Abstract
Nitrogen (N) is a limiting nutrient in vast regions of the world’s oceans, yet the sources of N available to various phytoplankton groups remain poorly understood. In this study, we investigated inorganic carbon (C) fixation rates and nitrate (NO3−), ammonium (NH4+) and urea uptake rates at the single cell level in photosynthetic pico-eukaryotes (PPE) and the cyanobacteria Prochlorococcus and Synechococcus. To that end, we used dual 15N and 13C-labeled incubation assays coupled to flow cytometry cell sorting and nanoSIMS analysis on samples collected in the North Pacific Subtropical Gyre (NPSG) and in the California Current System (CCS). Based on these analyses, we found that photosynthetic growth rates (based on C fixation) of PPE were higher in the CCS than in the NSPG, while the opposite was observed for Prochlorococcus. Reduced forms of N (NH4+ and urea) accounted for the majority of N acquisition for all the groups studied. NO3− represented a reduced fraction of total N uptake in all groups but was higher in PPE (17.4 ± 11.2% on average) than in Prochlorococcus and Synechococcus (4.5 ± 6.5 and 2.9 ± 2.1% on average, respectively). This may in part explain the contrasting biogeography of these picoplankton groups. Moreover, single cell analyses reveal that cell-to-cell heterogeneity within picoplankton groups was significantly greater for NO3− uptake than for C fixation and NH4+ uptake. We hypothesize that cellular heterogeneity in NO3− uptake within groups facilitates adaptation to the fluctuating availability of NO3− in the environment.
Similar content being viewed by others
Introduction
In many regions of the world’s oceans, particularly in the oligotrophic subtropical gyres, nitrogen (N) limits primary production and controls planktonic community composition [1, 2]. In these vast regions, planktonic biomass is dominated by picoplankton (size < 3 µm) whose high surface-area-to-volume ratio is believed to represent a competitive advantage under N scarcity [3]. While genetically diverse [4,5,6,7], photosynthetic picoplankton are generally grouped into the pico-eukaryotes (PPE) and the cyanobacterial prokaryotes Prochlorococcus and Synechococcus. As opposed to Synechococcus, which is widely distributed in the ocean between 50°S and 50°N, Prochlorococcus thrives mostly in nitrate-depleted subtropical and tropical waters [8], where it can contribute significantly to primary production [9,10,11]. In contrast, PPE are most abundant in productive waters such as upwelling or temperate regions [12, 13]. Despite their low abundance relative to pico-cyanobacteria in the open ocean, PPE can occasionally contribute as much as Prochlorococcus to primary production due to their larger biovolume [9, 10].
Differences in the geographical distributions of PPE, Prochlorococcus, and Synechococcus have been hypothesized to be driven by environmental factors such as light [8, 14,15,16], temperature [14, 17, 18], and the availability of different sources of N [19,20,21]. A recent study found, based on isotopic 15N signatures of cytometrically-sorted groups of picoplankton, that PPE obtain roughly half of their N from upwelled NO3−, while Synechococcus and Prochlorococcus rely on recycled N in the Deep Chlorophyll Maximum (DCM) of the North Atlantic Gyre [22]. However, the discovery of gene pathways for NO3− assimilation in the metagenome of Prochlorococcus [23] and the cultivation of isolates able to grow on NO3− as an exclusive source of N [24] have raised the possibility that NO3− may be a significant source of N in wild populations of Prochlorococcus under certain growth conditions. Measurements from the DCM in the Atlantic show measurable, albeit small, NO3− uptake by Prochlorococcus, accounting for ~5–10% of the group total N uptake [25]. Based on these observations, it can be argued that a subset of the genetically diverse Prochlorococcus genus may be capable of assimilating NO3− [24, 26,27,28].
Technological challenges have hindered progress in our understanding of group and cell specific N uptake. In the last two decades, isotope incubations combined with cell sorting by flow cytometry have been used to measure plankton activity at the group level [29]. However, stable 15N isotope analyses by conventional isotope ratio mass spectrometers require substantial amounts of material. As a result, a prohibitively large number of cells must be sorted to measure group-specific N utilization, which is especially problematic in oligotrophic systems. New generations of isotope ratio mass spectrometers, most particularly nano-scale secondary ion mass spectrometers (nanoSIMS), have opened new research directions by allowing analyses of isotopic composition at the single cell level [30, 31].
