a mean length of 410374 bp (±521792 bp). b The distribution of ITS lengths in the long-read sequence dataset. 10.1% of rRNA genes have an ITS >1500 bp. The majority of unlinked genes have an ITS of unknown length due to sequence length constraints in the long-read dataset (1470/1607). c Within our set of complete genomes from NCBI, 12 phyla had genomes containing at least one set of unlinked rRNA genes in >5% of members. Linked refers to genomes with exclusively linked rRNA genes, unlinked refers to genomes with exclusively unlinked rRNA genes, and mixed refers to genomes with at least one set each linked and unlinked rRNA genes. The number of genomes belonging to each phylum is indicated in parentheses. d By analyzing long-read metagenomic datasets, we confirmed that eight of the phyla with unlinked rRNA genes in the complete genome dataset also had unlinked rRNA genes in environmental samples (top portion), and found an additional 18 phyla in which >5% of reads that met our criteria for inclusion in downstream analyses (see “Methods”) contained unlinked rRNA genes. The number of sequences belonging to each phylum is indicated in parentheses
After classifying each rRNA gene pair as linked or unlinked based on the distance between the 16S and 23S rRNA genes, we found that 3.65% of the genomes in our dataset had exclusively unlinked rRNA genes, 0.62% had mixed rRNA gene status (i.e. genomes with multiple rRNA copies that had at least one set of unlinked rRNA genes and at least one canonical, linked rRNA operon), and 95.73% had exclusively linked operons (these numbers do not match up with the per rRNA gene dataset as each genome has a variable rRNA copy number). We found unlinked genomes to be relatively common (present in ≥5% of members) in taxa characterized as having an obligate intracellular lifestyle within the phyla Spirochetes (genus Borrelia), Epsilonbacteraeota (family Helicobacteraceae), Alphaproteobacteria (order Rickettsiales), and Tenericutes (species Mycoplasma gallisepticum). However, we also found high proportions of unlinked rRNA genes in phyla that are generally considered to be free-living, such as Deinococcus-Thermus (families Thermaceae and Deinococcaceae), Chloroflexi (family Dehalococcoidaceae), Planctomycetes (families Phycisphaeraceae and Planctomycetaceae), and Euryarchaeota (class Thermoplasmata). Phyla with at least 5% of genomes having exclusively unlinked rRNA genes are shown in Fig. 2c.
Unlinked rRNA genes are widespread in environmental metagenomic data
While the results from our complete genome dataset demonstrate that unlinked rRNA genes are common in some putatively free-living phyla, databases featuring complete genomes do not capture the full breadth of microbial diversity and are heavily biased towards cultivated organisms relevant to human health [13]. Just three phyla (Proteobacteria, Firmicutes, Actinobacteria) accounted for >83% of the genomes in our NCBI dataset—even though recent estimates of bacterial diversity total at least 99 unique phyla [39]. To investigate the ubiquity of unlinked rRNA genes among those taxa underrepresented in “complete’’ genome databases, we analyzed long-read metagenomic data from a range of distinct sample types. Focusing exclusively on long-read sequences allowed us to span the 1500 bp distance required for classification of rRNA genes without the need for assembly. This is important as the repetitive structure of rRNA genes makes it difficult to assemble a mix of nonidentical rRNA genes from the short reads typical of most current metagenomic sequencing projects [40].
From our initial long-read dataset encompassing 15 unique samples (~890,000 Illumina synthetic long reads (also known as Moleculo) and ~19 million Nanopore reads, with median read lengths of 8858 and 5398 bp, respectively), only 15855 sequences contained rRNA genes and met the criteria we established for the classification of rRNA genes as linked or unlinked (see “Methods”). Of these reads, we classified 1607 as unlinked, or 10.1% of the dataset (Fig. 2b). These long-read metagenomic analyses showed that unlinked rRNA genes are not equally distributed across environments—we found that up to 41% of the taxa in soil had unlinked rRNA genes, whereas other environments had much lower proportions, most notably the human gut, where all sequenced rRNA genes were linked (Fig. 3).
