Abstract
Lanthanides (Ln3+), known as rare earth elements, have recently emerged as enzyme cofactors, contrary to prior assumption of their biological inertia. Several bacterial alcohol dehydrogenases have been characterized so far that depend on Ln3+ for activity and expression, belonging to the methanol dehydrogenase clade XoxF and the ethanol dehydrogenase clade ExaF/PedH. Here we compile an inventory of genes potentially encoding Ln3+-dependent enzymes, closely related to the previously characterized XoxF and ExaF/PedH enzymes. We demonstrate their wide distribution among some of the most numerically abundant and environmentally important taxa, such as the phylogenetically disparate rhizobial species and metabolically versatile bacteria inhabiting world’s oceans, suggesting that reliance on Ln3+-mediated biochemistry is much more widespread in the microbial world than previously assumed. Through protein expression and analysis, we here more than double the extant collection of the biochemically characterized Ln3+-dependent enzymes, demonstrating a range of catalytic properties and substrate and cofactor specificities. Many of these enzymes reveal propensity for oxidation of methanol. This observation, in combination with genome-based reconstruction of methylotrophy pathways for select species suggests a much wider occurrence of this metabolic capability among bacterial species, and thus further suggests the importance of methylated compounds as parts of the global carbon cycling.
Similar content being viewed by others
Introduction
Metals are important elements in organic life, beside carbon, oxygen and nitrogen, as they serve as enzyme cofactors. Over years of research, a group of metals has been established of universal significance for life, and these became known as life metals. These include Na, K, Mg, Ca, Fe, Zn, Mn, Cu, Co and Mo [1]. At the same time, some metals, such as rare earth elements (REEs), while revealing attractive catalytic properties in in vitro manipulations, leading to their wide use in industrial and medical applications [2,3,4], have been assumed biologically inert [5]. This assumption has been dramatically reversed recently with the demonstration of the activity of REEs lanthanides (Ln3+) as cofactors of methanol dehydrogenases (MDH), enzymes essential in metabolism of single-carbon compounds such as methane and methanol [6,7,8,9,10,11,12,13,14]. An example of a Ln3+-dependent alcohol dehydrogenase (ADH) in a non-methylotroph has also been reported [15, 16]. In addition to their catalytic function, Ln3+ also act in regulation of gene expression, through a mechanism known as the ‘Ln3+ switch’ [10,11,12,13,14,15,16,17]. The understanding of the evolution of REE-linked enzymes is lacking, but it has been noted that they are more diverse than their Ca2+-dependent counterparts [18, 19]. It has been proposed that REE-linked enzymes may possess higher catalytic efficiencies compared to Ca2+-dependent counterparts [5], and some experimental data have been collected in support of this proposal [9, 20,21,22]. However, only a handful of REE-dependent enzymes have been characterized so far [7, 9, 15, 20, 23, 24], and no consensus exists at this point.
As the essential role of REE-linked ADH enzymes has been firmly established for the methylotrophy metabolism [25, 26], the important questions remain as to whether REEs are important in the context of other metabolisms and whether they are widespread beyond the known methylotrophs. In this study, we employed a recombinant system based on a Methylorubrum extorquens AM1 (formerly Methylobacterium extorquens AM1) mutant, to express and purify several REE-dependent enzymes from organisms representing some of the major taxa within Proteobacteria. These represent both, functional groups including methylotrophs and functional groups not known for methylotrophic life style. Our data demonstrate that, in general, these enzymes display similar properties across a range of organisms, while variations occur at the level of the downstream electron-accepting redox proteins. Straightforward bioinformatic inferences suggest that these enzymes, while appearing auxiliary in many taxa, are widely occurring in environmentally important and globally distributed microbes. The data we present here further suggest the importance of Ln3+ in microbial life, and potentially beyond, and firmly position these metals as true life metals.
Results and discussion
Lanthanide-dependent alcohol dehydrogenases of the XoxF5 and ExaF/PedH clades are widely distributed among Proteobacteria
While the role of Ln3+ has been already well established among the bona fide methylotrophs, it remains unclear how widespread are Ln3+-dependent biochemistries among bacteria in general, and, potentially, even beyond. Further investigations are required to understand the role of Ln3+ in life, and the significance of Ln3+ as substitutes for Ca2+ ions (or vice versa), including the evolutionarily aspects of the alternative biochemistries. To approach these questions through bioinformatics, we used sequences of the so far characterized XoxF5 enzymes [7, 23, 24], to query the genomes available in the IMG/M database, which includes finished and draft genomes of cultivated organisms, as well as genomic bins assembled from metagenomes (https://img. jgi.doe.gov/cgi-bin/m/main.cgi). We determined that representatives of 13 orders within Proteobacteria, a total of 698 genomes, encoded XoxF5-type enzymes that displayed amino acid identities over 65% with the characterized enzymes, placing them within the XoxF5 clade on XoxF phylogenetic trees [18, 19, 27], a total of 738 genes (some genomes encoding multiple homologs; Supplementary Tables 1 and 2). Of the 13 orders, only 1, the Methylococcales, was represented exclusively by the methylotrophs, organisms reliant on methanol oxidation for their major ecophysiological function [28] (Fig. 1). Five orders were represented by both species characterized as methylotrophs and species not known to be capable of methylotrophy, one by species capable of co-metabolism of methanol, while the remaining six orders were represented only by species not known for methylotrophic life styles (Fig. 1). This simple analysis demonstrates the wide distribution of XoxF5-type enzymes among alpha-, beta- and gammaproteobacteria, including some universally widespread, numerically abundant and ecologically significant functional guilds. These include the phylogenetically disparate rhizobia, the members of Vibrionales, known for parasitic relationships with multiple species, the metabolically versatile Rhodobacterales, abundant in world oceans, the ammonia-oxidizing Nitrosococcus, as well as other species within Chromatiales. In comparison, genes for the classic, Ca2+-dependent MDH large subunit, MxaF, were much less represented in the same database, occurring in species belonging to 11 orders, mostly known methylotrophs, these always present in a single copy per genome, a total of 254 genes (Fig. 1, Supplementary Tables 3 and 4). XoxF5 and MxaF co-occurred in 8 orders of Proteobacteria (Fig. 1). Note that MxaF proteins belonging to the methanotrophs of the NC10 phylum phylogenetically cluster together with proteobacterial MxaF proteins, and thus these were also counted (Supplementary Tables 3 and 4).
