Abstract
The acquisition of thermally tolerant algal symbionts by corals has been proposed as a natural or assisted mechanism of increasing coral reef resilience to anthropogenic climate change, but the cell-level processes determining the performance of new symbiotic associations are poorly understood. We used liquid chromatography–mass spectrometry to investigate the effects of an experimentally induced symbiosis on the host proteome of the model sea anemone Exaiptasia pallida. Aposymbiotic specimens were colonised by either the homologous dinoflagellate symbiont (Breviolum minutum) or a thermally tolerant, ecologically invasive heterologous symbiont (Durusdinium trenchii). Anemones containing D. trenchii exhibited minimal expression of Niemann-Pick C2 proteins, which have predicted biochemical roles in sterol transport and cell recognition, and glutamine synthetases, which are thought to be involved in nitrogen assimilation and recycling between partners. D. trenchii-colonised anemones had higher expression of methionine-synthesising betaine–homocysteine S-methyltransferases and proteins with predicted oxidative stress response functions. Multiple lysosome-associated proteins were less abundant in both symbiotic treatments compared with the aposymbiotic treatment. The differentially abundant proteins are predicted to represent pathways that may be involved in nutrient transport or resource allocation between partners. These results provide targets for specific experiments to elucidate the mechanisms underpinning compensatory physiology in the coral–dinoflagellate symbiosis.
Similar content being viewed by others
Introduction
The cnidarian–dinoflagellate symbiosis allows biodiverse coral reef ecosystems to thrive within nutrient-limited oceans [1]. A range of cnidarian taxa, such as corals, sea anemones, and jellyfish, are obligate hosts of dinoflagellate algal symbionts from the family Symbiodiniaceae, which support their hosts through the exchange of photosynthetic products [2]. Genetic diversity within the Symbiodiniaceae is extensive, and this family has been recently reclassified into seven genera containing a rapidly increasing number of formally described species [3,4,5]. Symbiodiniaceae exhibit variation in their physiologies, with a range of thermal tolerances and host specificities [6, 7]. Some Symbiodiniaceae can form symbioses with a diversity of hosts, whereas many hosts only associate with a single dominant symbiont species [8, 9]. The underlying mechanisms of host–symbiont specificity remain only partially described but include cellular recognition and nutrient exchange between the partners [2, 10, 11]. Symbiotic interactions between cnidarians and Symbiodiniaceae are thought to exist on a continuum from mutualism to parasitism [12, 13]. Symbionts can exhibit less beneficial qualities such as impairing host metabolism and reproduction, reducing the proportion of photosynthate translocated to the host, or increasing nitrogen assimilation at the expense of the host, particularly at elevated temperatures [13,14,15]. Symbiont genotypes that are found naturally in a given host species are considered to be homologous, while symbioses with heterologous symbionts can occur due to perturbation of the symbiosis by environmental stress, geographic invasion, or experimental manipulation [16, 17]. Some heterologous symbionts have been demonstrated to translocate lower amounts of photosynthetically fixed carbon to their hosts compared with homologous symbionts [16, 17].
Coral reefs worldwide are threatened by warming oceans inducing the breakdown of the coral–dinoflagellate symbiosis, also called coral bleaching [18,19,20]. Unless a symbiont population can be re-established quickly, coral bleaching may lead to host mortality [21]. Some Symbiodiniaceae are particularly tolerant of higher temperatures, both while free living and in symbiosis, potentially providing greater resistance to host bleaching [22,23,24]. These thermally-tolerant algae can act as opportunists, replacing the previously dominant symbiont genotype if its population declines, as occurs in a bleaching event [25, 26]. It has been proposed that corals may be able to adapt to climate change by ‘shuffling’ or ‘switching’ their symbiont assemblages, resulting in a more thermally tolerant symbiont species as the dominant partner, and a more thermally robust holobiont of coral, symbiont and associated microbes [7, 27,28,29]. Durusdinium trenchii (formerly Symbiodinium trenchii) is endemic to the Indo-Pacific, where it exists in mutualisms with multiple scleractinian coral genera, often in warm-water environments, and has been recently introduced into the Caribbean [16, 23, 30, 31]. In many Caribbean coral species, it exists as a background population that becomes temporarily dominant after bleaching, an example of shuffling [32, 33]; however, it is unknown if heterologous symbionts are capable of supplanting the homologous symbiont species and establishing a lasting true mutualism that increases host fitness and persists over multiple host generations. Given that D. trenchii has no coevolutionary history with Caribbean cnidarians but has become established, if at very low cell densities, in a wide diversity of Caribbean corals, it is an ideal genotype in which to test the effects of heterologous symbiont colonisation on a naive host.
In the anemone Exaiptasia pallida, a model for corals commonly known as Aiptasia [2, 34,35,36], colonisation by heterologous symbionts results in poorly understood differences in host phenotype [11, 17, 37]. Reduced, poorly regulated, or inefficient nutrient or metabolite exchange might reduce growth and fitness of either or both partners, resulting in a less mutualistic symbiosis. Additionally, heterologous symbionts may be less tolerated by host immune functions and consequently limited to a lower maximum density within host tissues, further restricting their potential contribution to the symbiosis. We hypothesise that nutrient transport and resource allocation between partners, as well as host immune tolerance of intracellular symbionts, is more optimal in established symbioses and that novel heterologous symbioses will exhibit deficiencies in these functions. Therefore, proteins predicted to be involved in host immune responses, nutrient regulation of symbionts, or transporting specific metabolites between partners are primary candidates for determining symbiosis success, and we anticipate differences in their abundances between symbioses.
To test these hypotheses, we compared the effects of colonisation with a heterologous symbiont (D. trenchii [23]), with those of colonisation with a homologous symbiont (Breviolum minutum, formerly Symbiodinium minutum [38]) on the Aiptasia host proteome. Aiptasia experimentally colonised by D. trenchii have been demonstrated to show greatly reduced growth and asexual reproductive output, despite displaying similar photosynthetic rates and symbiont densities to Aiptasia colonised by the homologous symbiont [17]. Furthermore, combined transcriptomic and metabolomic analyses of the D. trenchii–Aiptasia symbiosis showed an increased host catabolism of storage compounds and a differential expression of signalling and reactive oxygen species pathways, indicating a suboptimal symbiosis [11]. Here, we performed a proteomic characterisation of cnidarians hosting homologous and heterologous symbionts to clarify what cell-level changes are associated with the presence of different symbiont partners. This comparative approach was undertaken for its ability to identify for further study candidate proteins and functions that contribute to the successful establishment and persistence of the cnidarian–dinoflagellate symbiosis and to better understand the implications of symbioses with non-endemic dinoflagellate symbionts.