Unlinked rRNA genes have differential frequencies across environments. We found that soils (13–41% unlinked) and sediments (7.7–29%) have more unlinked rRNA genes on average than anaerobic digesters (8.1–8.8%) and the human gut (0%). Results obtained from analyses of Moleculo and Nanopore metagenomic data are indicated with (m) and (n), respectively
The results from our analyses of the long-read dataset generally mirrored the corresponding results from the complete genome dataset, in that many of the long reads classified as unlinked belonged to the same phyla where unlinked rRNA genes were prevalent in the complete genome dataset (Fig. 2). The long-read metagenomic dataset confirmed that members of the phyla Deinococcus-Thermus, Planctomycetes, Chloroflexi, Spirochetes, and Euryarchaeota frequently have unlinked rRNA genes (Fig. 2b). The long-read dataset also allowed us to provide additional evidence for the unlinked rRNA genes in poorly-studied phyla that were represented by only a handful of genomes in our complete genome dataset, such as candidate phyla Acetothermia (1 genome and 64 long-read sequences) and Patescibacteria (3 genomes and 330 long-read sequences). Using the long-read dataset, we identified 18 additional phyla where unlinked rRNA genes are common, including several candidate phyla (BRC1, GAL15, WS1, WS2) and members of the CPR (Patescibacteria, Fig. 2). We also found several clades with high proportions of unlinked rRNA genes that had no representation in our complete genome dataset, including Rikenellaceae RC9 gut group (334/624), Verrucomicrobia genus Candidatus Udaeobacter (80/80), Atribacteria order Caldatribacteriales (37/37), Cyanobacteria order Obscuribacterales (4/4), Acidobacteria Subgroup 2 (27/27), Planctomycetes order MSBL9 (40/40), and Chloroflexi class GIF9 (7/7). Overall, we found that 52% of the phyla covered in our combined datasets (37/71) have at least one representative with unlinked rRNA genes.
Unlinked rRNA genes are strongly conserved
We found that taxa with unlinked rRNA genes are not randomly distributed across bacterial and archaeal lineages—rather, we observed a strong phylogenetic signal for this trait, which we confirmed by calculating Pagel’s lambda (lambda = 0.96, p < 0.001). To highlight this point, we assembled a phylogenetic tree from full-length 16S rRNA gene sequences representing both the complete genome dataset and the long-read metagenomic dataset. We found clusters of related taxa with exclusively unlinked rRNA genes (Fig. 4) including: Euryarchaeota class Thermoplasmata, the vast majority of Deinococcus-Thermus, CPR division Patescibacteria, Verrucomicrobia DA101 group, Chloroflexi class Dehalococcoidia, and Alphaproteobacteria class Rickettsiales.
Unlinked rRNA genes occur in coherent phylogenetic clusters. This phylogenetic tree was created from full-length 16S rRNA sequences by combining both the NCBI complete genome and long-read metagenomic datasets (details in “Methods”). The outer ring indicates which dataset each sequence originated from, while the inner ring indicates the status of rRNA genes. Sequences originating from the long-read dataset cannot be mixed, as we could not distinguish multi-copy rRNA genes. Clades with high proportions of unlinked members and good representation in the tree are indicated in green: a Euryarchaeota class Thermoplasmata, b Spirochaetae classes Leptospirae and Spirochaetia, c Patescibacteria, d Chlorflexi class Dehalococcoidia, e Planctomycetes classes Phycisphaerae and Planctomycetacia, f Verrucomicrobia genus Candidatus Udaeobacter, g Tenericutes genus Mycoplasma, h Deinococcus-Thermus, i Epsilonbacteraeota genera Helicobacter and Campylobacter, j Alphaproteobacteria order Rickettsiales and k Gammaproteobacteria genus Buchnera
Genomic attributes associated with unlinked rRNA genes
Given that there are numerous bacterial and archaeal lineages where unlinked rRNA genes are commonly observed, we next sought to determine what other genomic features may be associated with this nonstandard rRNA gene arrangement. We treated the presence of unlinked rRNA genes as a binary trait—if a genome had at least one unlinked rRNA gene we counted the genome as “unlinked”. In our NCBI complete genome dataset, we found rRNA gene status to be conserved strongly at the species level—meaning that the majority of species had either exclusively linked or unlinked rRNA genes among their members (Supplementary Figure S2). Therefore, for the following tests, we used a subset of our NCBI complete genome dataset—retaining only a single representative of each species, unless the species had heterogeneous rRNA gene status (0.71% of species), in which case we retained one genome of each rRNA gene status. The analyses were corrected in order to account for the effect of phylogenetic structure in the data (see “Methods”).