Occurrence and distribution of REE-dependent alcohol dehydrogenases among Proteobacteria, compared to the occurrence and distribution of their calcium-dependent counterparts. Taxa are shown at the order level. In green, taxa that only contain methylotroph species [18]. In yellow, taxa that contain both methylotroph and non-mehylotroph species [18]. In gray, taxa that contain species capable of methylotrophic co-metabolism [55]. In purple, taxa that are not known to contain any methylotroph species. Blue, gene present, gray, gene absent
Importantly, a different form of XoxF, XoxF4, is encoded and expressed by Proteobacteria of the order Methylophilales, and XoxF4 and XoxF5 appear to be mutually exclusive [23]. Properties of the two of several subclasses of XoxF4 enzymes have recently been described, confirming them as Ln3+-dependent MDHs [23]. A total of 64 Methylophilales genomes/genomic bins have been identified in the IMG database, encoding 127 XoxF4 enzymes (multiple copies of XoxF4 are encoded in most Methylophilales genomes; Supplementary Tables 5 and 6).
Additional clades of XoxF enzymes (i.e., XoxF1, XoxF2 and XoxF3) are also known, but their distribution is limited to specific taxa [18, 19]. A single XoxF2 representative has been characterized as a bona fide Ln3+-dependent MDH, from a verrucomicrobial methanotroph Methylacidiphilum fumariolicum SolV [9]. Likely, all the verrucomicrobial methanotrophs encode and express this enzyme. However, only a handful of these organisms are available in cultures, and, thus, no statistical meaning can be gained from analysis of the current databases. The only study on the properties of a representative of XoxF1, from the NC10 phylum methanotroph Candidatus ‘Methylomirabilis oxyfera’, reported Ca2+ dependence for this enzyme, despite the presence of the motif for Ln3+ binding [29]. Thus, further data are needed to determine metal specificities for the enzymes of this clade. No biochemical data are available for XoxF3 clade enzymes.
We further queried the occurrence of the ExaF/PedH enzymes that have been characterized as Ln3+-ethanol dehydrogenases (EDH) [15, 20]. These were found in 14 orders of Proteobacteria (a total of 898 genes belonging to 888 genomes; Supplementary Tables 7 and 8), in 9 of these co-occurring with XoxF5 (Fig. 1). In comparison, the Ca2+-dependent counterparts ExaA/PedE/Mdh2 forms were detected in 16 orders of Proteobacteria (a total of 954 genes/951 genomes; Supplementary Tables 9 and 10), in the 14 orders ExaA/PedE co-occurring with ExaF/PedH/Mdh2 (Fig. 1). From these analyses, we conclude on the wide occurrence of REE-dependent ADH enzymes within Proteobacteria. Relative to Ca2+-dependent ADHs, they occur at similar frequencies. Importantly, in numerous species, enzymes with alternative inferred metal specificities are encoded within the same genome. Also of note, many genomes encode multiple predicted REE-dependent enzymes. All these observations suggest a much wider occurrence and distribution of REE-dependent enzymes than previously appreciated.
Methylorubrum extorquens mutant defective in methanol/ethanol utilization serves as a platform for expressing REE-dependent enzymes
The wide distribution of XoxF5 enzymes, especially among bacteria not known for methylotrophic life styles, made us question whether these XoxF5 enzymes would reveal properties similar to the properties of enzymes from bona fide methylotrophs. While it has been claimed that Ln3+ enzymes might be catalytically superior to their Ca2+ counterparts [9, 20, 22], the data on the properties of these enzymes are still very scarce. Moreover, a novel function has been recently proposed for XoxF5 from an Advenella species (Burkholderiales), in sulfur metabolism [30]. Remarkably, Ln3+ were not required for its activity in converting reduced sulfur compounds, as part of a newly proposed hypothetical metabolic cycle [30].
To resolve some of these controversies, we set up to purify and study properties of several XoxF5 enzymes, representing divergent alpha-, beta- and gammaproteobacteria. Methylorubrum extorquens AM1 was chosen as a major model methylotroph, in which the roles of different MDH enzymes have been previously investigated [17, 31]. Methyloversatilis discipulorum FAM1 was chosen as another bona fide methylotroph, representative of Rhodocyclales, known for the diversity of ADH enzymes encoded in their genomes, some with proven functions in methylotrophy [32, 33]. As the presumed non-methylotroph models, we employed Sinorhizobium and Bradyrhizobium species, representing phylogenetically disparate groups of rhizobia, known for their ecological function in nitrogen-fixing legume symbioses [34] (Table 1), a Rhodovulum species, a photosynthetic marine bacterium representing a broad diversity of Rhodobacterales [35] and a Tistlia species, another marine bacterium representing the metabolically versatile Rhodospirillales [36]. We also selected a representative of a broadly distributed order Vibrionales, a Grimontia (formerly Vibrio) species representing an important family within gammaproteobacteria containing both parasitic and free-living species [37]. To express representatives of the ExaF/PedH type enzymes, we elected to use the genes from M. discipulorum FAM1 and Methylopila sp. 107, the latter being a third methylotroph model [38].
As we have previously encountered problems with utilizing Escherichia coli as an expression host [23], we here utilized M. extorquens AM1 as an expression host, specifically the quadruple mutant variant with deletions in the mxaF, the two xoxF5 and the exaF genes, unable to grow on either methanol or ethanol [17]. The genes of interest were synthesized using the current JGI pipeline and cloned into a modified expression vector pCM80 (see Materials and methods), previously successfully used as an expression system in M. extorquens AM1 [39]. The respective plasmids were cloned into E. coli and subsequently conjugated into the quadruple mutant. The resulting progenies were selected for tetracycline resistance on succinate as a carbon source, and these were then tested for the ability to grow on methanol and ethanol, in the absence or in the presence of a range of Ln3+ (see Materials and methods).