Materials and methods
Experimental organisms
Individuals of the symbiotic sea anemone Aiptasia were collected from a clonal laboratory stock, strain NZ1. Anemones were rendered aposymbiotic (i.e., symbiont-free) using the menthol treatment method [39] and maintained in 0.22 µm-filtered seawater (FSW). Approximately 750 aposymbiotic individuals were divided amongst 18 containers and placed into 25 °C water baths designated for the three treatment groups: B. minutum-colonised, D. trenchii-colonised, and aposymbiotic anemones. Light was provided by fluorescent lamps at ~ 95 µmol photons m−2s −1 on a 12 h:12 h light:dark cycle. Dinoflagellate cultures were grown in f/2 medium at 25 °C under 100 µmol photons m−2 s−1 on a 12 h:12 h light:dark cycle for 6 weeks prior to inoculation. The B. minutum culture (strain ‘FLAp2’) was originally isolated from Exaiptasia pallida, while the D. trenchii culture (strain ‘Ap2’) was originally isolated from an unknown anemone in Okinawa. The genetic identity of each culture was verified by direct sequencing of ITS2 rDNA and cp23S [40].
Inoculation of Aiptasia
Anemones were inoculated during the midpoint of the light cycle by applying dinoflagellate cultures at a cell density of ~3 × 106 mL−1, combined with a dilute suspension of Artemia sp. nauplii to induce phagocytosis. This process was repeated weekly for three weeks (four weeks total). Full details of the inoculation procedure are provided in the Supplemental Materials.
Protein extraction
Five weeks after initial colonisation, anemones were flash frozen in liquid nitrogen five hours into the photoperiod and stored at −80 °C. Each biological replicate (n = 6) was composed of a pool of 20 individual anemones. Each biological replicate was thawed, mechanically homogenised, and an aliquot removed for symbiont cell density estimates. Symbiont cell densities were analysed as per Matthews et al. [11]. Protein content was quantified using the Bradford assay [41]. Low-speed centrifugation (2500 × g) was used to pellet the algae and the host fractions were lyophilised at –105 °C for 18 h. The genetic identity of dinoflagellates in each anemone sample was verified by ITS2 and cp23S sequencing as before [40]. 30 mg of lyophilised host material was extracted with methanol for a parallel metabolite study [37] before storage at –20 °C. These host samples were then solubilised, alkylated, digested with trypsin and desalted using the methods of Oakley et al. [42], which are fully detailed in the Supplemental Materials.
Identification of proteins using LC-ESI-MS/MS
Peptides were analysed by liquid chromatography–electrospray ionisation–tandem mass spectrometry (LC-ESI-MS/MS) using the instrumentation, settings and software described in [42]. The resulting spectra were searched against a custom Aiptasia database constructed primarily from the Aiptasia genome [43] and a contaminant database [44] using the Sequest algorithm in Proteome Discoverer (v2.1, Thermo Scientific). The X! Tandem algorithm in Scaffold (v4.4.8, Proteome Software, Inc.) was used to validate MS/MS-based peptide and protein identifications. Peptide and protein identifications required >99% probability by the Scaffold Local false discovery rate algorithm and a minimum of two peptides. Proteins sharing significant peptide evidence were grouped into protein clusters. Each biological replicate was analysed twice as technical replicates, and protein abundance calculated as the simple mean between technical replicates at the protein cluster level. To confirm that contaminating symbiont proteins were not present, all spectra were searched as above against a Symbiodinium microadriaticum protein sequence database [45]. This analysis identified minimal algal proteins (mean of 14 per sample), most of which are highly conserved among eukaryotes (data not shown). Full details are available in the Supplemental Materials. Mass spectrometry data are publically available at the PRIDE repository with the dataset identifier PXD009253.
Statistical analysis
Tests for significant differences in symbiont cell density between algal species were performed with SPSS (v. 20, IBM Corporation). Non-normal or heteroscedastic data were log10-transformed prior to analysis with Student’s t-tests (p < 0.05). Protein clusters were quantified by precursor intensity in Scaffold and statistical analyses were conducted in R v. 3.3.1 (www.r-project.org). To identify proteins with significantly different abundances between treatments, a custom R script for multivariate analysis with post hoc identifications and protein load normalisation was used, as described in Oakley et al. [42]. A linear model with treatment effects was fitted and compared against an intercept-only null model using an analysis of deviance χ2 test, where α = 0.05. This script is available online at https://zenodo.org/record/1327230#.W2Y-0FD-iUk. Protein clusters with significantly different concentrations between treatments were identified with the false discovery rate correction method of Benjamini and Hochberg [46], at a q-threshold of 0.05.
Proteins that were significantly different between treatments were searched against the UniProtKB database using BLAST in Geneious v10.0.2 (Biomatters Ltd, Auckland, New Zealand). The top manually-reviewed SwissProt match for each protein with an E-value ≤ 1 × 10−5 was used to assign an annotation, and any without a match were designated as hypothetical proteins. If the only match with an E-value ≤ 1 × 10−5 was an unannotated UniprotKB Nematostella vectensis sequence, that sequence identifier is listed. Principal component analysis (PCA) and heatmap plots were generated by ClustVis, using ln(x) transformed protein abundance values, Pearson’s correlations for row and column clustering, and average distances for row and column linkage [47]. Singular value decomposition with imputation was used to calculate principal components.
Comparison of transcript and protein data
A parallel study, using different biological samples, used RNASeq-based transcriptomics to assess changes in gene expression in B. minutum- and D. trenchii-colonised Aiptasia [11]. We attempted to assess the concordance of this dataset with the results of the proteomic analysis; full methods are available in the Supplemental Material. Briefly, transcripts from the original dataset (n = 80,703) were matched against the proteins identified (as above) using blastx with a threshold of 98% identity and an E-value of less than 1 × 10−20. Pearson’s correlation coefficient was calculated to test if the relationship between treatments was conserved between datasets by comparing the magnitude and direction of gene expression in protein abundances and mRNA counts of each matched gene.
Results and discussion
Colonisation success
Examination of anemones in the aposymbiotic control treatment revealed no re-establishment of background symbionts at the end of the experiment. Genotyping confirmed that symbionts at the end of the experiments were the same as those originally used for inoculation. Symbiont cell densities in anemones colonised by B. minutum (average of 1.12 × 107 ± 1.16 × 106 std. error) were 1.5-fold higher (p = 0.040) than those in anemones colonised by D. trenchii (average of 7.29 × 106 ± 1.19), which is in agreement with past studies [11, 17, 48]. D. trenchii cells are approximately 1.9-fold greater in volume than B. minutum cells, however, indicating similar total symbiont biomass density [5].
Proteins affected by symbiont presence and species
A total of 1608 proteins were identified among all treatments; 31 decoy (known false match) sequences were discarded, and 276 sequences were not analysed further due to their being detected in fewer than 25% of samples. Overall, 142 proteins were significantly differentially abundant between the three treatments (aposymbiotic, B. minutum and D. trenchii-colonised animals, q < 0.05). Of these, 129 sequences were annotated using sequence similarities from the SwissProt database (E-value ≤ 1 × 10−5), and the remaining 13 were categorised as hypothetical proteins; five of these 13 were homologues of predicted proteins in Nematostella vectensis. Between the B. minutum-colonised and aposymbiotic samples, 105 proteins were differentially expressed (Fig. S1, Tables S1, S2); 22 proteins were differentially expressed between B. minutum-colonised and D. trenchii-colonised anemones (Table S3), and 108 proteins were differentially expressed between D. trenchii-colonised and aposymbiotic anemones (Tables S1, S4). Anemones colonised by B. minutum and those colonised by D. trenchii shared 74 differentially expressed proteins when compared with aposymbiotic anemones (Table S1).