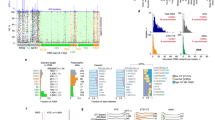
Historically, unlinked rRNA genes have been strongly associated with the reduced genomes of obligate intracellular bacteria, implying that this trait may merely be a side effect of the strong genetic drift and weak selection these taxa experience [10,11,12]. To test this hypothesis, we compared the genome sizes of species with linked and unlinked rRNA genes using Phylogenetic Generalized Linear Models (phyloglm). While we found that genomes with unlinked rRNA genes had smaller genomes on average, this difference was not significant (Fig. 5a, phyloglm p = 0.12, means of groups: 4.15 Mbp linked, 2.72 Mbp unlinked).
Genomic attributes of NCBI complete genomes based on their rRNA gene status. Linked genomes feature exclusively linked rRNA genes; unlinked genomes have at least one set of unlinked rRNA genes. We calculated these statistics using a subset of our complete genomes, including one genome per unique species and rRNA gene status. a Genomes with unlinked rRNA genes have smaller genomes on average, but this difference was not significant after accounting for phylogeny (phyloglm p = 0.12, means of groups: 4.15 Mbp linked, 2.72 Mbp unlinked). b On average, genomes with unlinked rRNA genes had significantly fewer rRNA copies (phyloglm p < 0.0001, means of groups: 4.25 copies linked, 2.72 copies unlinked). c Genomes with unlinked rRNA genes are predicted to have longer average generation times (phyloglm p = 0.028, means of groups: 0.23 linked, 0.18 unlinked; as a reference E. coli has an average ΔENC′ of 0.3). d We found that there were significantly fewer RNaseIII genes in genomes with unlinked rRNA genes (only PF00636 shown, for more detail see Supplementary Figure S3: phyloglm p < 0.001, means of groups: 1.0 linked, 0.71 unlinked)
The organization of rRNA genes within the same operon facilitates their joint regulation and co-expression at precise stoichiometric ratios. Selection for this trait is expected to be stronger in faster growing bacteria and archaea, where, at maximum growth rates, synthesis of the ribosome is the cell’s chief energy expenditure [4]. To test this hypothesis, we analyzed the association between the linkage of rRNA genes and traits related to rapid growth in bacteria and archaea. On average, genomes with unlinked rRNA genes had significantly fewer rRNA copies (Fig. 5b, phyloglm p < 0.0001, means of groups: 4.25 copies linked, 2.72 copies unlinked). We also calculated ΔENC′ for each complete genome—a measure of codon usage bias that is negatively correlated with minimum generation time in bacteria and archaea [35]. Interestingly, genomes with unlinked rRNA genes were predicted to have significantly longer minimal generation times (Fig. 5c, phyloglm p = 0.028, means of groups: 0.23 linked, 0.18 unlinked). In addition, in our long-read dataset we found that unlinked rRNA genes were more common in environments typified by slow growth rates; soil and sediment samples had higher proportions of unlinked rRNA genes than samples from anaerobic digesters and the human gut (Fig. 3).
RNaseIII separates the precursors of the 16S and 23S rRNA from their common transcript for subsequent maturation and inclusion in the ribosome [2]. RNaseIII is not an essential protein in most bacteria and archaea, and several phyla in which unlinked rRNA genes are common do not encode RNaseIII (e.g. Deinococcus-Thermus and Euryarchaeota; [41]). Therefore, we checked if there was a significant association between unlinked rRNA genes and the presence of RNaseIII genes. Interestingly, we found that genomes with unlinked rRNA genes were significantly less likely to encode the bacterial form of RNaseIII genes (Fig. 5d and Supplementary Figure S3, PF00636: phyloglm p < 0.001, means of groups: 1.0 linked, 0.71 unlinked; PF14622: phyloglm p = 0.007, means of groups: 0.86 linked, 0.66 unlinked). We were unable to check this relationship for archaeal RNaseIII, due to the size of our archaeal dataset (phyloglm failed to converge, only 39 genomes in our dataset had this gene). However, we note that the archaeal RNaseIII PF11469 was found in only two clades that feature exclusively linked rRNA genes (Euryarcheaota family Thermococcaceae and Crenarchaeota family Thermofilaceae).