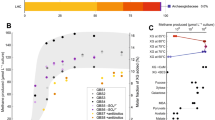
The constructs expressing the two native M. extorquens AM1 XoxF5 enzymes [17, 40], XoxF_M.e.1 and XoxF5_M.e.2, served as positive controls, as these were expressed in their native host. Both complemented growth of the mutant host on both methanol and ethanol, when REEs La3+ through Nd3+ were supplied, but not when the heavier REEs were supplied (Fig. 2, Supplementary Figure 1 and Supplementary Table 11). These results confirmed that both XoxF5 proteins encoded by the M. extorquens AM1 genome possessed the MDH/EDH enzymatic activities, even though these proteins have been reported to have differential roles in methylotrophy, XoxF5_M.e.1 appearing to be a more important enzyme compared to XoxF5_M.e.2 [17]. Seven of the heterologously expressed genes were also able to complement growth of the host on methanol and/or ethanol (Fig. 2 and Supplementary Fig. 1 and Supplementary Table 11). Plate and liquid culture growth experiments in the presence or in the absence of a range of Ln3+ revealed a total of four complementation patterns, as follows: (1) XoxF5 enzymes from Bradyrhizobium diazoefficiens, Grimontia marina, Rhodovulum kholense, Sinorhizobium meliloti and Tistlia consotensis showed the same pattern as the enzymes from M. extorquens AM1, supporting growth on both methanol and ethanol, in the presence of REEs ranging from La3+ to Nd3+. (2) XoxF5 from Methyloversatilis discipulorum did not complement growth of the host. (3) ExaF/PedH enzyme from M. discipulorum only complemented growth on ethanol, only when La3+ or Ce3+ were present. (4) Remarkably, ExaF/PedH from Methylopila sp. restored growth on both methanol and ethanol in all conditions, even in the absence of Ln3+. However, growth was significantly stimulated by REEs (La3+ through Sm3+).
Growth rates of constructs expressing XoxF or ExaF/PedH enzymes in the presence of La3+, Ce3+ and Nd3+. Metals were tested individually at concentration of 30 µM. As data for La3+, Ce3+ were very similar, they were joined together. Gray columns, growth on methanol (0.5% V/V); hatched columns, growth on ethanol (0.15% V/V). Cultures were incubated with shaking at 30 °C, with the exception of the construct expressing XoxF5_R.k., which was incubated at 20 °C. Data represent the means of two replicate tests, with error bars. Additional data are presented in Supplementary Table 11 and Supplementary Fig. 1
Effect of REEs on growth on methanol or ethanol has been reported before for several organisms [7, 9, 14, 15, 17, 20, 23, 41, 42], as well as for natural populations [43], and our data are in general in line with the published data. XoxF5, XoxF2 and XoxF4-2 enzymes display narrow specificity for Ln3+, selecting for the early Ln3+ (La3+–Nd3+), while others, such as XoxF4-1 and PedH display specificity toward a broader range of lanthanides (La3+–Gd3+). The decrease in catalytic efficiency with later Ln3+ has recently been experimentally demonstrated [14, 42]. While Ln3+ specificity of ExaF/PedH_M.s. we report here is similar to the one reported for PedH from Pseudomonas [15], it is not strictly dependent on Ln3+. This identifies it as a promiscuous REE enzyme that, in the absence of REEs, can apparently use Ca2+. Note that we exclude the possibility of contamination with REEs, as all the constructs were tested on the same plate for each condition, and thus nine of the strains served as negative controls for growth (Supplementary Fig. 2).
REE alcohol dehydrogenases reveal a range of catalytic properties
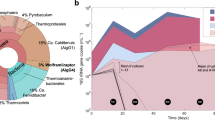
In this study, we focused on the kinetic properties of the XoxF5 enzymes, to test whether the enzymes from the presumed non-methylotrophs revealed propensity for oxidizing methanol. The eight XoxF5 proteins were expressed in the presence of Ce3+ (see Materials and methods). In crude extracts, MDH activities of 0.07 to 1.31 U were measured using the artificial dye assay (Fig. 3, Supplementary Fig. 3). Notably, the enzyme from M. discipulorum FAM1 showed activity within this range. Thus, the reason for negative complementation phenotype for XoxF5_M.d. is obviously not the lack of catalytic activity. More likely, the lack of complementation is due to the absence of compatible electron acceptors or other components of the electron transfer chain in the host strain. This speculation of course requires experimental testing, in the future. The two xoxF genes, present in a tandem on the chromosome of M. discipulorum FAM1 form a cluster with a xoxJ homolog, which frequently form operons with xoxF genes [9, 17], while there are no genes encoding cytochromes in the vicinity [33]. In M. extorquens AM1 and in many other organisms, a gene encoding a cytochrome c, named xoxG, is located between xoxF and xoxJ (the second xoxF gene in M. extorquens AM1 is an orphan [40]). We hypothesize that, while the XoxF5 proteins from M. extorquens AM1 and M. discipulorum FAM1 are highly homologous (~70% amino acid identity), the latter may not be able to transfer electrons to XoxG of M. extorquens AM1. Interestingly, G. marina also lacks recognizable xoxG in its genome. Instead, the xox gene cluster in this organism contains a gene encoding a plastocyanin, another type of a small redox protein, which likely serves as a natural electron acceptor from XoxF5_G.m. Nevertheless, XoxF5_G.m. appears to be able to transfer electrons to XoxG of M. extorquens AM1. The diversity of the putative electron acceptors from XoxF enzymes has been noted before [13, 44], and this study provides further evidence toward the complexity of XoxF/electron acceptor pairings. Moreover, an interaction between XoxF5 and particulate methane monooxygenase has been recently reported for a methanotrophic bacterium [24].