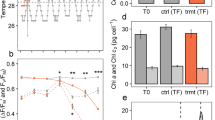
A principal component analysis of all detected proteins (Fig. 1a) and the differentially abundant proteins (Fig. 1b) showed that aposymbiotic Aiptasia were clearly distinguished from those colonised by either of the symbiont species. Anemones colonised by B. minutum or D. trenchii demonstrated overlap in their differentially expressed proteins, relative to the aposymbiotic anemones (Fig. 1b). Cluster analysis illustrated that the protein abundances of aposymbiotic anemones were separate from those of anemones colonised by B. minutum or D. trenchii (Fig. 1c). When compared to a previous transcriptomic analysis of the same symbioses [11], we found no consistent correlation between abundances of transcripts and proteins detected in both experiments (details in the Supplemental Materials). These data represent snapshots of independent experiments; thus, any correlation or lack thereof should be interpreted cautiously. Furthermore, correlations between transcript and protein abundances have been demonstrated to be low in other organisms [49], demonstrating the value of additional methodologies to complement transcript-based gene expression studies.
Principal component analysis plot and heatmap of proteins in aposymbiotic Aiptasia versus Aiptasia colonised by Breviolum minutum or Durusdinium trenchii. a Principal component analysis plot of all detected proteins, where each point represents one biological replicate (n = 6). Ellipses represent 95% confidence intervals. b Principal component analysis plot of differentially abundant proteins. c. Heatmap where each row represents one differentially abundant protein (n = 131) and each column represents one biological replicate. Proteins and biological replicates are clustered using average linkage and Pearson correlation distance. Protein abundances represent the mean of two unitless precursor intensities (as technical replicates) normalised to total protein content, ln(x) transformed and displayed relative to the mean. Proteins of note are indicated as follows: *NPC2; †glutamine synthetase; ‡betaine–homocysteine S-methyltransferase; §carbonic anhydrase
Symbiont presence, regardless of species, was associated with changes in a suite of Aiptasia proteins (summarised in Table 1), which we interpret as primarily representing changes to host cell physiology in accommodating symbiont cells and the gain of photosynthetic capability via symbiosis. These proteins have predicted functions in inorganic carbon acquisition, host immune processes, and catabolism of fatty acids, carbohydrates, and proteins (see below). The homologous and heterologous symbiont associations displayed differences in their abundances of proteins with predicted lipid transport, methionine metabolism, and immune regulation functions (Table 2). These reflect host cell-level differences in response to the nature of the symbiosis. The potential roles of these proteins and their relevance to the physiological performance of the different symbioses, based on known homologue functions, are discussed below.
Dissolved inorganic carbon (DIC) transport
Two carbonic anhydrase homologues were found to be highly upregulated (4-fold) in both symbiotic associations when compared with the aposymbiotic state (q < 1.3 × 10−2, Tables 1, 2). Carbonic anhydrase is an integral part of the carbon concentrating mechanism used by symbiotic cnidarians to facilitate the interconversion of DIC species, which, in turn, makes the transport of DIC from seawater to symbiont plastids for carbon fixation more efficient [50,51,52]. The significantly higher abundance of carbonic anhydrase homologues suggests that DIC transport functions are induced in both homologous and heterologous symbiont associations. When comparing the physiology of multiple Symbiodiniaceae in the coral Orbicella spp., Pettay et al. [16] noted that D. trenchii-colonised hosts had similar rates of photosynthetic oxygen evolution but much lower rates of host calcification, indicating that algal DIC acquisition was uninhibited but that the host was not benefiting substantially from the produced photosynthate. Photosynthetic activity alters DIC and pH within host tissues [53]; however, whether the apparent carbonic anhydrase expression in hosts containing symbionts is a homeostatic response to regulate these parameters or is induced by recognition of the symbionts is unknown.
Endolysosomal processes
Of the 74 shared proteins differentially expressed between anemones colonised by either of the symbiont species and aposymbiotic anemones (Fig. S1), many are homologues of proteins typically localised to the lysosome (Table 1). Most of these differentially expressed and presumably endolysosomal proteins were less abundant in either of the symbiotic associations relative to the aposymbiotic state. The symbiosome membrane, which surrounds intracellular symbiont cells, is an arrested endosomal membrane that avoids lysosomal fusion [2, 54].
The protein that most significantly differentiates Aiptasia hosting the homologous B. minutum is Niemann-Pick 2 (NPC2); two homologues of NPC2 were detected exclusively in anemones colonised by B. minutum, and they were undetectable in aposymbiotic anemones or those hosting D. trenchii (q < 2 × 10−8, Table 2). In the broader UniProtKB database, these proteins were most similar to the NPC2-d protein of Anemonia viridis [55]. Although a third NPC2 protein, most similar to NPC2-a, was detected at low levels in aposymbiotic anemones, it was significantly more abundant following colonisation with B. minutum (q = 1.9 × 10−2, Table 2). None of the NPC2 homologues had significantly different abundances between aposymbiotic and D. trenchii-colonised anemones. Four NPC2 genes have been found in the symbiotic sea anemone Anemonia viridis, where NPC2-d is localised to the symbiosome compartment [55, 56], and six NPC2 genes have been identified in the Aiptasia genome [43]. In mammals, NPC2 is a soluble lysosomal protein that rapidly binds and shuttles cholesterol to the membrane-bound NPC1 protein for transfer and export across the lysosomal membrane [57, 58]. The cnidarian NPC2 protein may act in symbiont recognition and persistence, based on its role in the immune signalling pathways of other invertebrates [55, 59], and it has been proposed that NPC2 participates in the transport of sterols between partners [43, 55, 56, 60, 61], but this hypothesised function requires further investigation. Homologues of acid ceramidase and peroxiredoxin, both of which have predicted lysosomal lipid degradation functions [62, 63], were also upregulated in both symbioses (Table 1).