Discussion
While unlinked rRNA genes have been documented previously, we have demonstrated that they are far more widespread among bacteria and archaea than expected. We found that unlinked rRNA genes consistently occur in 12 phyla using a dataset of complete genomes (Fig. 2c), and 18 additional phyla using a dataset of long-read metagenomic sequences obtained from environmental samples (Fig. 2d). Interestingly, some phyla were classified as exclusively linked in our complete genome dataset, yet had many members with unlinked rRNA genes in our long-read dataset. For example, while there were no complete genomes in the phylum Verrucomicrobia with unlinked rRNA genes (0/32), 38% of verrucomicrobial rRNA sequences were unlinked in our long-read dataset (82/217), with the majority of this group closely related to the bacterium Ca. Udaeobacter copiosus from the DA101 soil group [42]. This imbalance is likely due to the strong bias towards faster-growing organisms when using traditional cultivation methods [43], and the fact that cultivated bacteria and archaea still make up the majority of high-quality genomes in public databases [13]. Our results highlight the importance of using a combination of complete genomes, where genetic organization and traits can be assessed rigorously, with metagenomic data that allows us to sample the diversity found in selected environments in an unbiased manner. Together, these independent datasets show that unlinked rRNA genes occur across many bacterial and archaeal phyla.
The widespread prevalence of unlinked rRNA genes in many environmental samples has important implications for the use of community analysis methods that require the 16S and 23S rRNA genes to be in close proximity. For instance, before 16S rRNA gene sequencing became common practice, the ITS region of the 16S and 23S rRNA operon was routinely used to fingerprint microbial communities [44]. Likewise, the increasing popularity of long-read sequencing technologies has led to bacterial genotyping methods that target the full rRNA operon. While sequencing from the 16S rRNA gene into the 23S rRNA gene (thus including the ITS region of the rRNA operon) can increase taxonomic resolution and allow strain level identification [45], our work shows that amplicon-based studies dependent on 16S and 23S rRNA genes being located in close proximity may miss a large portion of bacterial and archaeal diversity. We found the average distance between unlinked 16S and 23S rRNA genes in our complete genome dataset to be ~410 Kbp, a rather impractical distance to amplify by PCR. While strategies which use reads spanning the 16S and 23S rRNA genes to improve taxonomic resolution (e.g. [45, 46]) are less likely to introduce biases in some environments (e.g. human gut), they will miss many phylogenetic groups in other environments like soil and sediment, where a significant fraction of taxa have unlinked rRNA genes (Fig. 3).
We used our long-read metagenomic dataset to not only bypass the cultivation bias of our complete genome dataset, but to also estimate the abundance of unlinked rRNA genes in a range of microbial community types. Our analyses of the long-read metagenomic dataset show that taxa with unlinked rRNA genes are far more abundant in some environments than others. Most notably, unlinked rRNA genes were far more common in soil (where as many as 41% of rRNA genes detected were unlinked) than the human gut (where no unlinked rRNA genes were detected, Fig. 3). The environments with higher proportions of unlinked rRNA genes (soil and sediment) are generally thought to be populated by slower-growing taxa [35, 47]. Likewise, we found that genomes with unlinked rRNA genes have significantly fewer rRNA copies than genomes with exclusively linked rRNA genes, a trait which is correlated with the maximum potential growth rate [35, 48]. We also found that genomes with unlinked rRNA genes are predicted to have significantly longer generation times (using codon usage bias in ribosomal proteins as a proxy for maximal growth rates) compared with genomes with exclusively linked rRNA genes. These lines of evidence suggest that unlinked rRNA genes are more common in the genomes of taxa with slower potential growth rates.