Denaturing polyacrylamide gel electrophoresis of crude extracts (lanes with odd numbers) and respective purified enzymes (lanes with even numbers), and respective specific activities measured with methanol as a substrate for XoxF5 enzymes and with ethanol for ExaF/PedH enzymes (Units/mg, defined as μM substrate/min/mg protein). 1, 2, XoxF5_T.c.; 3, 4, XoxF5_S.m.; 5,6, XoxF5_M.d.; 7, 8, XoxF5_G.m.; 9, 10, ExaF/PedH_M.d.; 11, 12, XoxF5_M.e.2; 13, 14, XoxF5_M.e.1; 15, 16, XoxF5_R.k.; 17, 18, XoxF5_B.d.; 19, 20, ExaF/PedH_M.s. M denotes marker protein lanes. Respective molecular masses are indicated. Full length gels are presented in Supplementary Fig. 3
Pure XoxF5 proteins revealed specific activities in the range of 1.68 to 18.51 U with methanol as a substrate (Fig. 3, Supplementary Fig. 3). The enzymes were relatively unstable even in the presence of glycerol and dithiothreitol, losing 12–80% of initial activity within 24 h when kept on ice (Supplementary Table 12). The enzymes showed typical UV-Vis absorption spectra, with a maximum of 355 nm and a slight shoulder at 400 nm, previously reported for redox cofactor pyrroloquinoline quinone (PQQ) in XoxF type enymes [9, 14]. The PQQ peak decreased somewhat over time, potentially indicative of protein denaturation (Supplementary Fig. 4). We determined pH optima and ammonia requirements for the eight enzymes, and found that all required ammonia for the in vitro activity assay, and all had a pH optimum between 9.0 and 9.5 (Supplementary Fig. 5), similarly to the properties of the previously described XoxF5 from Methylomonas sp. LW13 [23]. The artificially high pH and the ammonia ion requirements have been known for the Ca2+-dependent MDHs and for other PQQ-dependent dehydrogenases for decades, but the biochemical bases for these requirements remained a mystery [45].
We further determined substrate specificities for the eight enzymes, and they also fell into similar ranges, all enzymes revealing high activities with the saturating concentrations of methanol, ethanol, 1-propanol, 1-butanol, 1-hexanol and formaldehyde (Supplementary Fig. 6), again, similar to the properties determined for XoxF5 from Methylomonas sp. LW13 [23]. The kinetic constants were determined for a range of alcohols and for formaldehyde (Table 2, Fig. 4, Supplementary Fig. 6), and these, again, were in line with the constants previously determined for XoxF5 enzymes, demonstrating that these enzymes had affinities for a broad range of alcohols, and that the affinities for formaldehyde were significantly lower than for alcohols. Km values for alcohols generally varied between 0.012 ± 0.002 and 0.130 ± 0.015 mM, while Km values for formaldehyde were in general higher (0.072 ± 0.07 to 0.182 ± 0.041 mM), as previously observed for XoxF5 from Methylomonas sp. LW13 [23]. One exception was XoxF5_R.k., which revealed poor affinities for propanol, butanol and hexanol (Supplementary Fig. 6). Respectively, Keff values (the proxy for enzyme catalytic efficiency, defined as Kcat/Km), while varied enzyme to enzyme, were four- to eleven-fold higher for methanol compared to formaldehyde (Supplementary Fig. 6).
Interestingly, the two XoxF5 enzymes from the host organism, while 88% identical and 94% similar, demonstrated different kinetic properties. XoxF5_M.e.1 revealed a much higher maximal velocity (Vmax) rates with all substrates employed in this study, as well as much higher Keff values compared to XoxF5_M.e.2, and it showed highest affinity toward methanol and hexanol as substrates, while XoxF5_M.e.2 revealed highest affinity for methanol (Supplementary Fig. 6). While this manuscript was in revision, kinetic parameters have been presented for XoxF5_M.e.1 by another group [46]. While similar Km values were reported for methanol and formaldehyde, Km for ethanol appeared 16-fold higher compared to the value we obtained. In addition, much lower Vmax values were reported, resulting in Keff values 4.5 to 27 lower than the ones we are reporting here [46]. These discrepancies highlight the difficulty of comparing kinetic data from different studies.
As a XoxF5 type enzyme from Advenella kashmirensis has been implicated recently in dehydrogenation of thiosulfate [30], we measured this activity with the eight XoxF5 enzymes purified in this study, utilizing the published assay protocol [30]. No thiosulfate dehydrogenase activity was detected for any of the eight enzymes (Supplementary Fig. 7).
ExaF/PedH_M.s. revealed higher affinities for ethanol and propanol compared to other alcohols, differing this enzyme from the XoxF5 enzymes, while the affinity for formaldehyde, again, was much lower than for alcohols (Supplementary Fig. 6). However, the difference in catalytic efficiency displayed with methanol versus ethanol was not nearly as dramatic as has been reported for a similar enzyme from M. extorquens AM1 (7250-fold) [20]. The Keff value for methanol was only three-fold lower compared to ethanol, in agreement with the robust growth of the host strain on both substrates (Supplementary Table 11). Differently from the XoxF5 enzymes, ExaF/PedH_M.s. did not reveal strict requirement for ammonia, but its activity was significantly stimulated by it (Supplementary Figs. 1 and 2). We were not able to determine kinetic constants for the ExaF/PedH_M.d. due to low specific activity and rapid enzyme activity loss over the course of the purification protocol (Fig. 3, Supplementary Fig. 3; Incubation with either Ln3+ or both Ln3+ and PQQ could not improve the performance of any of the enzymes we purified in this or in our previous study [23], contrary to reports by others [15, 20]). However, from the complementation experiments and from activity measurements with crude cell extracts, this enzyme revealed properties very different from the ones of other characterized ExaF/PedH enzymes, most importantly, showing no affinity for methanol (Supplementary Table 13). Remarkably, of all the ExaF/PedH enzymes described so far, only ExaF/PedH_M.s. revealed a promiscuous metal dependence, showing (reduced) growth on methanol/ethanol (Supplementary Fig. 1) and (reduced) enzymatic activity (Supplementary Fig. 8) in the absence of Ln3+. Low activity in the absence of Ln3+ has previously been reported for purified XoxF5_Me1 [47], and low activity of XoxF4 enzymes in Methylotenera species was implicated from growth experiments [48].
We measured metal contents in protein preparations from cultures grown in the presence of both Ln3+ and Ca2+, and found that all proteins bound Ln3+ (up to 0.9 mol per mol of the enzyme protomer). Several enzymes were purified from cultures grown with cocktails of lighter Ln3+, and in each of these cases, the lightest of the Ln3+ were preferentially bound (Supplementary Table 14).
