Abstract
Many soil bacteria and fungi remain unclassified at the highest taxonomic ranks (e.g. phyla level), which hampers our ability to assess the ecology and functional capabilities of these soil organisms in terrestrial ecosystems globally. The first logical step toward the classification of these unknown soil taxa is to identify potential locations on Earth where these unclassified bacteria and fungi are feasibly most prevalent. To do this, here I used data from a global soil survey across 235 locations, including amplicon sequencing information for fungal and bacterial communities, and generated global atlases highlighting those soils where the percentages of taxa of bacteria and fungi with an unknown phyla are expected to be more prevalent. Results indicate that soil samples with the largest percentage of fungal taxa with an unknown phyla can be found in dry forests and grasslands, while those with the largest percentage of bacterial taxa with an unknown phyla are found in boreal and tropical forests. This information can be used by taxonomists and microbiologists to target these potentially new soil taxa.
Similar content being viewed by others
Introduction
Soil microbial communities play an essential role in maintaining important soil processes such as nutrient cycling, waste decomposition, climate regulation, and pollution degradation [1, 2]. Today, sequencing technologies are well established and broadly used [3]. As such, producing large amounts of data on the composition and diversity of bacterial and fungal communities is no longer so challenging. Moreover, the major ecological drivers of the variation in these microbial communities are becoming increasingly visible [4, 5]. The spotlight is now on the soil taxonomists. Although progress has been made in the past few years [6, 7], culturing, isolating, and classifying soil microbes are still a difficult task. For most soil bacterial and fungal species, we know very little about their identity or the tasks performed even by the most dominant microbial taxa [8]. More concerning, in some cases, we lack the most basic taxonomic information to classify these bacterial and fungal taxa as they do not match the latest data within taxonomic databases (e.g. [9] and [10]; Zomer et al. [11]) even at the highest taxonomic ranks (e.g. phyla level).
The first logical step toward the classification of these unknown microbial taxa is to identify potential locations where they could be found across the globe. This information can then be used by taxonomists and microbiologists to target these new soil taxa. Here, I used data from a global soil survey [8] across 235 locations (Fig. S1), and including amplicon sequencing information on fungal (ITS gene) and bacterial (16S rRNA gene) communities from around the world, to highlight those locations on Earth where taxa of bacteria and fungi with an unknown phyla are feasibly most prevalent. The database in Delgado-Baquerizo et al. [8] has been used previously to identify the dominant taxa of bacteria globally, and more recently, the major ecological predictors of bacterial diversity [12]. I used the bioinformatics pipeline described in Delgado-Baquerizo et al. [8], and two of the most commonly used microbial databases for taxonomic identification (Greengenes and UNITE), to estimate, at the global scale, the percentage of phylotypes of bacteria and fungi with an unknown phyla in soils across the globe. These taxa are classified as fungi or bacteria using taxonomic databases, but do not match any known phyla. As such, they are expected to be potential new phyla of fungi or bacteria.
Results and Discussion
As expected, the taxonomic information at the “species” (OTU, phylotypes) level could not be found for 99% of bacterial and 63% of fungal phylotypes (clustered at 97% similarity). Notably, up to 1.36% and 9.37% of the retrieved phylotypes classified as bacteria or fungi remained unclassified at the phyla level in soils across the globe. For these microbes, we do not know the phylum to which they belong. In other words, for some soils, almost 10% of taxa within bacteria and fungi are totally unknown to us. These taxa represent between 0.01–1.86% (average of 0.12%) of all 16S rRNA sequences, and between 0.00–22.11% (average of 3.98%) of all ITS retrieved sequences. On average, soil samples with the largest percentage of phylotypes of bacteria with an unknown phyla can be found in boreal and tropical forests (Fig. 1), while those with the largest percentage of phylotypes of fungi with an unknown phyla are found in dry forests and grasslands (Fig. 1).
I then generated a global atlas highlighting those global soils where bacterial and fungal phylotypes with an unknown phyla are expected to be more prevalent. Building these global maps is possible for three main reasons; firstly, the percentages of phylotypes of bacteria and fungi with an unknown phyla are highly correlated with key environmental factors at the global scale (Table 1). This result suggests that environmental data can be used to predict the distribution of phylotypes of fungi and bacteria unclassified at the phyla level. Secondly, the database used here covers a wide gradient of environmental conditions and soil properties found on Earth, being highly representative for globally distributed terrestrial ecosystems. For example, mean annual precipitation and temperature in these locations ranged from 67 to 3085 mm and −11.4 to 26.5 °C, respectively. Moreover, soil pH ranged from 4.04 to 9.21; soil C from 0.15 to 34.77%; and fine texture fraction (% clay + silt) from 1.40 to 92.00%. Finally, high resolution maps for key environmental factors predicting the percentage of unclassified taxa (Table 1) are available at the global scale. Therefore, globally available information on environmental factors can potentially be used to predict global hotspots for phylotypes of bacteria and fungi with an unknown phyla. These three important points allowed me to generate global atlases for the potential distribution of percentages of phylotypes of bacteria and fungi with an unknown phyla (Fig. 2). These global atlases were cross-validated as explained in Appendix 1 (Supplementary Materials).