The existence of numerous genomes that have unlinked 16S and 23S rRNA genes and the differential frequency of these genomes across environments raise the question of the role and implications of this genetic organization. Upon first consideration, having unlinked 16S and 23S rRNA genes would seem to be disadvantageous given that both rRNA molecules are needed in equal proportions to yield a functioning ribosome. The importance of linkage for identical expression of both rRNA genes should be greater in faster growing taxa, where a higher rate of ribosome synthesis is key to rapid growth and accounts for a large proportion of the cell energy budget [4]. Studies in the fast-growing species E.coli have shown that, while unbalanced rRNA gene dosage has a slight negative effect on doubling times, balanced synthesis of ribosomal proteins still occurs in most cases [49]. If unequal expression of rRNA subunits is associated with unlinked rRNA genes, it may not confer a selective disadvantage in many environments (like soils and sediments) where longer generation times are the norm, not the exception. For slower-growing taxa, the selection coefficient associated with the effect of linked rRNA genes on growth may be small, because rRNAs are less expressed and rapid growth is a trait under weaker selection. Under these circumstances, unlinked rRNA genes may become fixed in populations by genetic drift. This is more likely to occur in species with small effective population sizes, i.e. few effectively reproducing individuals, where natural selection is not efficient enough to avoid the loss of genes or the degradation of genome organizational traits that are under weak selection [50]. This is the most common explanation for the occurrence of unlinked 16S and 23S rRNA genes [10,11,12]. It fits our observations that many of the taxa we identified with unlinked rRNA genes are restricted to obligate intracellular lifestyles (including members of the phyla Spirochetes, Epsilonbacteraeota, Alphaproteobacteria, and Tenericutes) or contain signatures of symbiotic lifestyles (CPR phyla; [51, 52]).
However, fixation of mutations due to genetic drift is much less likely to explain the presence of unlinked rRNA genes among the large proportion of free-living taxa that we have identified (including members of the phyla Deinococcus-Thermus, Euryarchaeota, Chloroflexi, Planctomycetes, and Verrucomicrobia). Some of these taxa are abundant and ubiquitous in their respective environments, e.g. the Verrucomicrobia Ca. U. copiosus [42] and members of the Rikenellaceae RC9 gut group [53]. These genomes do not show traits typically associated with genome reduction caused by small effective population sizes, i.e. abundant pseudogenes, transposable elements, or small genomes. While we found that, on average, the genomes of taxa with unlinked rRNA genes were smaller than those with linked rRNA genes, this difference was not significant after accounting for phylogeny. Thus, there is little evidence that the highly conserved trait of unlinked rRNA genes is caused exclusively by genetic drift—especially in free-living taxa.
Unlinked rRNA genes could provide a selective advantage in certain circumstances, which may explain their existence in free-living taxa. Transcribing the 16S and 23S rRNA genes separately may eliminate or reduce the need for RNaseIII, which we found to occur in lower frequencies in taxa with unlinked rRNA genes (Supplementary Figure S3). We also found RNaseIII to be completely absent in the phyla Deinococcus-Thermus and Gemmatimonadetes, both phyla with high proportions of unlinked rRNA genes. Interestingly, two recent studies have investigated the function of RNaseIII in Borrelia burgdorferi [54] and Helicobacter pylori [55], two intracellular bacteria with exclusively unlinked rRNA genes. When RNaseIII was knocked out both bacteria remained viable, but accumulated unprocessed rRNA intermediates and exhibited decreased growth rates [54, 55]. On the other hand, some bacteriophages hijack host RNaseIII to process their own mRNA [56]—in some cases, host RNaseIII can stimulate the translation of infecting phage mRNA by several orders of magnitude [57] (although other phage appear indifferent to the presence of RNaseIII; [58]). Regardless, increased resistance to predation at the cost of reduced maximum potential growth rates is a widely observed ecological trade-off [59]. Lastly, recent work has shown that some rRNA loci specialize in the translation of genes involved in adaption to temperature and nutrient shifts [60]. It is thus tempting to speculate that unlinked rRNA genes could facilitate the production of heterogeneous ribosomes with a diverse range of characteristics.
Conclusions
Unlinked rRNA genes are far more prevalent than expected, especially among those bacteria and archaea found in environmental samples for which complete genomes are not yet available. While this rearrangement appears to occur more frequently in slower-growing taxa and may be related to the presence of RNaseIII, it remains to be determined if unlinked rRNA genes confer any specific advantages. Regardless, we have shown that 52% of the phyla included in our combined datasets (37/71) have at least one member with unlinked rRNA genes, that unlinked rRNA genes occur in taxa that are abundant and ubiquitous, and that up to 41% of rRNA genes in some environments are unlinked—meaning unlinked rRNA genes are far from atypical anomalies. Indeed, unlinked rRNA genes function as a reminder that the metabolisms of poorly-studied environmental bacteria and archaea sometimes differ from conventions derived from model organisms. We have developed hypotheses about the potential advantages of unlinked rRNA genes, hypotheses which could be tested experimentally and represent a promising direction for future research—especially as some taxa with unlinked rRNA genes are relatively easy to manipulate in culture [61, 62].