In this study, we use nanoSIMS coupled with flow cytometry cell sorting to measure the metabolic activity of cells after incubating the natural plankton community with stable isotopes of C and N (13C and 15N). To the best of our knowledge, this is the first study reporting cell-specific contribution of NO3−, NH4+, and N-urea to the photosynthetic growth requirements of the Prochlorococcus, Synechococcus, and PPE groups. The large number of cells analyzed (~16 000) from distinct biomes of the North Pacific highlights clear functional differences between picoplankton groups in terms of N uptake and cell-to-cell metabolic heterogeneity.
Materials and methods
Sampling location and hydrological context
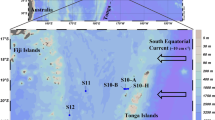
Our study was carried out during Schmidt Ocean Institute and NASA’s Sea-to-Space cruise aboard the R/V Falkor in January/February 2017. Two stations in the North Pacific Subtropical Gyre (NPSG, stations 1, 2) and one station in the California Current System (CCS, station 3) were investigated (Fig. 1). Seawater samples were collected using Niskin bottles mounted on a rosette equipped with a CTD probe. Samples were collected at the surface (5 m) and at the 1% surface light level, which corresponded roughly to the DCM at stations 1 and 2. Chlorophyll a (Chl a), NO3−, NH4+ and urea were measured in triplicate according to standard procedures (supplementary information). Picoplankton abundances were determined in triplicate by flow cytometry (supplementary information).
Experimental setup and sampling
At each station and depth, C fixation, NO3−, NH4+ and urea uptake were measured during daylight. In addition, the same experiments were performed at night from surface samples at stations 1 and 3. Dual 13C-15N isotope incubations were performed with the addition of either dissolved inorganic 13C (DI13C, NaHCO3, 98% Sigma-Aldrich) in combination with 15NO3− (KNO3, 99%, Euriso-top) or 15 NH4+ (NH4Cl, 99%, Euriso-top) or with the addition of dual labelled 15N-13C-urea (98% 15N, 99% 13C, Euriso-top). Seawater was collected directly from the Niskin bottles in a set of five HCl-cleaned polycarbonate 1.3 or 2.3 L bottles for each isotopic treatment. Isotopes were added within ~2 h of sample collection, with additions targeted at ≤ 10% of ambient concentrations estimated from the literature for DIC, NO3− and urea and from onboard measurements of NH4+. To define the initial 13C and 15N enrichments in the particulate matter, one bottle from each set was immediately filtered after isotope inoculation on a combusted (4 h, 450 °C) GF/F filter using a vacuum pump ( < 150 mbar). The filters were rinsed using non-labeled 0.2 µm filtered seawater and stored at −20 °C. The other four bottles of each set were placed in an on-deck incubator reproducing the light intensity at the sampling depth using blue light screens (Lee Filter) and cooled with circulating sea surface water. The incubations, centered around local noon for daylight incubations, were short (~5 h on average) in order to minimize bottle artifacts, including isotope dilution effects and recycling of 15NO3− that could make 15N available as reduced N [25, 32]. After incubation, triplicate bottles from each set were filtered onto GF/F filters as described above. The remaining bottle from each set was used to concentrate, recover and store the cells as described in Fawcett et al. [22]. An additional bottle was added to the set of bottles collected from sea surface water at station 2, then incubated without isotope addition to measure the natural 13C and 15N abundances of recovered cells. Back onshore, cells were sorted using an Influx flow cytometer (BD Biosciences, San Jose, CA, USA) according to Duhamel et al. [33] (supplementary information). Sorted cells were directly collected on a 0.2 µm polycarbonate membrane connected to a low-vacuum pump at the exit of the cell sorter, in order to limit dispersion of cells on the filter. The filters were then stored at −20 °C until further analyses.
Single cell analysis using nanoSIMS
Filters with sorted cells were mounted on a 5 mm spherical aluminum plot using double-sided adhesive copper tape. The regions of interests (ROIs) containing the cells were located using an epifluorescence microscope. To facilitate further identification of the ROIs under the nanoSIMS, the fractions of the filters surrounding the ROIs were removed, retaining ~1–2 mm2 of filter with a high density of cells. The preparation was subsequently metalized by sputter deposition of a gold film (20 nm thickness) before introduction into a nanoSIMS 50 (Cameca, Gennevillier, France).