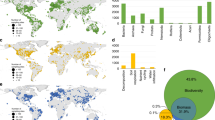
Global atlas including the potential distribution of % of phylotypes of bacteria and fungi with an unknown phyla (unclassified bacteria and fungi) based on their natural co-occurrence with climatic (aridity index, maximum and minimum temperature, precipitation seasonality and mean diurnal temperature range), primary productivity, dominant ecosystem type (forest and grasslands), soil properties (total organic carbon, pH and texture) and UV light in 235 locations. See Fig. S1 for the locations of the 235 in this study. See Appendix S1 for a cross-validation of these maps. A colour version of this figure is available in Fig. S2
The global maps included in this study indicate the potential distribution of unclassified taxa within bacteria and fungi. Interestingly, locations where bacteria with an unknown phyla are more prevalent are distinct from those of fungi. This global atlas suggests that soils from Brazil, Chile, Russia, Indonesia, Iceland, Northern Europe, and the coastlines of North America contain a relatively high percentage of bacteria with an unknown phyla. On the other hand, deserts from Peru, China, Australia, South Africa, the Middle East, the Saharan region, and the western coast of North America contain a relatively high percentage of unclassified taxa within fungi. Soil taxonomists and microbiologists should target soils from these environments and global locations to increase our chances of isolating and classifying these elusive yet significant soil taxa, and thus, increase our knowledge of who they are and what they are doing in our soils.
Methods
Soil sampling
Soils were collected from 235 locations across 18 countries and six continents. Soil samples (top ~7.5 cm depth) were collected under the most common vegetation across a wide range of ecosystem (forests, grasslands, and shrublands) and climatic (arid, temperate, tropical, continental, and polar ecosystems) types. The locations sampled represent wide gradients in environmental factors, which is critical for mapping predictions. Detailed information about this survey can be found in Delgado-Baquerizo et al. [8].
Molecular analyses
Soil DNA was extracted using the Powersoil® DNA Isolation Kit (MoBio Laboratories, Carlsbad, CA, USA) according to the manufacturer’s instructions. Amplicons targeting the bacterial 16S rRNA gene (341F-805R; [13]) and the fungal ITS region (FITS7-ITS4R; [14]) were sequenced at Western Sydney University’s NGS facility (Sydney, Australia) using the Illumina MiSeq platform. Bioinformatic processing was performed using a combination of QIIME [3], USEARCH [15], and UPARSE [16]. Operational taxonomic units—OTUs—(phylotypes hereafter), were identified at the ≥97% identity level. Taxonomy for bacteria and fungi was assigned using the Greengenes and UNITE databases, respectively. OTU abundance tables were constructed from these analyses. 16 s rRNA reads classified as Archaea, chloroplasts, or mitochondria were removed. The percentage of phylotypes of bacteria and fungi with an unknown phyla for each sample were calculated from these OTU tables. These phylotypes are classified as fungi or bacteria, but do not match data within taxonomic databases at the phyla level (unclassified bacteria and fungi hereafter). Given that soil and DNA samples were collected, extracted, and analysed following the same standardised protocol and within the same laboratory, any biases (e.g. sequencing error) would be consistent across analyses.
Environmental factors
For each location, information for twelve environmental factors was obtained: climate (maximum and minimum temperatures, precipitation seasonality; mean diurnal temperature range and Aridity Index); soil properties (pH, texture and total organic carbon); dominant ecosystem type (forest and grasslands); plant productivity; and UV light intensity. Information on soil pH, texture and total organic carbon (soil C) was obtained using standard laboratory methods [17, 18] in the laboratories from the Universidad Rey Juan Carlos (Spain). Climatic information (1 km resolution) for all sampling locations was obtained from the Worldclim database (www.worldclim.org; [11, 19]). The dominant ecosystem types (forest and grasslands) were determined in the field. Plant productivity (net primary productivity) data were obtained using the Normalized Difference Vegetation Index (NDVI) from the Moderate Resolution Imaging Spectroradiometer (MODIS) aboard NASA’s Terra satellites (http://neo.sci.gsfc.nasa.gov/). The monthly average value for this variable was calculated between 2003–2015 (~10 km resolution), when all soil samplings were conducted. Information on the annual ultraviolet index (UV index) was obtained from the NASA’s Aura satellite (https://neo.sci.gsfc.nasa.gov).
Mapping the global distribution of unclassified soil taxa
The prediction-oriented regression model Cubist [20] was used to predict the percentage of phylotypes of bacteria and fungi with an unknown phyla across the globe. Mapping analyses were independently done to find the percentage of unclassified taxa within bacteria and fungi. The Cubist algorithm uses a regression tree analysis to generate a set of hierarchical rules using information on environmental covariates, based on real data (235 locations), which are later used for spatial prediction [21]. Covariates in our models include the above described 12 environmental factors as well as space (latitude and longitude). Global predictions on the distribution of the percentage of unclassified taxa within bacteria and fungi were done on a 25 km resolution grid, which resulted in a grid including 225530 locations. Environmental information for each of these locations, including soil properties, climatic information, plant production, ecosystem types, and UV light, was obtained from global databases available online. Global information on soil properties for this grid was obtained using the ISRIC (global gridded soil information) Soil Grids (https://soilgrids.org/#!/?layer=geonode:taxnwrb_250m). Global information on the major vegetation types in this study (grasslands and forests) was obtained using the Globcover2009 map from the European Space Agency (http://due.esrin.esa.int/page_globcover.php). Global information on climate, UV radiation, and net primary productivity were obtained from the WorldClim database (www.worldclim.org) and NASA satellites (https://neo.sci.gsfc.nasa.gov), as explained above. The R package Cubist was used to conduct these analyses [21].