A successful intracellular mutualism requires host recognition of potential symbiont cells, tolerating their presence while regulating their growth, and this process begins with the formation of the symbiosome [2]. To persist, the symbiosome must then avoid host lysosomal fusion, which may be a potential mechanism for symbiont population regulation [2]. A homologue of the Ras-related protein Rab2, a signalling protein that is required for the fusion of late endosomes and lysosomes in Drosophila [64], was more abundant in anemones colonised by either B. minutum or D. trenchii when compared with aposymbiotic anemones (q = 1.4 × 10−2, Table 1). We detected differential abundances of several proteins with predicted immune-related protease functions depending on symbiotic state and symbiont species, including homologues of legumain, neurolysin, and several cathepsins. A legumain homologue was 4.7-fold more abundant in anemones colonised by B. minutum versus D. trenchii (q < 3 × 10−3, Table 2), while anemones hosting either B. minutum or D. trenchii expressed much lower abundances of homologues of cathepsins L, Z, and the lysosomal dipeptidyl peptidase 2 than did aposymbiotic anemones (q < 2 × 10−2, Tables 1, 2). Legumain is primarily localised to acidic compartments of the endolysosomal system [65]. It is has a role in antigen processing and Toll-like receptor (TLR) signalling via the highly-specific cleavage of TLR receptors [66]. TLR signalling is thought to be a primary means of immune regulation in the cnidarian innate immune system [67]. A homologue of the peptidase neurolysin was also upregulated (3.0-fold, q = 1.2 × 10−2) in B. minutum versus D. trenchii-colonised anemones. Like legumain, the cathepsins are cysteine proteases of the endolysosomal system that play roles in TLR signalling and immunity; they also have protein catabolic functions in autophagic and endosomal vacuoles [68]. Legumain and neurolysin are unlike the other peptidases in that their homologues were upregulated in the B. minutum symbiosis relative to the D. trenchii symbiosis (Table 2), and this may be a result of host immune responses that differ in the presence of different symbiont species. Furthermore, both cathepsin L and Z homologues exhibited much lower expression when Aiptasia hosted either algal species than when the anemones were devoid of symbionts (Table 1). Interestingly, cathepsin L deficiency in mice results in the generation of large, long-lived autophagolysosomes [69]. As a hypothesis, a potential role of cathepsins in symbiosome maturation merits further study.
Potential immune responses to symbiont presence
The role of the coral immune response in symbiont specificity is an area of active research [70]. Compared with aposymbiotic anemones, a homologue of the 40 S ribosomal protein S19 was elevated 4-fold and 7.6-fold in anemones colonised by B. minutum and D. trenchii, respectively (q < 1.7 × 10−2, Table 1). As this pattern was not found for other ribosomal protein homologues, we considered the possibility that cnidarian S19 has roles in regulation of the symbiosis. In addition to its ribosomal functions, S19 attenuates the activity of the pro-inflammatory cytokine macrophage migration inhibitory factor (MIF) in vertebrates [71]. Apicomplexans, a sister taxon to the dinoflagellates, appear to exploit this to facilitate parasitic infection of vertebrates [72, 73], and MIF is also involved in regulating parasite populations in other invertebrates [74]. Given the increased abundance of an S19 homologue in anemones colonised by dinoflagellates, especially D. trenchii, S19 and its potential role in the cnidarian host immune response to dinoflagellate colonisation should be a topic for further targeted study. A homologue of the circumsporozoite protein (CSP) was highly upregulated (4.5-fold) in B. minutum-colonised anemones compared with D. trenchii-colonised or aposymbiotic anemones (q < 2.1 × 10−2, Table 2). CSP is the primary surface antigen produced by the apicomplexan Plasmodium, and it plays a role in the invasion of host cells by receptor-mediated binding to the host plasma membrane [75]. A potential role of the CSP homologue in the cnidarian–dinoflagellate symbiosis as a host surface ligand is interesting given the close evolutionary relationship between apicomplexans and dinoflagellates, but more directed investigation is needed to determine both the function and localisation of this protein.
Nitrogen and methionine metabolism
Nitrogen exchange between partners is central to the cnidarian–dinoflagellate symbiosis, particularly in oligotrophic reef waters, and glutamine synthetase is thought to be the primary enzyme responsible for inorganic nitrogen assimilation in both host and symbiont [76,77,78]. Two glutamine synthetase homologues were more abundant in B. minutum- versus D. trenchii-colonised anemones (q < 1.2 × 10−3, Table 2). Increased glutamine synthetase activity has previously been reported to be characteristic of symbiotic Aiptasia and other cnidarians [60, 76, 79]. Ammonium can be assimilated by both the host and symbiont through the glutamine synthetase/glutamine oxoglutarate aminotransferase cycle [78], which is used to catalyse the conversion of ammonia and glutamate into the amino acid glutamine [80]. Parallel metabolomic analysis of the Aiptasia specimens used in this study measured greater abundances of labelled glutamine in D. trenchii-colonised hosts relative to B. minutum-colonised hosts [37], suggesting either an increased demand or a reduced synthesis of glutamine in anemones hosting B. minutum. When combined, the glutamine synthetase protein and glutamine metabolite data seem to be consistent with, but cannot confirm, simultaneous increased host glutamine synthesis and consumption when hosting homologous symbionts. This may simply be a direct result of the higher density of B. minutum cells than D. trenchii; alternately, an enhanced abundance of glutamine synthetase may serve as a mechanism for avoiding nitrogen limitation of the holobiont.
Given the importance of nitrogen and amino acid exchange in a productive coral–algal symbiosis, the observed differential expression of homologues of the methionine-synthesising enzyme betaine–homocysteine S-methyltransferase (BHMT) between symbiont species is also notable. BHMT synthesises methionine from homocysteine and betaine and acts as a central enzyme in the methionine cycle responsible for molecular methylation [81, 82]. The expression levels of two and three different BHMT protein homologues, respectively, were lower in anemones colonised by B. minutum and in those hosting D. trenchii, relative to aposymbiotic anemones (q < 3.3 × 10−2, Table 2). Aposymbiotic Aiptasia have previously been demonstrated to both synthesise methionine and exhibit high abundances of BHMT compared with Aiptasia colonised by homologous symbionts [60, 83, 84]. There is evidence that, in symbiotic corals, half of the methionine pathway enzymes are expressed by the host while the other half are expressed by the symbiont, suggesting that partner coordination may be important for the proper functioning of this pathway [85]. The observed changes in BHMT homologue abundances suggest a change in either the methionine demand by the host or its supply by the symbiont. In a parallel study, metabolomic analysis detected doubled concentrations of methionine in Aiptasia colonised by D. trenchii rather than B. minutum [11]. One hypothesis consistent with these data is that although aposymbiotic Aiptasia are capable of de novo methionine synthesis via the BHMT pathway, they may receive sufficient methionine (or methionine precursors) from homologous B. minutum, but not heterologous D. trenchii, thus necessitating an upregulation of BHMT in D. trenchii-colonised anemones; this topic requires further research.