ROIs containing sorted cells were recovered using the optical camera of the nanoSIMS, and analyses were conducted on field size of 40 × 40, 20 × 20 or 10 × 10 µm with an image size of 245 × 256 pixels. The areas were pre-sputtered with a primary Cs+ ion beam of 300 pA for ~0.5–2 min, targeting a cumulative charge of ~22 pC cm−2. Analyses were performed using a 1.2 pA Cs+ primary ion beam focused to a spot size of 60–120 nm for 1000 µs px−1. At least 20 planes were accumulated for each analysis, alternatively recording the masses 12C−, 13C−, 12C14N−, 12C15N−, 30P−, and 12C−, 13C−, 12C14N−, 13C14N−, 30P−. The mass resolution for CN− ions was > 7000. Switching between the ions 12C15N− and 13C14N− was performed using the “peak jumping” mode, without changing the magnetic field but varying the deflectors’ voltage. This allowed the quasi-simultaneous measurement of the 15N/14N and 13C/12C ratios using the 12C15N−/12C14N− and 13C14N−/12C14N− ions. The ions 13C14N−/12C14N− were preferred over the 13C−/12C− ions because of the higher ionization efficiency of CN− compared to C−, leading to a greater precision in the determination of the 13C/12C isotopic ratios (Fig S1).
Due to the high number of cells analyzed, the cells were automatically (instead of manually) outlined using the particle detection mode of the LIMAGE software, based on the total 12C14N− ion counts. Each identified cell was individually checked for quality based on its shape, size, and ion counts. Poor quality cells were withdrawn from further analyses. In each assay and for each group, 218 cells were analyzed on average (range: 12–1040) which was achieved in one to four ion images randomly chosen within the previously defined ROIs. This led to a total of ~16,000 cells analyzed for this study. The average sizes of the analyzed Prochlorococcus, Synechococcus, and PPE cells were 0.6 ± 0.1, 1.2 ± 0.2, and 1.7 ± 0.2 µm, respectively.
Rate calculations and statistical analyses
For each cell analyzed with nanoSIMS, CN− ion isotopes were recorded and fractional abundances of A13C \(\left( {\frac{{{\,}^{13}}{{\mathrm{C}}{\,}^{14}}{{\mathrm{N}}^{-}}}{{{\,}^{13}}{{\mathrm{C}}^{14}}{{\mathrm{N}^-}+{{\,}^{12}}{{\mathrm{C}}^{14}}{{\mathrm{N}}^{-}}}} \ast 100} \right)\) and A15N \(\left( {\frac{{{\,}^{12}{{\mathrm{C}}^{15}}{\mathrm{N}}^{-} }}{{{\,}^{12}{{\mathrm{C}}^{14}}{\mathrm{N}}^{-} + {\,}^{12}{{\mathrm{C}}^{15}}{\mathrm{N}}^{-} }} \ast 100} \right)\) were computed (Fig. 2).
The cellular C- and N-specific uptake rates (h−1) were calculated as follows:
where Acell, \(\overline A _{nat}\), and Asource reflect the isotopic fractional abundances of the cell after incubation (A13C or A15N), of the cells (mean) prior to incubation (Fig S2) and of the source pool, respectively. t is the incubation time. Specific uptake rates were converted to group absolute uptake rates (fmol L−1 h−1), by multiplying the specific rates by the cell C or N content and by the cell abundances. Cell C content was computed from cell biovolume (derived from its equivalent spherical diameter) and a volumetric C cell content of 237 fg C µm−3 [34]. The cell N content was derived from the cell C content, assuming C/N ratios of 7.7, 9.7 and 11 for Prochlorococcus, Synechococcus and PPE, respectively [35].
In the case of DI13C incubations, C-based specific division rates were calculated as follows, assuming that DIC was the only C source for cellular growth:
Hourly specific division rates were scaled to daily rates using the model developed by Moutin et al. [36].
Total community uptake rates (nmol L−1 h−1) were estimated from the material collected on GF/F filters and analyzed with an elemental analyzer coupled to an isotope ratio mass spectrometer (EA-IRMS), and calculated as follows:
where Anat,GFF, APOM,GFF, and POM represent the isotopic abundances prior to and after incubations and the particulate organic matter C or N concentrations (nmol L−1), respectively.