Metabolic reconstruction implicates methylotrophy capabilities for the numerically abundant and environmentally important microbial guilds
As the properties of XoxF5 enzymes from non-methylotrophs were determined here to be rather similar to the properties of XoxF5 enzymes from true methylotrophs, with some variations on catalytic activities and substrate specificities, their function as bona fide MDH is likely, despite the fact that some of these enzymes do not necessarily reveal specificity toward methanol. We analyzed the genomes of B. diazoefficiens, G. marina, R. kholense, S. meliloti, T. consotensis, as well as the genome of A. kashmirensis, in attempts to reconstruct the potential methylotrophy metabolic pathways, and these could be easily identified. All six genomes encoded, in addition to XoxF5 MDH enzymes, enzymes for formaldehyde oxidation to formate, via the glutathione-linked pathway and further to CO2, via formate dehydrogenases, and a pathway for CO2 assimilation via the Calvin–Benson–Bassham cycle, the metabolic scheme typical of some bona fide methylotrophs, such as certain Paracoccus species [18] (Table 3). Genes for degradation of an array of other methylated compounds (choline, glycine betaine, dimethylglycine, sarcosine, trimethylamine, trimethylamine N-oxide, dimethylamine, methylamine) were identified in some of the genomes (Table 3), these pathways producing either formaldehyde or methyl-groups as intermediates. This suggests a substantial potential for these organisms in gaining both energy and carbon from single-carbon compounds, thus significantly expanding our understanding of the occurrence of methylotrophy in abundant, globally distributed microbes and the significance of Ln3+ in their environmental fitness.
Conclusion
In this study, we more than doubled the number of extant biochemically characterized enzymes that require REEs for their activity and expression, demonstrating that enzymes representing diverse taxa of globally distributed, abundant and environmentally important bacteria likely rely on REEs for their central metabolisms that involve but may not be limited to conversions of alcohols. The wide distribution of REE-dependent enzymes, along with the recently discovered high-affinity mechanisms involved in their acquisition by bacterial cells [49,50,51] position REEs as true life metals in, and potentially beyond, the bacterial domain of life.
Materials and methods
Bioinformatics
To assess the presence of alternative alcohol dehydrogenases in bacteria, enzymes with previously characterized properties were employed [7, 9, 15, 20, 23, 24]. The IMG/MER database (https://img.jgi.doe.gov/cgi-bin/mer/main.cgi) was searched using the Homolog Display tool, in an iterative fashion as follows. First an enzyme of interest was used to display top homologs in the IMG/M database, and sequences with more than 65% amino acid identity were selected. Based on prior phylogenetic analyses, this cutoff selects members of specific clades of REE enzymes [18, 19, 27]. Then, the most dissimilar sequence was used to display its top homologs and additional sequences were selected, iterated until the database for each homology group was saturated and no new sequences were identified.
Phylogenetic analysis
Protein sequences were aligned using the CLUSTAL W algorithm, and phylogenetic trees were constructed using the Maximum-Likelihood method, as implemented in the MEGA7 software [52]. Statistical support was obtained from 1000 bootstrap replicates.
Gene synthesis and expression vector construction
The vector used for protein expression in this study was a modified pCM80 vector previously successfully used for expressing genes in M. extorquens AM1 [39]. The modified vector, pCM80-PxoxF, contains a 411 bp synthetic DNA fragment inserted between the mxa operon promoter (PmxaF) and lacZa. This DNA fragment contains a presumed promoter for the native xoxF gene in M. extorquens AM1, a ribosome-binding site, a unique NcoI restriction site for cloning genes of interest, a TEV cleavage site, and 8x His-tags (Supplementary File 1). The selected gene homologs were codon-optimized for expression in M. extorquens AM1 using an empirically derived codon usage table. Codon optimization and vendor defined synthesis constraints removal were performed using BOOST [53]. Synthetic DNA were obtained from Twist Biosciences and cloned into the NcoI site of the pCM80-PxoxF vector using the Gibson Assembly method (NEBuilder HiFi, NEB). The Escherichia coli Top10 transformants were plated on lysogeny broth (LB) agar plates supplemented with tetracycline (10μgml−1). All gene constructs were sequence-verified using the PacBio RSII sequencing platform (Pacific Biosciences).
Plasmid transfer into host strain
As the expression host strain, we utilized the quadruple mutant of. M. extorquens AM1, unable to grow on either methanol or ethanol [17]. Plasmid constructs were transferred From E. coli Top10 to the host via three-way conjugation, employing a helper strain E. coli 2073. Matings were carried out on nutrient agar plates as described before [54], and progenies were selected on minimal medium plates supplemented with succinate (0.2% W/V) as a substrate and tetracycline (10 μgml−1) and rifampicin (50 μgml−1) for, respectively, selecting for the plasmid-borne function and counter-selecting for E. coli presence.
Growth complementation experiments
The ability of the host strain to grow on methanol (0.5% V/V) or ethanol (0.15% V/V) was tested on plates and in liquid medium, comparing growth on standard minimal media and media supplemented with 30 μM of either of the following Ln3+: La3+, Ce3+, Nd3+, Sm3+, Gd3+, Dy3+, Yb3+. Plates were incubated at room temperature (~25 °C), and growth patterns were monitored daily for up to 6 weeks. Liquid cultures were incubated in tubes (20 × 150 mm) filled with 6 ml cultures, and these were shaken on a rotating shaker at 200 rpm at a 45° angle, at 30 ℃, with the exception of the construct expressing XoxF5_R.k., which was incubated at 20 °C, as it demonstrated better growth and higher MDH activity at lower temperatures.