Besides being required for protein synthesis, methionine is a substrate for synthesis of the methyl donor s-adenosylmethionine and the antioxidants dimethylsulfoniopropionate (DMSP) and glutathione. DMSP is a strong reactive oxygen species scavenger, and its impact on mediating coral bleaching is of interest given that DMSP concentrations in coral tissues are greatly elevated during acute thermal stress concurrent with photoinhibition and reactive oxygen species generation [86], as well as being dependent on symbiont genotype [87]. Increased BHMT transcript expression corresponded with increased DMSP concentrations in Acropora under osmotic stress [88]. A persulfide dioxygenase homologue was much less abundant in symbiosis, regardless of symbiont species (q < 2.8 × 10−6, Table 1). This enzyme catabolizes toxic hydrogen sulphide, which is generated from the interconversion of homocysteine to cysteine, to glutathione [89]. The upregulation of BHMT homologues and of other proteins with predicted functions in oxidative stress responses in anemones hosting D. trenchii, together with previously reported methionine accumulation, suggest that symbioses may differ in their redox state. Direct measurements of host reactive oxygen species concentrations, and studies of the metabolic origins of antioxidants in the host, are needed to test whether hosts harbouring heterologous symbionts suffer elevated oxidative stress.
ER and oxidative stress
Among the proteins that were more abundantly expressed in the D. trenchii-colonised Aiptasia, two have predicted functions which suggest changes in protein assembly or endoplasmic reticulum (ER) performance. A homologue of the neurotropic factor protein Armet was upregulated 3.3-fold in these anemones relative to those hosting B. minutum (q < 1.8 × 10−2, Table 2). Armet is secreted in response to misfolded protein accumulation in the ER, protecting cells against stress-induced cell death [90]. Additionally, BiP, a 78 kDa glucose-regulating protein that also plays a role in the ER protein folding stress-response [91], was previously found to be both constitutively abundant and upregulated in Aiptasia during heat shock [42]. Here, a homologue of BiP was more abundant in D. trenchii-colonised than in B. minutum-colonised anemones (q < 4.2 × 10−3). The upregulation of BiP and Armet homologues suggests an elevated Aiptasia host stress response when colonised by D. trenchii, as supported by previously reported gene transcript and metabolite data [11].
Pirin is a highly conserved, redox-sensitive regulator of the transcription factor NF-κB that is upregulated in response to oxidative stress [92]. A pirin homologue was significantly more abundant in aposymbiotic anemones than in those colonised by either B. minutum or D. trenchii (q < 3.9 × 10−3) and was also more abundant in B. minutum- than in D. trenchii-colonised anemones (q < 3.8 × 10−2, Table 2). In its oxidised form, pirin is proposed to bind and activate NF-κB [92], which is highly conserved, moderates immune function, and has been shown to be suppressed by dinoflagellate colonisation in Aiptasia [93].
Conclusion
This study identified proteins (summarised in Fig. 2) that differed in abundance between Aiptasia hosting different dinoflagellate symbiont species, and the predicted functions of these proteins give rise to hypotheses regarding the establishment and regulation of effective cnidarian–dinoflagellate symbioses. A NPC2-d homologue distinguished the homologous symbiosis and expression of a glutamine synthetase homologue varied between symbioses. Anemones colonised by the homologous B. minutum exhibited elevated levels of proteins with predicted nitrogen and lipid metabolism functions, whereas those colonised by the heterologous D. trenchii had greater abundances of proteins with predicted methionine pathway and oxidative stress functions. Homologues of carbonic anhydrases were expressed in both symbioses, emphasising the importance of DIC availability and photosynthetic performance for the symbiont. Only those anemones colonised by B. minutum expressed detectable levels of a homologue for NPC2-d protein, for which there is increasing evidence of a role in successful cnidarian–dinoflagellate associations [55, 56]. If this finding is confirmed in corals, it may offer a potential mechanism to explain why D. trenchii rarely dominates cnidarian hosts in its invasive range despite its ubiquity and thermal resilience [16]. Even in hosts in which it is homologous, D. trenchii is known to trade fitness for thermal tolerance [94]. D. trenchii colonisation also increases the methionine concentration [37] and BHMT homologue abundance in Aiptasia. These data identify new targets for investigating the cell biology of the cnidarian–dinoflagellate symbiosis as well as host metabolic processes that may be affected by symbiosis with heterologous symbionts. If consistent among novel cnidarian–dinoflagellate pairings, these responses may serve as physiological barriers to the establishment of successful, ecologically fit coral symbioses unless they are mutable over timescales relevant to the increasing frequency of coral bleaching events.
Summary of differentially abundant proteins and their predicted functions in Aiptasia colonised by Breviolum minutum, Durusdinium trenchii, or lacking symbionts (aposymbiotic). Coloured proteins were significantly more abundant in the indicated treatment(s) than in the remaining treatment(s), and proteins shown in white were unchanged. Proteins have predicted functions in the indicated compartments, or their localisation was inferred by homology. Metabolite translocations that have not been experimentally confirmed are shown with dotted lines. Dashed boxes indicate proteins grouped by predicted protein metabolic function. ER endoplasmic reticulum, Golgi Golgi apparatus. Ar armet, ASAH1 acid ceramidase, BiP 78 kDa glucose-regulated protein, BHMT betaine-homocysteine S-methyltransferase, CA carbonic anhydrase, CatL/Z cathepsin L/Z, CSP circumsporozoite protein, DPP2 dipeptidyl peptidase 2, ETHE1 persulfide dioxygenase, GS glutamine synthetase, HEXA beta-hexosaminidase subunit alpha, LGMN legumain, MAN2B1 lysosomal alpha-mannosidase, NAGA alpha-n-acetylgalactosaminidase, NAGLU alpha-n-acetylglucosaminidase, NPC2 Niemann-Pick type C2 protein, PRDX6 peroxiredoxin-6, S19 40 S ribosomal protein S19. GOGAT glutamine synthetase/glutamine oxoglutarate aminotransferase cycle, Gln glutamine, NH3 ammonia, NH4+ ammonium, Hcy homocysteine, Met methionine. CA catalyses the interconversion of inorganic carbon species to facilitate diffusion into the algal cell for photosynthetic carbon fixation. CSP is a potential surface antigen, whereas NPC2 is localised to the symbiosome in other anemone symbioses. GS reduces ammonia into glutamine while also acting in the GOGAT cycle. BHMT reacts with homocysteine to produce methionine. Ar and BiP have predicted roles in protein misfolding in the ER and Golgi. Lysosomal proteins are grouped by the metabolite classes they catabolise
References
Porter JW, Muscatine L. Reef corals: mutualistic symbioses adapted to nutrient-poor environments. Bioscience. 1977;27:454–60.
Davy SK, Allemand D, Weis VM. Cell biology of cnidarian-dinoflagellate symbiosis. Microbiol Mol Biol Rev. 2012;76:229–61.
Lee SY, Jeong HJ, Kang NS, Jang TY, Jang SH, Lajeunesse TC. Symbiodinium tridacnidorum sp. nov., a dinoflagellate common to Indo-Pacific giant clams, and a revised morphological description of Symbiodinium microadriaticum Freudenthal, amended Trench & Blank. Eur J Phycol. 2015;50:155–72.
Wham DC, Ning G, LaJeunesse TC. Symbiodinium glynnii sp. nov., a species of stress tolerant symbiotic dinoflagellates from pocilloporid and montiporid corals in the Pacific Ocean. Phycologia. 2017;56:396–409.