Ambient NO3−, NH4+, and urea concentrations were often low, and the addition of isotopes at tracer level (i.e. < 10% of initial concentration) was not always possible, leading to a potential overestimation of N uptake rates [37]. To overcome this issue, kinetics experiments consisting of increasing additions of N substrates were conducted and N uptake rates were corrected using the equation described in Rees et al. [38], as detailed in the supplementary information.
The large number of cells analyzed for each group in each assay (218 on average) allowed for an assessment of the intra-group cell-to-cell rate heterogeneity or “metabolic heterogeneity”, defined as the coefficient of variation in isotope uptake rate. However, a fraction of the measured variability stems from analytical errors associated with the determination of cellular isotopic ratios. This uncertainty, resulting from the limited signal of CN− ions detected with the nanoSIMS, follows a Poisson distribution (Fig. S2). The metabolic heterogeneity was thus corrected for the influence of the Poisson dispersion (Po) as follows:
std(Agroup) is the standard deviation of the isotopic abundance of the group, \({\widetilde{A}}_{\mathrm{group}}\) the mean isotopic abundance of the group, \({\widetilde{A}}_{\mathrm{nat}}\), the mean natural isotopic abundance of the group, and std(Po) is the standard deviation associated with the Poisson distribution modeled for each group. The Poisson distribution was modeled using an average number of events (λ):
where \({\widetilde{N}}_{{{\rm{CN}}^-,{\rm{group}}}}\) is the average total number of CN− ions detected in the cells of the group. On average, std(Po) represented 15 ± 21 and 27 ± 18% of the measured standard deviation of cell N uptake and C fixation, respectively. To err on the side of caution, we only show results where std(Po) contributed to less than 50% of std(Agroup).
Cellular rates were considered to be detected when their fractional abundance enrichment \(A_{\mathrm{cell}} - {\widetilde{A}}_{\mathrm{nat}}\) was higher than two times the standard deviation associated with the Poisson distribution parameterized by \({\mathrm{\lambda }} = A_{\mathrm{cell}} \ast N_{{{\rm{CN}}^-,{\rm{cell}}}}\), where \({\widetilde{N}}_{{{\rm{CN}}^-,{\rm{cell}}}}\) is the CN− ion count of the cell. We note that the latter metric should be interpreted cautiously, as it depends not only on the cellular activity itself but also on the isotopic fractional abundance in the initial pool, the duration of the incubation, and the number of ions detected in the cells, which are variable between groups and assays. Both detected and undetected cellular rates were considered in the calculation of average group rates. The groups were considered as active when the mean cellular fractional isotopic abundances of the groups were significantly higher than those of non-incubated cells belonging to the same group (unpaired Mann-Whitney test, p < 0.001). Differences in C or N uptake rates between stations, depths or groups were tested using unpaired Mann-Whitney test and considered significant if p < 0.05.
Results
Hydrography and Biogeochemistry of the sampled regions
Stations one and two were characteristic of the oligotrophic NPSG, with low NO3− ( < 2 nmol L−1) and Chl a concentrations ( < 0.2 µg L−1) in surface waters and a DCM deeper than 100 m (Table 1). In contrast, station three showed some evidence of coastal upwelling associated with the CCS, with higher NO3− (>100 nmol L−1) and Chl a (0.71 µg L−1) concentrations in surface waters. NH4+ concentrations ranged between < 3 to 21 nmol L−1 in the NPSG, and were higher in the CCS (55–59 nmol L−1) (Table 1). Urea concentrations were generally higher but followed the same pattern, ranging between 50–109 nmol N L−1 in the NPSG and from 162 to 165 nmol N L−1 in the CCS. While Prochlorococcus numerically dominated the picoplankton community over Synechococcus and PPE, both at the surface and at the DCM in the NPSG, the opposite was observed in the CCS (Table 1).
C fixation rates and N uptake at the community level
In the NPSG, the low NO3− concentrations at the surface led to significantly lower total community NO3− uptake ( < 0.1 nmol L−1 h−1) compared to at the DCM (0.1–0.6 nmol L−1 h−1) (p < 0.05). In contrast, rates of total community NH4+ uptake were significantly higher in the surface than in the DCM (p < 0.001) (Table 1). Rates of community C fixation, and NO3− and NH4+ uptake were lower in the NPSG than in the CCS (p < 0.05) (Table 1). Community N-urea uptake rates ranged from 0.7–3.3 nmol N L−1 h−1, without clear trends between the two oceanic regions and at rates similar or higher than NO3− and NH4+ uptake in the NPSG. In contrast to the relatively high N-urea uptake, C-urea uptake was not detected in the sampled planktonic communities.