Data availability
The primary data used in this paper have been deposited in figshare: https://figshare.com/s/f7f34a772b316cdbda24 (https://doi.org/10.6084/m9.figshare.7895675).
References
Bardgett RD, van der Putten WH. Belowground biodiversity and ecosystem functioning. Nature. 2014;515:505–511.
Delgado-Baquerizo M, Maestre FT, Reich PB, Jeffries TC, Gaitan JJ, Encinar D, et al. Microbial diversity drives multifunctionality in terrestrial ecosystems. Nat Commun. 2016;28:10541.
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Method. 2010;7:335.
Tedersoo L, Bahram M, Põlme S, Kõljalg U, Yorou NS, Wijesundera R, et al. Fungal biogeography. Global diversity and geography of soil fungi. Science. 2014;346:1256688.
Fierer N. Embracing the unknown: disentangling the complexities of the soil microbiome. Nat Rev Microbiol. 2017;15:579–590.
Marx V. Microbiology: the return of culture. Nat Methods. 2017;14:37–40.
York A. Next-generation bacterial taxonomy. Nat Rev Microbiol. 2018;16:583.
Delgado-Baquerizo M, Oliverio AM, Brewer TE, Benavent-González A, Eldridge DJ, Bardgett RD, et al. A global atlas of the dominant bacteria found in soil. Science. 2018;19:320–325.
DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microb 2006;72:5069–5072.
Nilsson RH, Larsson K-H, Taylor AFS, Bengtsson-Palme J, Jeppesen TS, Schigel D, Kennedy P, Picard K, Glöckner FO, Tedersoo L, Saar I, Kõljalg U, Abarenkov K. The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Research 2018;47:259–264.
Zomer RJ, Trabucco A, Bossio DA, Verchot LV. Climate change mitigation: a spatial analysis of global land suitability for clean development mechanism afforestation and reforestation. Agric Ecosyst Envir. 2008;126:67–80.
Delgado-Baquerizo M., Eldridge DJ. Ecosystems. 2019. https://doi.org/10.1007/s10021-018-0333-2.
Herlemann DP, Labrenz M, Jürgens K, Bertilsson S, Waniek JJ, Andersson AF. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 2011;5:1571–1579.
Ihrmark K, Bödeker IT, Cruz-Martinez K, Friberg H, Kubartova A, Schenck J, et al. New primers to amplify the fungal ITS2 region-evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol Ecol. 2012;82:666–677.
Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460.
Edgar RG. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–998.
Anderson JM. JSI, Ingramm, Tropical Soil Biology and Fertility: A Handbook of Methods. Wallingford: CABI; 1993.
Kettler TA, Doran JW, Gilbert TL. Simplified method for soil particle-size determination to accompany soil-quality analyses. Soil Sci Soc Am J. 2001;65:849.
Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. Very high resolution interpolated climate surfaces for global land areas. Int J Climatol. 2005;25:1965–1978.
Quinlan JR. C4.5: Programs for Machine Learning. San Mateo: Morgan Kaufmann Publishers; 1993.
M. Kuhn, S. Weston, C. Keefer, N. Coulter (2016) Cubist: Rule- And Instance-Based Regression Modeling. R package version 0.0.19.
Acknowledgements
This project has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No 702057. I would like to thank Melissa S. Martín, David J. Eldridge, and Fernando T. Maestre for their comments and suggestions, which have helped to improve this piece. I would also like to thank Brajesh K. Singh, Noah Fierer, Richard Bardgett, Alberto Benavent-González, David J. Eldridge, and Fernando T. Maestre for their original contribution to the databases included in this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The author declares that he has no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Delgado-Baquerizo, M. Obscure soil microbes and where to find them. ISME J 13, 2120–2124 (2019). https://doi.org/10.1038/s41396-019-0405-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-019-0405-0
This article is cited by
-
A Comprehensive Insight of Current and Future Challenges in Large-Scale Soil Microbiome Analyses
Microbial Ecology (2023)
-
Composition, distribution, health risks, and drivers of phthalates in typical red paddy soils
Environmental Science and Pollution Research (2023)
-
Soil microbial diversity–biomass relationships are driven by soil carbon content across global biomes
The ISME Journal (2021)
-
The profile of the soil microbiota in the Cerrado is influenced by land use
Applied Microbiology and Biotechnology (2021)
-
Fungal diversity and occurrence of mycotoxin producing fungi in tropical vineyards
World Journal of Microbiology and Biotechnology (2021)