LaJeunesse TC, Parkinson JE, Gabrielson PW, Jeong HJ, Reimer JD, Voolstra CR, et al. Systematic revision of Symbiodiniaceae highlights the antiquity and diversity of coral endosymbionts. Curr Biol. 2018;28:2570–80.
Rowan R, Knowlton N, Baker A, Jara J. Landscape ecology of algal symbionts creates variation in episodes of coral bleaching. Nature. 1997;388:265–9.
Berkelmans R, van Oppen MJH. The role of zooxanthellae in the thermal tolerance of corals: a ‘nugget of hope’ for coral reefs in an era of climate change. Proc Biol Sci. 2006;273:2305–12.
Xiang T, Hambleton EA, DeNofrio JC, Pringle JR, Grossman AR. Isolation of clonal axenic strains of the symbiotic dinoflagellate Symbiodinium and their growth and host specificity. J Phycol. 2013;49:447–58.
Santos SR, Shearer TL, Hannes AR, Coffroth MA. Fine-scale diversity and specificity in the most prevalent lineage of symbiotic dinoflagellates (Symbiodinium, Dinophyceae) of the Caribbean. Mol Ecol. 2004;13:459–69.
Bay LK, Cumbo VR, Abrego D, Kool JT, Ainsworth TD, Willis BL. Infection dynamics vary between Symbiodinium types and cell surface treatments during establishment of endosymbiosis with coral larvae. Diversity. 2011;3:356–74.
Matthews JL, Crowder CM, Oakley CA, Lutz A, Roessner U, Meyer E, et al. Optimal nutrient exchange and immune responses operate in partner specificity in the cnidarian-dinoflagellate symbiosis. Proc Natl Acad Sci. 2017;114:13194–9.
Lesser M, Stat M, Gates RD. The endosymbiotic dinoflagellates (Symbiodinium sp.) of corals are parasites and mutualists. Coral Reefs. 2013;32:603–11.
Baker DM, Freeman CJ, Wong JCY, Fogel ML, Knowlton N. Climate change promotes parasitism in a coral symbiosis. ISME J. 2018;12:921–30.
Jones AM, Berkelmans R. Tradeoffs to thermal acclimation: energetics and reproduction of a reef coral with heat tolerant Symbiodinium type-D. J Mar Biol. 2011;2011:1–12.
Stat M, Morris E, Gates RD. Functional diversity in coral-dinoflagellate symbiosis. Proc Natl Acad Sci USA. 2008;105:9256–61.
Pettay DT, Wham DC, Smith RT, Iglesias-Prieto R, LaJeunesse TC. Microbial invasion of the Caribbean by an Indo-Pacific coral zooxanthella. Proc Natl Acad Sci. 2015;112:201502283.
Gabay Y, Weis VM, Davy SK. Symbiont identity influences patterns of symbiosis establishment, host growth, and asexual reproduction in a model cnidarian-dinoflagellate symbiosis. Biol Bull. 2018;234:1–10.
Hoegh-Guldberg O, Mumby PJ, Hooten AJ, Steneck RS, Greenfield P, Gomez E, et al. Coral reefs under rapid climate change and ocean acidification. Science. 2007;318:1737–42.
Hughes TP, Kerry J, Álvarez-Noriega M, Álvarez-Romero J, Anderson K, Baird A, et al. Global warming and recurrent mass bleaching of corals. Nature. 2017;543:373–8.
Hughes TP, Anderson KD, Connolly SR, Heron SF, Kerry JT, Lough JM, et al. Spatial and temporal patterns of mass bleaching of corals in the Anthropocene. Science. 2018;359:80–83.
Bosch TCG, Miller DJ. Bleaching as an obvious dysbiosis in corals. In: The holobiont imperative: perspectives from early emerging animals. Springer: Vienna; 2016. pp. 113–25.
Hume BCC, Angelo CD, Smith EG, Stevens JR, Burt J, Wiedenmann J. Symbiodinium thermophilum sp. nov., a thermotolerant symbiotic alga prevalent in corals of the world’s hottest sea, the Persian/Arabian Gulf. Sci Rep. 2015;5:8562.
LaJeunesse TC, Wham DC, Pettay DT, Parkinson JE, Keshavmurthy S, Chen CA. Ecologically differentiated stress-tolerant endosymbionts in the dinoflagellate genus Symbiodinium (Dinophyceae) Clade D are different species. Phycologia. 2014;53:305–19.
Swain TD, Chandler J, Backman V, Marcelino L. Consensus thermotolerance ranking for 110 Symbiodinium phylotypes: an exemplar utilization of a novel iterative partial-rank aggregation tool with broad application potential. Funct Ecol. 2017;31:172–83.
LaJeunesse TC, Smith RT, Finney J, Oxenford H. Outbreak and persistence of opportunistic symbiotic dinoflagellates during the 2005 Caribbean mass coral ‘bleaching’ event. Proc Biol Sci. 2009;276:4139–48.
Kemp DW, Hernandez-Pech X, Iglesias-Prieto R, Fitt WK, Schmidt GW. Community dynamics and physiology of Symbiodinium spp. before, during, and after a coral beaching event. Limnol Oceanogr. 2014;59:788–97.
Baskett ML, Gaines SD, Nisbet RM. Symbiont diversity may help coral reefs survive moderateclimate change. Ecol Appl. 2009;19:3–17.
Baker AC. Flexibility and specificity in coral-algal symbiosis: diversity, ecology, and biogeography of Symbiodinium. Annu Rev Ecol Evol Syst. 2003;34:661–89.
Buddemeier RW, Fautin DG. Coral bleaching as an adaptative mechanism. Bioscience. 1993;43:320–6.
Hoadley KD, Pettay DT, Grottoli AG, Cai W-J, Melman TF, Levas S, et al. High-temperature acclimation strategies within the thermally tolerant endosymbiont Symbiodinium trenchii and its coral host, Turbinaria reniformis, differ with changing pCO2 and nutrients. Mar Biol. 2016;163:134.
Howells EJ, Abrego D, Meyer E, Kirk NL, Burt JA. Host adaptation and unexpected symbiont partners enable reef-building corals to tolerate extreme temperatures. Glob Chang Biol. 2016;22:2702–14.
Grottoli G, Warner ME, Levas SJ, Matthew D. The cumulative impact of annual coral bleaching can turn some coral species winners into losers. Glob Chang Biol. 2014;20:3823–33.
Kennedy EV, Foster NL, Mumby PJ, Stevens JR. Widespread prevalence of cryptic Symbiodinium D in the key Caribbean reef builder, Orbicella annularis. Coral Reefs. 2015;34:519–31.
Grajales A, Rodríguez E. Morphological revision of the genus Aiptasia and the family Aiptasiidae (Cnidaria, Actiniaria, Etridioidea). Zootaxa. 2014;3826:55–100.
Weis VM, Davy SK, Hoegh-Guldberg O, Rodriguez-Lanetty M, Pringle JR. Cell biology in model systems as the key to understanding corals. Trends Ecol Evol. 2008;23:369–76.