C fixation rates and N uptake at the single-cell and group levels
13C-enrichment measurements at the cellular level following DI13C incubations allowed a direct assessment of specific C-based division rates of wild groups of Prochlorococcus, Synechococcus, and PPE. At the single-cell level, division rates varied widely, ranging from undetectable to 1 d−1 (Fig. 3) with clear group-level differences in C-based division rates. Prochlorococcus groups were active at all the stations (Fig. 3) but mean division rates were significantly higher in the NPSG (ranging between 0.32 and 0.43 d−1) than in the CCS ( < 0.1 d−1) (p < 0.001) which is in agreement with the findings of Ribalet et al. [39] along a similar transect. Mean PPE division rates showed an opposite pattern, with significantly higher rates in the CCS (0.42–0.50 d−1) than in the NPSG (0.15–0.26 d−1) (p < 0.001) highlighting the contrasting biogeography of these two groups. Mean Synechococcus division rates ranged from 0.21 to 0.46 d−1, without a clear pattern between the NPSG and the CCS (p > 0.05). Using cellular volume-to-biomass conversion factors, the measured C fixation by picoplankton (sum of the C fixation attributed to PPE, Prochlorococcus and Synechococcus) was estimated to account for 43 to 67% of the total community C fixation (55% on average), without a clear geographical pattern, demonstrating the important contribution of picoplankton to oceanic C fixation (Table S1). In the NPSG, Prochlorococcus accounted for the largest fraction of community C fixation (25–56%), followed by PPE (7–18%), and Synechococcus ( < 1–3%). In the CCS, PPE and Synechococcus accounted for most of the community C fixation (42–50 and 11–19%, respectively), while Prochlorococcus accounted for < 1%.
Whisker plot of C-based specific division rate (d−1) for each group (Prochlorococcus, Synechococcus, and PPE) in each daylight assay. Each dot represents an analyzed cell. Grey dots denote cells with rates not significantly different from zero. Colored lines denote mean division rates and standard deviations (plain horizontal and dashed vertical, respectively) in each assay. The percentages indicate proportions of detected active cells. Grey lines denote mean division rates and standard deviations (horizontal plain and vertical dashed, respectively) in the North Pacific Subtropical Gyre (NPSG) and in the California Current System (CCS) regions
N-specific uptake rates at the single-cell level were also highly variable (Fig. 4). NH4+ and urea uptake rates were detected in most cells (86 ± 20 and 86 ± 23%, respectively) at rates on average twice higher (p < 0.001) for Synechococcus (0.0130 and 0.0151 h−1, respectively) than for Prochlorococcus (0.0099 and 0.0077 h−1, respectively) and PPE (0.0073 and 0.0067 h−1, respectively). In contrast, group specific NO3− uptake rates were significantly (p < 0.001) higher for PPE (0.0041 h−1) than for Prochlorococcus (0.0004 h−1) and Synechococcus (0.0010 h−1) and were detected in a greater proportion of PPE cells (81 ± 23%) than for Prochlorococcus (37 ± 32%) or Synechococcus (56 ± 42%) (Fig. 4). As a result, in most of the assays, the average contribution of NO3− to group N uptake (sum of NH4+, urea and NO3− uptake rates) was significantly higher for PPE (17.4 ± 11.2%) than for Prochlorococcus (4.5 ± 6.5%) or Synechococcus (2.9 ± 2.1%) (p < 0.001, Fig. 5c). The contribution of NO3− to group N uptake was, however, highly variable between assays, ranging from 2.4to 31.8%, undetectableto17% and undetectable to 5.3% for PPE, Prochlorococcus and Synechococcus, respectively. The contributions of NO3− to group N uptake were higher in the CCS than in the NPSG for Prochlorococcus (10.1 ± 7.5 vs. 1.5 ± 0.4%, p < 0.05) and PPE (22.3 ± 10.2 vs. 14.5 ± 11%, p > 0.05), albeit not significantly in the latter case. For Synechococcus, variability was lower and no clear spatial trends were observed. In agreement with the results at the community-level, C-urea uptake was undetectable at the group level.