Data availability
All genomes used in this study were downloaded from NCBI, with assembly IDs listed in Supplementary Dataset S1. All Nanopore data are available at the Sequence Read Archive (SRA) under Bioproject ID PRJNA553237 or the European Nucleotide Archive (ENA) under PRJEB33278. All Moleculo data has been published previously, with publications listed in methods. Classifications and details of both the complete genome and long-read datasets are included in Supplementary Dataset S1 and S2, respectively.
References
Raoult D, Forterre P. Redefining viruses: lessons from Mimivirus. Nat Rev Microbiol. 2008;6:315–9.
Srivastava AK, Schlessinger D. Mechanism and regulation of bacterial ribosomal RNA processing. Annu Rev Microbiol. 1990;44:105–29.
Condon C, Squires C, Squires CL. Control of rRNA transcription in Escherichia coli. Microbiol Rev. 1995;59:623–45.
Gourse RL, Gaal T, Bartlett MS, Appleman JA, Ross W. rRNA transcription and growth rate–dependent regulation of ribosome synthesis in Escherichia coli. Annu Rev Microbiol. 1996;50:645–77.
Klappenbach JA, Dunbar JM, Schmidt TM. rRNA operon copy number reflects ecological strategies of bacteria. Appl Environ Microbiol. 2000;66:1328–33.
Hartmann RK, Ulbrich N, Erdmann VA. An unusual rRNA operon constellation: in Thermus thermophilus HB8 the 23S/5S rRNA operon is a separate entity from the 16S rRNA operon. Biochimie. 1987;69:1097–104.
Liesack W, Stackebrandt E. Evidence for unlinked rrn operons in the Planctomycete Pirellula marina. J Bacteriol. 1989;171:5025–30.
Munson MA, Baumann L, Baumann P. Buchnera aphidicola (a prokaryotic endosymbiont of aphids) contains a putative 16S rRNA operon unlinked to the 23s rRNA-encoding gene: sequence determination, and promoter and terminator analysis. Gene. 1993;137:171–8.
Andersson SGE, Zomorodipour A, Winkler HH, Kurland CG. Unusual organization of the rRNA genes in Rickettsia prowazekii. J Bacteriol. 1995;177:4171–5.
Rurangirwa FR, Brayton KA, McGuire TC, Knowles DP, Palmer GH. Conservation of the unique rickettsial rRNA gene arrangement in Anaplasma. Int J Syst Evolut Microbiol. 2002;52:1405–9.
Merhej V, Royer-Carenzi M, Pontarotti P, Raoult D. Massive comparative genomic analysis reveals convergent evolution of specialized bacteria. Biol Direct. 2009;4:13–25.
Andersson JO, Andersson SGE. Genome degradation is an ongoing process in Rickettsia. Mol Biol Evol. 1999;16:1178–91.
Zhi X-Y, Zhao W, Li W-J, Zhao G-P. Prokaryotic systematics in the genomics era. Antonie van Leeuwenhoek. 2012;101:21–34.
O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44:D733–45.
R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria; 2018.
Kuleshov V, Xie D, Chen R, Pushkarev D, Ma Z, Blauwkamp T, et al. Whole-genome haplotyping using long reads and statistical methods. Nat Biotechnol. 2014;32:261–6.
Kuleshov V, Jiang C, Zhou W, Jahanbani F, Batzoglou S, Snyder M. Synthetic long-read sequencing reveals intraspecies diversity in the human microbiome. Nat Biotechnol. 2016;34:64–9.
White RA, Bottos EM, Roy Chowdhury T, Zucker JD, Brislawn CJ, Nicora CD, et al. Moleculo long-read sequencing facilitates assembly and genomic binning from complex soil metagenomes. Am Soc Microbiol J. 2016;1:309–15.
Sharon I, Kertesz M, Hug LA, Pushkarev D, Blauwkamp TA, Castelle CJ, et al. Accurate, multi-kb reads resolve complex populations and detect rare microorganisms. Genome Res. 2015;25:534–43.