Goldstein B, King N. The future of cell biology: emerging model organisms. Trends Cell Biol. 2016;26:818–24.
Matthews JL, Oakley CA, Lutz A, Hillyer KE, Roessner U, Grossman AR, et al. Partner switching and metabolic flux in a model cnidarian—dinoflagellate symbiosis. Proc R Soc L B. 2018;285:20182336.
LaJeunesse TC, Parkinson JE, Reimer JD. A genetics-based description of Symbiodinium minutum sp. nov. and S. psygmophilum sp. nov. (Dinophyceae), two dinoflagellates symbiotic with cnidaria. J Phycol. 2012;48:1380–91.
Matthews JL, Sproles AE, Oakley CA, Grossman AR, Weis VM, Davy SK. Menthol-induced bleaching rapidly and effectively provides experimental aposymbiotic sea anemones (Aiptasia sp.) for symbiosis investigations. J Exp Biol. 2016;219:306–10.
Hill R, Fernance C, Wilkinson SP, Davy SK, Scott A. Symbiont shuffling during thermal bleaching and recovery in the sea anemone Entacmaea quadricolor. Mar Biol. 2014;161:2931–7.
Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54.
Oakley CA, Durand E, Wilkinson SP, Peng L, Weis VM, Grossman AR, et al. Thermal shock induces host proteostasis disruption and endoplasmic reticulum stress in the model symbiotic cnidarian Aiptasia. J Proteome Res. 2017;16:2121–34.
Baumgarten S, Simakov O, Esherick LY, Liew YJ, Lehnert EM, Michell CT, et al. The genome of Aiptasia, a sea anemone model for coral symbiosis. Proc Natl Acad Sci USA. 2015;112:11893–8.
Mellacheruvu D, Wright Z, Couzens AL, Lambert J, St-Denis N, Li T, et al. The CRAPome: A contaminant repository for affinity purification mass spectrometry data. Nat Methods. 2014;10:730–6.
Aranda M, Li Y, Liew YJ, Baumgarten S, Sim O, Wilson MC, et al. Genomes of coral dinoflagellate symbionts highlight evolutionary adaptations conducive to a symbiotic lifestyle. Sci Rep. 2016; 1–15.
Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat B. 1995;57:289–300.
Metsalu T, Vilo J. ClustVis: a web tool for visualizing clustering of multivariate data using principal component analysis and heatmap. Nucleic Acids Res. 2015;43:W566–W570.
Starzak DE, Quinnell RG, Nitschke MR, Davy SK. The influence of symbiont type on photosynthetic carbon flux in a model cnidarian—dinoflagellate symbiosis. Mar Biol. 2014;161:711–24.
Taniguchi Y, Choi PJ, Li G, Chen H, Babu M, Hearn J, et al. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010;329:533–58.
Bertucci A, Moya A, Tambutte S, Allemand D, Supuran CT, Zoccola D. Carbonic anhydrases in anthozoan corals—a review. Bioorganic Med Chem. 2013;21:1437–50.
Weis VM. The induction of carbonic anhydrase in the symbiotic sea anemone Aiptasia pulchella. Biol Bull. 1991;180:496–504.
Tansik AL, Fitt WK, Hopkinson BM. Inorganic carbon is scarce for symbionts in scleractinian corals. Limnol Oceanogr. 2017;62:2045–55.
Kuhl M, Cohen Y, Dalsgaard T, Jergensenl BB, Revsbech NP. Microenvironment and photosynthesis of zooxanthellae in scleractinian corals studied pH and light with microsensors for O2, pH and light. Mar Ecol Prog Ser. 1995;117:159–72.
Mohamed AR, Cumbo V, Harii S, Shinzato C, Chan CX, Ragan MA, et al. The transcriptomic response of the coral Acropora digitifera to a competent Symbiodinium strain: The symbiosome as an arrested early phagosome. Mol Ecol. 2016;25:3127–41.
Dani V, Ganot P, Priouzeau F, Furla P, Sabourault C. Are Niemann-Pick type C proteins key players in cnidarian-dinoflagellate endosymbioses? Mol Ecol. 2014;23:4527–40.
Dani V, Priouzeau F, Mertz M, Mondin M, Pagnotta S, Lacas-Gervais S, et al. Expression patterns of sterol transporters NPC1 and NPC2 in the cnidarian–dinoflagellate symbiosis. Cell Microbiol. 2017;19:1–13.
Storch J, Xu Z. Niemann-Pick C2 (NPC2) and intracellular cholesterol trafficking. Biochim Biophys Acta Mol Cell Biol Lipids. 2009;1791:671–8.
Infante RE, Wang ML, Radhakrishnan A, Kwon HJ, Brown MS, Goldstein JL. NPC2 facilitates bidirectional transfer of cholesterol between NPC1 and lipid bilayers, a step in cholesterol egress from lysosomes. Proc Natl Acad Sci USA. 2008;105:15287–92.
wShi X-Z, Zhong X, Yu X-Q. Drosophila melanogaster NPC2 proteins bind bacterial cell wall components and may function in immune signal pathways. Insect Biochem Mol Biol. 2012;42:545–56.
Oakley CA, Ameismeier MF, Peng L, Weis VM, Grossman AR, Davy SK. Symbiosis induces widespread changes in the proteome of the model cnidarian Aiptasia. Cell Microbiol. 2016;18:1009–23.
Lehnert EM, Mouchka ME, Burriesci MS, Gallo ND, Schwarz JA, Pringle JR. Extensive differences in gene expression between symbiotic and aposymbiotic cnidarians. Genes Genomes Genet. 2014;4:277–95.
Fisher AB. Peroxiredoxin 6: A bifunctional enzyme with glutathione peroxidase and phospholipase A2 activities. Antioxid Redox Signal. 2011;15:831–44.
Gebai A, Gorelik A, Li Z, Illes K, Nagar B. Structural basis for the activation of acid ceramidase. Nat Commun. 2018;9:1621.
Lörincz P, Tóth S, Benkö P, Lakatos Z, Boda A, Glatz G, et al. Rab2 promotes autophagic and endocytic lysosomal degradation. J Cell Biol. 2017;216:1937–47.
Dall E, Brandstetter H. Structure and function of legumain in health and disease. Biochimie. 2016;122:126–50.
Sepulveda FE, Maschalidi S, Colisson R, Heslop L, Ghirelli C, Sakka E, et al. Critical role for asparagine endopeptidase in endocytic toll-like receptor signaling in dendritic cells. Immunity. 2009;31:737–48.
Miller DJ, Hemmrich G, Ball EE, Hayward DC, Khalturin K, Funayama N, et al. The innate immune repertoire in Cnidaria—ancestral complexity and stochastic gene loss. Genome Biol. 2007;8:R59.1–R59.13.
Müller S, Dennemärker J, Reinheckel T. Specific functions of lysosomal proteases in endocytic and autophagic pathways. Biochim Biophys Acta Proteins Proteomics. 2012;1824:34–43.