NH4+, urea, and NO3− specific uptake rates (h−1) of photosynthetic picoeukaryotes (PPE), Prochlorococcus, and Synechococcus in each assay. Each dot represents an analyzed cell. Colored and grey dots denote cells with detected and undetected activities, respectively. The percentages are the proportions of detected active cells in each assay
a Group N specific uptake (sum of NO3−, NH4+, and urea specific uptake, h−1) of photosynthetic picoeukaryotes (PPE), Prochlorococcus (pro), and Synechococcus (syn) in each assay. b Contribution of the different N sources to group N uptake (%) in each assay. c Contribution of the different N sources averaged over all assays for each population investigated (%)
Comparisons of specific C fixation to the sum of specific N uptakes from the daylight incubations were close to the 1:1 unity line (except for surface Prochlorococcus and Synechococcus at station 1) without clear patterns between groups or depths, suggesting that cells meet their N requirement during their photosynthetic C acquisition period (Fig. S3 and further discussion in the SI information).
Single-cell analyses revealed heterogeneity in C and N uptake of individuals within each planktonic group. The metabolic heterogeneity is qualitatively estimated using the coefficient of variation of cellular rates corrected for the influence of instrumental noise. In all groups, heterogeneity was the lowest for C fixation and NH4+ uptake (0.2 to 0.8), slightly higher for urea (0.3 to 1.3) and highest for NO3− uptake (0.5 to 2.6) (Fig. 6). In most of the assays and for most of the measured rates, Synechococcus was the most homogeneous group, followed by Prochlorococcus and PPE, respectively (Fig. 6).
Box-and-whisker plot of the metabolic heterogeneity for C fixation and NH4+, NO3−, and N-urea uptake for photosynthetic pico-eukaryote (PPE), Prochlorococcus, and Synechococcus groups. Each dot represents an assay. Only daylight assays were considered for the C metabolic heterogeneity. For each group, metabolic heterogeneity medians that are not statistically different are indicated by the same letters (unpaired Kruskal-Wallis test, p > 0.05)
In assays where Prochlorococcus and Synechococcus contributed to a significant fraction of community C fixation (>5%), these groups did not rely on NO3− for their growth (<2% of the group N uptake) (Fig. 7). In contrast, in assays where PPE contributed significantly to C fixation, this group relied substantially on NO3− for its growth (11–36% of the group N uptake, Fig. 7), again highlighting the contrasting biogeography and physiologies of these plankton groups.
Discussion
In this study, we determined the forms of N taken up by the most abundant groups of marine phytoplankton: PPE, Prochlorococcus, and Synechococcus. We report the first measurements of the contribution of NO3−, NH4+, and urea to N uptake, estimated at the single-cell level, in relation to C-based division rates by individual cells. Our results, based on ~16 000 cells collected in contrasting biomes of the North Pacific Ocean, clearly show substantial intra- and inter-group heterogeneity.
Inter-group similarities and differences in N uptake
NH4+ and urea species are generally scarce in oligotrophic regions, but are believed to represent the main sources of N for phytoplankton because of their high turnover rate of < 1–5 days [40,41,42,43,44]. Our measurements confirm these results at the scale of the entire plankton community (Table 1), but also for each plankton group investigated in the two regions sampled (Fig. 5). In line with earlier studies ([45] and references therein), we find that urea is an important source of N for open ocean photosynthetic picoplankton populations, accounting for the largest fraction of N uptake for all groups tested (38–68%; on average greater than that of NH4+). However, the absence of 13C-enrichment following the 13C-15N-urea incubations indicates that the C derived from urea is not used by the groups investigated (see supplementary information for further discussion).
While NH4+ and urea uptake rates were systematically significant for all the groups tested, the contribution of NO3− uptake showed greater contrasts between plankton groups. At all the study sites, the prokaryotes Prochlorococcus and Synechococcus relied ~4–6 times less on NO3− than PPE on average, indicating clear functional diversity between photosynthetic prokaryotes and PPE (Fig. 5). This is in line with the contrasting δ15N signatures of prokaryotes and eukaryotes in the subtropical Atlantic [22, 46]. If we exclude groups that contributed to an insignificant fraction of C fixation (<5%), N strategies are even more clear, with an order of magnitude discrepancy in the NO3− contribution to N uptake between prokaryotes and eukaryotes (Fig. 7). However, our results do show significant, albeit small, NO3− uptake by photosynthetic prokaryotes including Prochlorococcus. This confirms the potential for NO3− uptake by wild populations of Prochlorococcus as suggested by genomic and culture studies [23, 24, 47].