Flynn TM, Koval JC, Greenwald SM, Owens SM, Kemner KM, Antonopoulos DA. Parallelized, aerobic, single carbon-source enrichments from different natural environments contain divergent microbial communities. Front Microbiol. 2017;8:1540–14.
Bengtsson-Palme J, Hartmann M, Eriksson KM, Pal C, Thorell K, Larsson DGJ, et al. Metaxa2: improved identification and taxonomic classification of small and large subunit rRNA in metagenomic data. Mol Ecol Resour. 2015;15:1403–14.
Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–7.
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2012;41:D590–6.
Brown CT, Hug LA, Thomas BC, Sharon I, Castelle CJ, Singh A, et al. Unusual biology across a group comprising more than 15% of domain Bacteria. Nature. 2015;523:208–11.
Pei A, Nossa CW, Chokshi P, Blaser MJ, Yang L, Rosmarin DM, et al. Diversity of 23S rRNA genes within individual prokaryotic genomes. PLoS ONE. 2009;4:1–9.
Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–1.
Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26:266–7.
Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26:1641–50.
Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–5.
Pagel M. Inferring the historical patterns of biological evolution. Nature. 1999;401:877–84.
Revell LJ. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol Evol. 2011;3:217–23.
Tung HoLS, Ané C. A linear-time algorithm for gaussian and non-gaussian trait evolution models. Syst Biol. 2014;63:397–408.
Novembre JA. Accounting for background nucleotide composition when measuring codon usage bias. Mol Biol Evol. 2002;19:1390–4.
Rocha E. Codon usage bias from tRNA’s point of view: redundancy, specialization, and efficient decoding for translation optimization. Genome Res. 2004;14:2279–86.
Vieira-Silva S, Rocha E. The systemic imprint of growth and its uses in ecological (meta)genomics. PLOS Genet. 2009;6:1–15.
Eddy SR. Accelerated profile HMM searches. PLoS Comput Biol. 2011;7:e1002195–16.
Moreno-Hagelsieb G, Collado-Vides J. A powerful non-homology method for the prediction of operons in prokaryotes. Bioinformatics. 2002;18:S329–36.
Shepherd J, Ibba M. Bacterial transfer RNAs. FEMS Microbiol Rev. 2015;39:280–300.
Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, Chaumeil P-A, et al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol. 2018;36:996–1004.
Yuan C, Lei J, Cole J, Sun Y. Reconstructing 16S rRNA genes in metagenomic data. Bioinformatics. 2015;31:i35–i43.
Durand S, Gilet L, Condon C. The Essential Function of B. subtilis RNase III is to silence foreign toxin genes. PLOS Genet. 2012;8:e1003181–11.
Brewer TE, Handley KM, Carini P, Gilbert JA, Fierer N. Genome reduction in an abundant and ubiquitous soil bacterium “Candidatus Udaeobacter copiosus.” Nat Microbiol. 2016;2:16198.
Vartoukian SR, Palmer RM, Wade WG. Strategies for culture of “unculturable” bacteria. FEMS Microbiol Lett. 2010;309:1–7.
Garcia-Martinez J, Acinas SG, Anton AI, Rodriguez-Valera F. Use of the 16S-23S ribosomal genes spacer region in studies of prokaryotic diversity. J Microbiol Methods. 1999;36:55–64.
Zeng YH, Koblížek M, Li YX, Liu YP, Feng FY, Ji JD, et al. Long PCR-RFLP of 16S-ITS-23S rRNA genes: a high-resolution molecular tool for bacterial genotyping. J Appl Microbiol. 2012;114:433–47.
Cuscó A, Catozzi C, Viñes J, Sanchez A, Francino O. Microbiota profiling with long amplicons using Nanopore sequencing: full-length 16S rRNA gene and whole rrn operon. F1000Res. 2018;7:1755–25.
Brown CT, Olm MR, Thomas BC, Banfield JF. Measurement of bacterial replication rates in microbial communities. Nat Biotechnol. 2016;34:1256–63.
Roller BRK, Stoddard SF, Schmidt TM. Exploiting rRNA operon copy number to investigate bacterial reproductive strategies. Nat Microbiol. 2016;1:1–7.