Protein expression and purification
For protein expression, 300-ml cultures were grown in shake flasks with succinate to late exponential phase and collected by centrifugation at 4700 × g for 10 min. Pellets were transferred to 100 ml of fresh minimal medium supplemented with methanol (0.5% V/V) and Ce3+ (30 μM), and these were incubated overnight at 30 ℃ with shaking at 200 rpm. Cells were harvested by centrifugation, as above, and used immediately or kept at −80 ℃. For XoxF5_R.k., best results were achieved when expression was carried out as above, but at 20 °C, for 2 days. Protein purification was carried out using the previously described protocol [23]. Briefly, cell pellets were resuspended in 0.1 M Tris-HCl buffer pH 9.0 and broken using the French Pressure Cell (Sim-Aminco), at 20,000 psi. His-tagged proteins were purified by Ni-NTA agarose by successive wash steps, finishing with elution step, using standard buffers (start buffer: 0.1 M Tris–HCl pH 9.0, 150 mM NaCl, 5 mM imidazole, 1 mM β-mercaptoethanol (β-ME), a PierceTM protease inhibitor tablet (Thermo Fisher Scientific); wash buffer: 0.1 M Tris–HCl pH 9.0, 150 mM NaCl, 30 mM imidazole, 1 mM β-ME; elution buffer: 0.1 M Tris–HCl pH 9.0, 150 mM NaCl, 250 mM imidazole, 1 mM β-ME.) with one exception. A higher concentration of sodium salts, up to 350 mM, was required for XoxF5_R.k., in all the purification buffers. Proteins were desalted and concentrated via a series of dilution/concentration steps until the concentration of imidazole reached below 1 µM. Purified proteins denatured by sodium dodecyl sulfate (SDS) were analyzed by electrophoresis in 12.5% polyacrylamide gel run with a standard buffer (1× Tris/Glycine/SDS; Bio-Rad Laboratories, Hercules, CA, USA). Protein concentrations were determined using the bicinchonic acid Protein Assay kit (Pierce, Thermo Fisher Scientific), with serum albumin as a standard. Proteins were kept in a storage buffer (0.1 M Tris–HCl pH 9.0, 1 mM dithiothreitol, 10% glycerol) on ice. Kinetic analyses were carried out within hours of purification.
Methanol dehydrogenase assay
Methanol dehydrogenase activity was measured by monitoring the phenazine methosulfate (PMS; 1 mM)-mediated reduction of 2,6-dichlorophenol-indophenol (DCPIP; 150 μM; ε600 = 21.0 mM−1cm−1) as described as previously [23]. Assays were performed at room temperature (~25 ℃ in a total volume of 0.8 ml in plastic cuvettes (1 cm path length). One unit (U) of specific enzyme activity was defined as 1 µM DCPIP reduced per minute (determined at 600 nm) and was expressed as U per milligram of protein. To determine optimum pH values, standard buffers were used as follows: 100 mM sodium/potassium phosphate (pH range 6.0–8.0), 100 mM Tris-HCl (pH range 7.5–9.0), 100 mM sodium carbonate/ sodium bicarbonate (pH range 9.0–10.0). The effect of ammonia was examined in the standard assay including or omitting 45 mM NH4Cl. Substrate specificities were determined for several alcohols and aldehydes using optimal assay conditions for each enzyme, as follows: 0.1 M Tris–HCl buffer pH 9.0 was used for XoxF5_T.c., XoxF5_M.e.1, XoxF5_R.k., XoxF5_B.d., XoxF5_G.m. and ExaF/PedH_M.s.; 0.1 M Na2CO3/ NaHCO3 buffer pH 9.0 was used for XoxF5_M.e.2 and XoxF5_M.d.; and 0.1 M Tris–HCl buffer pH 9.5 was used for XoxF5_S.m. All substrates were purchased from Sigma Aldrich. Formaldehyde was prepared by incubating paraformaldehyde in water at 95 ℃ for approximately 1 h, until all the powder has been completely dissolved. Enzyme activity was determined at three time points, at 0, 4 and 24 h, using freshly prepared PMS and DCPIP.
Enzyme kinetics
Kinetic parameters were determined using the respective optimal assay conditions, as above, using varying concentrations of select substrates (0.005 to 1 mM). The values of Km and Vmax were obtained using the Michaelis–Menten equation, with a GraphPad Software (GraphPad Inc., USA). Averages of kinetic constants were determined for each enzyme based on two to four biologically independent experiments.
Metal analysis
Metals were analyzed by inductively coupled plasma mass spectrometry (ICP-MS) on an ELAN DRC-e (PerkinElmer, Shelton, CT). Standard curves were generated with a dilution series of elements (Ce3+, La3+, Nd3+, Ca2+), with concentrations of 0, 500, 1000, 1500, and 3000 nM in 1% nitric acid (Thermo Fisher Scientific, Waltham, MA, USA). Ammonia was used in the DRC mode to measure Ca2+. For metal content in proteins, 50–300 µl of purified enzyme (0.5 to 4 mgml−1) was destructed with nitric acid (5 to 10%) at 90 °C for 1 h and diluted to 5 ml with water.
References
Maret W. The metals in the biological periodic system of the elements: concepts and conjectures. Int J Mol Sci. 2016;17:66.
Zhao SN, Wang G, Poelman D, Voort PV. Luminescent lanthanide MOFs: a unique platform for chemical sensing. Materials. 2018;11:E572.
Écija D, Urgel JI, Seitsonen AP, Auwärter W, Barth JV. Lanthanide-directed assembly of interfacial coordination architectures-from complex networks to functional nanosystems. Acc Chem Res. 2018;51:365–75.
Biju S, Parac-Vogt T. Recent advances in lanthanide based nano-architectures as probes for ultra high-field magnetic resonance imaging. Curr Med Chem. 2018. https://doi.org/10.2174/0929867325666180201110244.
Lim S, Franklin SJ. Lanthanide-binding peptides and the enzymes that might have been. Cell Mol Life Sci. 2004;61:2184–8.
Hibi Y, Asai K, Arafuka H, Hamajima M, Iwama T, Kawai K. Molecular structure of La3+-induced methanol dehydrogenase-like protein in Methylobacterium radiotolerans. J Biosci Bioeng. 2011;111:547–9.
Fitriyanto NA, Fushimi M, Matsunaga M, Pertiwiningrum A, Iwama T, Kawai K. Molecular structure and gene analysis of Ce3+-induced methanol dehydrogenase of Bradyrhizobium sp. MAFF211645. J Biosci Bioeng. 2011;111:613–7.
Nakagawa T, Mitsui R, Tani A, Sasa K, Tashiro S, Iwama T, et al. A catalytic role of XoxF1 as La3+-dependent methanol dehydrogenase in Methylobacterium extorquens strain AM1. PLoS ONE. 2012;7:e50480.
Pol A, Barends TR, Dietl A, Khadem AF, Eygensteyn J, Jetten MS, et al. Rare earth metals are essential for methanotrophic life in volcanic mudpots. Environ Microbiol. 2014;16:255–64.