Intra-group differences in N uptake between stations and sampled depths
In addition to the inter-group differences in N uptake strategies, we also found large group variations in Prochlorococcus and PPE in the contribution of NO3− to the N uptake between locations, with NO3− uptake increasing with NO3− availability (Fig. 5). This suggests adaptive strategies in response to the N species available, which might result from genetic selection and/or phenotypic plasticity. Prochlorococcus and PPE groups harbor genetically diverse populations [4, 6, 7, 28, 48,49,50,51]. These populations display varying affinities for the different sources of N [23, 24, 52, 53] and do not always possess the necessary pathways for NO3− assimilation [47, 54] which could result in ecological adaptation as a function of NO3− availability.
Our results could also be explained by phenotypic plasticity, since photosynthetic organisms can regulate the expression of genes involved in N assimilation as a function of the availability of different forms of N [54,55,56]. NO3− is energetically more expensive than NH4+ to assimilate ([57] and references therein), leading to NO3−uptake inhibition with increasing NH4+ availability [52, 58, 59]. One would therefore expect the contribution of NO3− to the total N uptake would be controlled by NH4+ availability. This is not what we observed, presumably because the NH4+ concentrations observed in this study ( < 60 nmol L−1) were not high enough to fully inhibit NO3− uptake [37]. In other words, at the ambient concentrations measured in this study, both sources of N could efficiently be used by the phytoplankton community and may explain the adaptation of groups to N availability.
Intra-group heterogeneity in C and N uptake within assays
While genetic diversity of plankton in the ocean has received increasing attention in the past two decades [60, 61], the heterogeneity of cellular metabolisms remains poorly characterized. However, there is growing evidence that phenotypic heterogeneity, or the diversity of metabolisms between genetically identical cells living in the same environment, plays a role in population dynamics and consequently in ecosystem functions [62]. Through the specialization of subsets of cells in metabolic processes, phenotypic heterogeneity represents an adaptive advantage in fluctuating environments [63,64,65] and can enhance populations growth rates [66]. Here we shed light on cell-to-cell C and N uptake and metabolic heterogeneity of pico-phytoplankton groups in the ocean using a single cell approach. Within our cytometrically sorted groups we cannot quantitatively assess the relative influences of genetic and phenotypic variabilities on the measured metabolic heterogeneity. Nevertheless, our results of heterogeneity for C fixation and NH4+ uptake (0.2–0.9) are within the range of those measured from C fixation in pure cultures of the cyanobacterial diazotroph Crocosphaera (~0.5) and from NH4+ uptake in the heterotrophic bacteria Staphylococcus aureus and Pseudomonas aeruginosa (0.4–0.9) [67, 68]. This suggests that, at the group level, C-fixation and NH4+ uptake are within the range of the phenotypic noise, despite potentially high genetic diversity.
Interestingly, for each group investigated, the metabolic heterogeneity was systematically higher for NO3− uptake compared to NH4+ and urea uptake or to C fixation (Fig. 6). The lack of data documenting phenotypic diversity in NO3− uptake from pure cultures makes it difficult to contextualize our values. However, NO3− assimilation is an energetically costly process and NO3− availability can be highly dynamic throughout the year in the studied area. It can be postulated that within planktonic populations, a subset of cells invests in NO3− assimilation machinery in prevision of higher NO3− availability, which might facilitate population’s adaptation to new nutritional environment. Similar strategies for N2 fixation, another energetically costly N assimilation process, have been reported in multicellular filamentous cyanobacteria [69, 70] and in unicellular cyanobacteria [63, 71]. However, we cannot completely rule out the influence of genetic diversity on the metabolic heterogeneity measured here. While the genetic potential for NO3− assimilation is widespread in PPE and Synechococcus [54, 72], this is not the case for Prochlorococcus, for which only some ecotypes present the full set of genes involved in the NO3− assimilation process [47]. Thus, further studies characterizing the relative influence of genetic and phenotypic factors in metabolic heterogeneity are needed to improve our understanding of the role of cell-to-cell heterogeneity in the adaption of plankton populations to their highly dynamic environments.