Siehnel RJ, Morgan EA. Unbalanced rRNA gene dosage and its effects on rRNA and ribosomal-protein synthesis. J Bacteriol. 1985;163:476–86.
Moran NA. Microbial minimalism: genome reduction in bacterial pathogens. Cell. 2002;108:583–6.
Nelson WC, Stegen JC. The reduced genomes of Parcubacteria (OD1) contain signatures of a symbiotic lifestyle. Front Microbiol. 2015;6:693–14.
Burstein D, Sun CL, Brown CT, Sharon I, Anantharaman K, Probst AJ, et al. Major bacterial lineages are essentially devoid of CRISPR-Cas viral defence systems. Nat Commun. 2016;7:1–8.
Holman DB, Brunelle BW, Trachsel J, Allen HK. Meta-analysis to define a core microbiota in the swine gut. mSystems. 2017;2:676–14.
Anacker ML, Drecktrah D, LeCoultre RD, Lybecker M, Samuels DS. RNase III processing of rRNA in the Lyme disease Spirochete Borrelia burgdorferi. Journal of Bacteriology. Am Soc Microbiol J. 2018;200:1–11.
Iost I, Chabas S, Darfeuille F. Maturation of atypical ribosomal RNA precursors in Helicobacter pylori. Nucleic Acids Res. 2019;47:5906–21.
Gone S, Alfonso-Prieto M, Paudyal S, Nicholson AW. Mechanism of ribonuclease III catalytic regulation by serine phosphorylation. Nature. 2016;6:1–9.
Wilcon HR, Yu D, Peters HK III, Zhou J-G, Court DL. The global regulator RNase III modulates translation repression by the transcription elongation factor N. EMBO J. 2002;21:4154–61.
Hagen FS, Young ET. Effect of RNase III on efficiency of translation of bacteriophage T7 lysozyme mRNA. J Virol. 1978;26:793–804.
Bohannan BJM, Lenski RE. Linking genetic change to community evolution: insights from studies of bacteria and bacteriophage. Ecol Lett. 2000;3:362–77.
Song W, Joo M, Yeom J-H, Shin E, Lee M, Choi H-K, et al. Divergent rRNAs as regulators of gene expression at the ribosome level. Nat Microbiol. 2019;4:515–26.
Holland AD, Rothfuss HM, Lidstrom ME. Development of a defined medium supporting rapid growth for Deinococcus radiodurans and analysis of metabolic capacities. Appl Microbiol Biotechnol. 2006;72:1074–82.
Devos DP. Gemmata obscuriglobus. Curr Biol. 2013;23:R705–7.
Acknowledgements
This research was supported in part by the Chateaubriand Fellowship awarded to TEB from the Office for Science & Technology of the Embassy of France in the United States and a grant to NF from the U.S. National Science Foundation (EAR1331828). MA was supported by a research grant (15510) from Villum Fonden. AE gratefully acknowledges the support of a Leverhulme Trust Research Fellowship (RF-2017–652\2). ER was supported by the INCEPTION project (PIA/ANR-16-CONV-0005). We thank Will Trimble for assistance tracking down publicly available Moleculo sequences, Michael Engel for figure design input, and Eric Johnston for early discussions on unlinked rRNA genes.
Author information
Authors and Affiliations
Contributions
TEB, ER, and NF conceived and designed the project and wrote the paper with input from all co-authors. AE, MA, and RK performed the Nanopore sequencing. TEB performed all analyses.
Corresponding author
Ethics declarations
Conflict of interest
MA and RK own a portion of the company DNASense.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Brewer, T.E., Albertsen, M., Edwards, A. et al. Unlinked rRNA genes are widespread among bacteria and archaea. ISME J 14, 597–608 (2020). https://doi.org/10.1038/s41396-019-0552-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-019-0552-3
This article is cited by
-
High-resolution phylogenetic and population genetic analysis of microbial communities with RoC-ITS
ISME Communications (2022)
-
Microdiversity and phylogeographic diversification of bacterioplankton in pelagic freshwater systems revealed through long-read amplicon sequencing
Microbiome (2021)
-
Establishment and assessment of an amplicon sequencing method targeting the 16S-ITS-23S rRNA operon for analysis of the equine gut microbiome
Scientific Reports (2021)