Chu F, Lidstrom ME. XoxF acts as the predominant methanol dehydrogenase in the type I methanotroph Methylomicrobium buryatense. J Bacteriol. 2016;198:1317–25.
Farhan Ul Haque M, Gu W, DiSpirito AA, Semrau JD. Marker exchange mutagenesis of mxaF, encoding the large subunit of the Mxa methanol dehydrogenase, in Methylosinus trichosporium OB3b. Appl Environ Microbiol. 2015;82:1549–55.
Farhan Ul Haque M, Kalidass B, Bandow N, Turpin EA, DiSpirito AA, Semrau JD, et al. Cerium regulates expression of alternative methanol dehydrogenases in Methylosinus trichosporium OB3b. Appl Environ Microbiol. 2015;81:7546–52.
Zheng Y, Huang J, Zhao F, Chistoserdova L. Physiological effect of XoxG(4) on lanthanide-dependent methanotrophy. mBio. 2018;9:e02430-17.
Jahn B, Pol A, Lumpe H, Barends TRM, Dietl A, Hogendoorn C, et al. Similar but not the same: first kinetic and structural analyses of a methanol dehydrogenase containing a europium ion in the active site. Chembiochem. 2018;19:1147–53.
Wehrmann M, Billard P, Martin-Meriadec A, Zegeye A, Klebensberger J. Functional role of lanthanides in enzymatic activity and transcriptional regulation of pyrroloquinoline quinone-dependent alcohol dehydrogenases in Pseudomonas putida KT2440. mBio. 2017;8:e00570-17.
Wehrmann M, Berthelot C, Billard P, Klebensberger J. The PedS2/PedR2 two-component system Is crucial for the rare earth element switch in Pseudomonas putida KT2440. mSphere. 2018;3:e00376-18.
Vu HN, Subuyuj GA, Vijayakumar S, Good NM, Martinez-Gomez NC, Skovran E. Lanthanide-dependent regulation of methanol oxidation systems in Methylobacterium extorquens AM1 and their contribution to methanol growth. J Bacteriol. 2016;98:1250–9.
Chistoserdova L. Modularity of methylotrophy, revisited. Environ Microbiol. 2011;13:2603–22.
Keltjens JT, Pol A, Reimann J, Op den Camp HJ. PQQ-dependent methanol dehydrogenases: rare-earth elements make a difference. Appl Microbiol Biotechnol. 2014;98:6163–83.
Good NM, Vu HN, Suriano CJ, Subuyuj GA, Skovran E, Martinez-Gomez NC. Pyrroloquinoline quinone ethanol dehydrogenase in Methylobacterium extorquens AM1 extends lanthanide-dependent metabolism to multicarbon substrates. J Bacteriol. 2016;198:3109–18.
Picone N, Op den Camp HJ. Role of rare earth elements in methanol oxidation. Curr Opin Chem Biol. 2018;49:39–44.
Bogart JA, Lewis AJ, Schelter EJ. DFT study of the active site of the XoxF-type natural, cerium-dependent methanol dehydrogenase enzyme. Chemistry 2015;21:1743–8.
Huang J, Yu Z, Chistoserdova L. Lanthanide-dependent methanol dehydrogenases of XoxF4 and XoxF5 clades are differentially distributed among methylotrophic bacteria and they reveal different biochemical properties. Front Microbiol. 2018;9:1366.
Deng YW, Ro SY, Rosenzweig AC. Structure and function of the lanthanide-dependent methanol dehydrogenase XoxF from the methanotroph Methylomicrobium buryatense 5GB1C. J Biol Inorg Chem. 2018;23:1037–47.
Chistoserdova L. Lanthanides: new life metals? World J. Microbiol. Biotechnol. 2016;32:138.
Chistoserdova L, Kalyuzhnaya MG. Current trends in methylotrophy. Trends Microbiol. 2018;26:703–14.
Taubert M, Grob C, Howat AM, Burns OJ, Dixon JL, Chen Y, et al. XoxF encoding an alternative methanol dehydrogenase is widespread in coastal marine environments. Environ Microbiol. 2015;17:3937–48.
Chistoserdova L, Lidstrom ME. Aerobic methylotrophic prokaryotes. In Rosenberg E, DeLong EF, Thompson F, Lory S, Stackebrandt E, editors. The Prokaryotes. 3rd ed. Berlin, Springer; 2013. pp. 267–85.
Wu ML, Wessels JC, Pol A, Op den Camp HJ, Jetten MS, van Niftrik L. XoxF-type methanol dehydrogenase from the anaerobic methanotroph “Candidatus Methylomirabilis oxyfera”. App. Environ. Microbiol. 2015;81:1442–51.
Pyne P, Alam M, Rameez MJ, Mandal S, Sar A, Mondal N, et al. Homologs from sulfur oxidation (Sox) and methanol dehydrogenation (Xox) enzyme systems collaborate to give rise to a novel pathway of chemolithotrophic tetrathionate oxidation. Mol Microbiol. 2018. https://doi.org/10.1111/mmi.13972.
Skovran E, Palmer AD, Rountree AM, Good NM, Lidstrom ME. XoxF is required for expression of methanol dehydrogenase in Methylobacterium extorquens AM1. J Bacteriol. 2011;193:6032–8.
Kalyuzhnaya MG, Hristova KR, Lidstrom ME, Chistoserdova L. Characterization of a novel methanol dehydrogenase in representatives of Burkholderiales: implications for environmental detection of methylotrophy and evidence for convergent evolution. J Bacteriol. 2008;190:3817–23.
Smalley NE, Taipale S, De Marco P, Doronina NV, Kyrpides N, Shapiro N, et al. Functional and genomic diversity of methylotrophic Rhodocyclaceae: description of Methyloversatilis discipulorum sp. nov. Int J Syst Evol Microbiol. 2015;65:2227–33.
Masson-Boivin C, Giraud E, Perrer X, Batut J. Establishing nitrogen-fixing symbiosis with legumes: how many rhizobium species? Trends Microbiol. 2009;17:458–66.
Petersen J, Frank O, Göker M, Pradella S. Extrachromosomal, extraordinary and essential--the plasmids of the Roseobacter clade. Appl Microbiol Biotechnol. 2013;97:2805–15.