Dennemärker J, Lohmüller T, Müller S, Aguilar SV, Tobin DJ, Peters C, et al. Impaired turnover of autophagolysosomes in cathepsin L deficiency. Biol Chem. 2010;391:913–22.
Parkinson JE, Tivey TR, Mandelare PE, Adpressa DA, Loesgen S, Weis VM. Subtle differences in symbiont cell surface glycan profiles do not explain species-specific colonization rates in a model cnidarian-algal symbiosis. Front Microbiol. 2018;9:842.
Filip AM, Klug J, Cayli S, Fröhlich S, Henke T, Lacher P, et al. Ribosomal protein S19 interacts with macrophage migration inhibitory factor and attenuates its pro-inflammatory function. J Biol Chem. 2009;284:7977–85.
Moore RB, Oborník M, Janouškovec J, Chrudimský T, Vancová M, Green DH, et al. A photosynthetic alveolate closely related to apicomplexan parasites. Nature. 2008;451:959–63.
Holowka T, Bucala R. MIF family cytokines in innate immunity and homeostasis. In: Bernhagen J, Bucala R (eds). Progress in inflammation research. Cham: Springer International Publishing; 2017.
Baeza Garcia A, Pierce RJ, Gourbal B, Werkmeister E, Reichhart J-M, Dissous C, et al. Involvement of the cytokine MIF in the snail host immune response to the parasite Schistosoma mansoni. PLoS ONE. 2010;6:e1001115.
Ferguson DJP, Balaban AE, Patzewitz EM, Wall RJ, Hopp CS, Poulin B, et al. The repeat region of the circumsporozoite protein is critical for sporozoite formation and maturation in Plasmodium. PLoS ONE. 2014;9:1–25.
Wang JT, Douglas AE. Nitrogen recycling or nitrogen conservation in an alga-invertebrate symbiosis? J Exp Biol. 1998;201:2445–53.
Miller D, Yellowlees D. Inorganic nitrogen uptake by symbiotic marine cnidarians: a critical review. Proc R Soc B Biol Sci. 1989;237:109–25.
Pernice M, Meibom A, Van Den Heuvel A, Kopp C, Domart-Coulon I, Hoegh-Guldberg O, et al. A single-cell view of ammonium assimilation in coral-dinoflagellate symbiosis. ISME J. 2012;6:1314–24.
Lipschultz F, Cook CB. Uptake and assimilation of 15N-ammonium by the symbiotic sea anemones Bartholomea annulata and Aiptasia pallida: Conservation versus recycling of nitrogen. Mar Biol. 2002;140:489–502.
Syrett APJ, Peplinska AM. Effects of nitrogen-deprivation, and recovery from it, on the metabolism of microalgae. New Phytol. 1988;109:289–96.
Finkelstein JD, Martin JJ. Methionine metabolism in mammals. J Biol Chem. 1984;259:9508–13.
Szegedi SS, Castro CC, Koutmos M, Garrow TA. Betaine-homocysteine S -methyltransferase-2 is an S−methylmethionine-homocysteine methyltransferase. J Biol Chem. 2008;283:8939–45.
Rodriguez-Lanetty M, Phillips WS, Weis VM. Transcriptome analysis of a cnidarian-dinoflagellate mutualism reveals complex modulation of host gene expression. BMC Genomics. 2006;7:23.
Wang JT, Douglas AE. Essential amino acid synthesis and nitrogen recycling in an alga-invertebrate symbiosis. Mar Biol. 1999;135:219–22.
Shinzato C, Inoue M, Kusakabe M. A snapshot of a coral ‘holobiont’: a transcriptome assembly of the scleractinian coral, Porites, captures a wide variety of genes from both the host and symbiotic zooxanthellae. PLoS ONE. 2014;9:e85182.
Raina J-B, Tapiolas DM, Forêt S, Lutz A, Abrego D, Ceh J, et al. DMSP biosynthesis by an animal and its role in coral thermal stress response. Nature. 2013;502:677–80.
Borell EM, Pettay DT, Steinke M, Warner M, Fine M. Symbiosis-specific changes in dimethylsulphoniopropionate concentrations in Stylophora pistillata along a depth gradient. Coral Reefs. 2016;35:1383–92.
Aguilar C, Raina JB, Motti CA, Fôret S, Hayward DC, Lapeyre B, et al. Transcriptomic analysis of the response of Acropora millepora to hypo-osmotic stress provides insights into DMSP biosynthesis by corals. BMC Genomics. 2017;18:1–14.
Kabil O, Banerjee R. Characterization of patient mutations in human persulfide dioxygenase (ETHE1) involved in H2S catabolism. J Biol Chem. 2012;287:44561–667.
Apostolou A, Shen Y, Liang Y, Luo J, Fang S. Armet, a UPR-upregulated protein, inhibits cell proliferation and ER stress-induced cell death. Exp Cell Res. 2008;314:2454–67.
Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2:326–32.
Liu F, Rehmani I, Esaki S, Fu R, Chen L, de Serrano V, et al. Pirin is an iron-dependent redox regulator of NF-B. Proc Natl Acad Sci. 2013;110:9722–7.
Mansfield KM, Carter NM, Nguyen L, Cleves PA, Alshanbayeva A, Williams LM, et al. Transcription factor NF-κB is modulated by symbiotic status in a sea anemone model of cnidarian bleaching. Sci Rep. 2017;7:1–14.
Stat M, Gates RD, Clade D. Symbiodinium in scleractinian corals: a “nugget” of hope, a selfish opportunist, an ominous sign, or all of the above? J Mar Biol. 2011;2011:1–9.
Acknowledgements
Financial support for this research was provided by the Marsden Fund of the Royal Society Te Apārangi, grant number 1202, to SKD, VMW and ARG. The authors thank S. Wilkinson for the script and the three anonymous reviewers for their comments.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Sproles, A.E., Oakley, C.A., Matthews, J.L. et al. Proteomics quantifies protein expression changes in a model cnidarian colonised by a thermally tolerant but suboptimal symbiont. ISME J 13, 2334–2345 (2019). https://doi.org/10.1038/s41396-019-0437-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-019-0437-5
This article is cited by
-
Seascape genomics of common dolphins (Delphinus delphis) reveals adaptive diversity linked to regional and local oceanography
BMC Ecology and Evolution (2022)
-
Colonization and metabolite profiles of homologous, heterologous and experimentally evolved algal symbionts in the sea anemone Exaiptasia diaphana
ISME Communications (2022)
-
Thylakoid fatty acid composition and response to short-term cold and heat stress in high-latitude Symbiodiniaceae
Coral Reefs (2022)
-
Broad scale proteomic analysis of heat-destabilised symbiosis in the hard coral Acropora millepora
Scientific Reports (2021)
-
Integrating novel tools to elucidate the metabolic basis of microbial symbiosis in reef holobionts
Marine Biology (2021)