Rubiano-Labrador C, Bland C, Miotello G, Armengaud J, Baena S. Salt stress induced changes in the exoproteome of the halotolerant bacterium Tistlia consotensis deciphered by proteogenomics. PLoS ONE. 2015;10:e0135065.
Thompson FL, Iida T, Swings J. Biodiversity of vibrios. Microbiol Mol Biol Rev. 2004;68:403–31.
Beck DA, McTaggart TL, Setboonsarng U, Vorobev A, Goodwin L, Shapiro N, et al. Multiphyletic origins of methylotrophy in Alphaproteobacteria, exemplified by comparative genomics of Lake Washington isolates. Environ Microbiol. 2015;17:547–54.
Marx CJ, Lidstrom ME. Development of improved versatile broad-host-range vectors for use in methylotrophs and other Gram-negative bacteria. Microbiology. 2001;147:2065–75.
Vuilleumier S, Chistoserdova L, Lee MC, Bringel F, Lajus A, Zhou Y, et al. Methylobacterium genome sequences: a reference blueprint to investigate microbial metabolism of C1 compounds from natural and industrial sources. PLoS ONE. 2009;4:e5584.
Masuda S, Suzuki Y, Fujitani Y, Mitsui R, Nakagawa T, Shintani M, et al. Lanthanide-dependent regulation of methylotrophy in Methylobacterium aquaticum strain 22A. mSphere. 2018;3:e00462–17.
Lumpe H, Pol A, Op den Camp HJM, Daumann LJ. Impact of the lanthanide contraction on the activity of a lanthanide-dependent methanol dehydrogenase - a kinetic and DFT study. Dalton Trans. 2018;47:10463–72.
Shiller AM, Chan EW, Joung DJ, Redmond MC, Kessler JD. Light rare earth element depletion during Deepwater Horizon blowout methanotrophy. Sci Rep. 2017;7:1–9.
Yu Z, Beck DAC, Chistoserdova L. Natural selection in synthetic communities highlights the roles of Methylococcaceae and Methylophilaceae and suggests differential roles for alternative methanol dehydrogenases in methane consumption. Front Microbiol. 2017;8:2392.
Anthony C. Methanol dehydrogenase, a PQQ-containing quinoprotein dehydrogenase. Subcell Biochem. 2000;35:73–117.
Good NM, Moore RS, Suriano C, Martinez-Gomez NC. Contrasting in vitro and in vivo methanol oxidation activities of lanthanide-dependent alcohol dehydrogenases XoxF1 and ExaF from Methylobacterium extorquens AM1. Sci Rep. 2019;9:4248.
Schmidt S, Christen P, Kiefer P, Vorholt JA. Functional investigation of methanol dehydrogenase-like protein XoxF in Methylobacterium extorquens AM1. Microbiology. 2010;156:2575–86.
Mustakhimov I, Kalyuzhnaya MG, Lidstrom ME, Chistoserdova L. Insights into denitrification in Methylotenera mobilis from denitrification pathway and methanol metabolism mutants. J Bacteriol. 2013;195:2207–11.
Cook EC, Featherston ER, Showalter SA, Cotruvo JA Jr. Structural basis for rare Earth element recognition by Methylobacterium extorquens lanmodulin. Biochem. 2018;58:120–5.
Cotruvo JA Jr., Featherston ER, Mattocks JA, Ho JV, Laremore TN. Lanmodulin: a highly selective lanthanide-binding protein from a lanthanide-utilizing bacterium. J Am Chem Soc. 2018;140:15056–61.
Ochsner AM, Hemmerle L, Vonderach T, Nüssli R, Bortfeld-Miller M, Hattendorf B, et al. Use of rare-earth elements in the phyllosphere colonizer Methylobacterium extorquens PA1. Mol Microbiol. 2019. https://doi.org/10.1111/mmi.14208.
Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–4.
Oberortner E, Cheng J-F, Hillson NJ, Deutsch S. Streamlining the design-to-build transition with build-optimization software tools. ACS Synth. Biol. 2017;6:485–96.
Chistoserdova L, Crowther GJ, Vorholt JA, Skovran E, Portais J-C, Lidstrom ME. Identification of a fourth formate dehydrogenase in Methylobacterium extorquens AM1 and confirmation of the essential role of formate oxidation in methylotrophy. J Bacteriol. 2007;189:9076–81.
Quayle JR, Pfennig N. Utilization of methanol by Rhodospirillaceae. Arch Microbiol. 1975;102:193–8.
Acknowledgements
The quadruple mutant of M. extorquens AM1 was a generous gift by Dr. Elizabeth Skovran. This material is based upon work supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research under Award Number DE-SC-0016224. The work conducted by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, was supported under Contract No. DE-AC02-05CH11231.
Author information
Authors and Affiliations
Contributions
LC and JH conceived the study. LC compiled inventories of the ADH genes and carried out plasmid transfer, LC and JH carried out complementation tests. JH and ZY carried out enzyme expression, purification and analysis tasks. ZY carried out phylogenetic analyses. JG carried out metal content quantification. YY supervised the synthetic component of the study. JFC carried out vector and synthetic DNA design. AT generated the expression constructs and carried out sequence verification. LC and JH wrote the manuscript, and all authors approved the final version.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Huang, J., Yu, Z., Groom, J. et al. Rare earth element alcohol dehydrogenases widely occur among globally distributed, numerically abundant and environmentally important microbes. ISME J 13, 2005–2017 (2019). https://doi.org/10.1038/s41396-019-0414-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-019-0414-z
This article is cited by
-
Weathered granites and soils harbour microbes with lanthanide-dependent methylotrophic enzymes
BMC Biology (2024)
-
Queer in Chem: Q&A with Professor Polly Arnold
Communications Chemistry (2023)
-
The effect of barium and strontium on activity of glucoamylase QsGH97a from Qipengyuania seohaensis SW-135
Scientific Reports (2023)
-
Gene products and processes contributing to lanthanide homeostasis and methanol metabolism in Methylorubrum extorquens AM1
Scientific Reports (2020)
-
Understanding the chemistry of the artificial electron acceptors PES, PMS, DCPIP and Wurster’s Blue in methanol dehydrogenase assays
JBIC Journal of Biological Inorganic Chemistry (2020)