Potential links between the biogeography of plankton groups and their N uptake strategies
A multitude of factors can limit plankton growth and biomass accumulation, including temperature, light, and N availability. For example, the decrease in Prochlorococcus abundances and growth from the NPSG to the CCS (Fig. 3, Table 1), as reported in previous studies [39], has been attributed to low sea surface temperatures in the CCS (8–12 °C) inhibiting growth [15, 16, 73]. Similarly, the slightly lower division rate of PPE at the bottom of the euphotic layer compared to the surface (Fig. 3) suggests regulation by light availability, as previously hypothesized [10, 74]. In addition, the nature of the N sources available are suspected to affect planktonic community structure, particularly in regions where N limits primary production such as in the NPSG [75,76,77,78]. Our results highlight group specific N strategies that are in line with this theory. However, with the present dataset it is difficult to disentangle the combined effects of temperature, light and N availability on this contrasting biogeography. Combined with controlled perturbation experiments (e.g. light, temperature, nutrients), the present approach will help to better resolve the relative influence of environmental parameters on the biogeography of marine plankton.
Conclusion
In this study, we investigated the C and N uptake strategies of marine picophytoplankton. For this purpose, we incubated our samples with isotopically-labelled substrates, sorted cells by flow cytometry, and analyzed their isotopic signatures using nanoSIMS technology. The analysis of a large number of cells by nanoSIMS provided a robust characterization of the average nutrient uptake strategy for each group as well as the cell-to-cell heterogeneity in each analyzed group. Our results confirm the differing N acquisition strategies of eukaryotes and prokaryotes. While both groups use reduced sources of N such as NH4+ and urea, eukaryotes also rely on NO3− to fulfill their N demand, a finding which may drive the contrasting biogeography of these pico-phytoplankton groups. However, the relatively large heterogeneity in NO3− uptake for all groups implies that only subsets of cells are involved in this process. Molecular [23, 79] and biogeochemical approaches [22] have reported conflicting evidence on whether prokaryotic picoplankton assimilate NO3−. Our results reconcile these approaches by showing that NO3− can represent a significant proportion of N nutrition in some Prochlorococcus and Synechococcus cells, but that this process is small at the group level. We postulate that variability in cell-to-cell uptake of NO3−, possibly resulting from genetic and/or phenotypic heterogeneity, may provide wild populations a competitive advantage in fluctuating environments. Our study thus raises numerous questions concerning diversity in the cellular metabolic activities of oceanic phytoplankton, which may be best investigated with a combination of culture, field and modeling experiments.
Change history
23 October 2020
A Correction to this paper has been published: https://doi.org/10.1038/s41396-020-00813-w
References
Tyrrell T. The relative influences of nitrogen and phosphorus on oceanic primary production. Nature. 1999;400:525–31.
Moore JK, Geider RJ, Guieu C, Jaccard SL, Jickells TD, LaRoche J, et al. Processes and patterns of nutrient limitation. Nat Geosci. 2013;6:701–10.
Chisholm SW. Phytoplankton size. In: Falkowski PG, Woodhead AD, Vivirito K, eds. Primary productivity and biogeochemical cycles in the sea. Boston, MA: Springer US; 1992. p. 213–37.
de Vargas C, Audic S, Henry N, Decelle J, Mahe F, Logares R, et al. Eukaryotic plankton diversity in the sunlit ocean. Science. 2015;348:1261605.
Kirkham AR, Lepère C, Jardillier LE, Not F, Bouman H, Mead A, et al. A global perspective on marine photosynthetic picoeukaryote community structure. ISME J. 2013;7:922–36.
Sohm JA, Ahlgren NA, Thomson ZJ, Williams C, Moffett JW, Saito MA, et al. Co-occurring Synechococcus ecotypes occupy four major oceanic regimes defined by temperature, macronutrients and iron. ISME J. 2016;10:333–45.
Biller SJ, Berube PM, Lindell D, Chisholm SW. Prochlorococcus: the structure and function of collective diversity. Nat Rev Microbiol. 2014;13:13–27.
Flombaum P, Gallegos JL, Gordillo RA, Rincon J, Zabala LL, Jiao N, et al. Present and future global distributions of the marine Cyanobacteria Prochlorococcus and Synechococcus. Proc Natl Acad Sci. 2013;110:9824–9.
Rii YM, Duhamel S, Bidigare RR, Karl DM, Repeta DJ, Church MJ. Diversity and productivity of photosynthetic picoeukaryotes in biogeochemically distinct regions of the South East Pacific Ocean. Limnol Oceanogr. 2016;61:806–24.
Rii YM, Karl DM, Church MJ. Temporal and vertical variability in picophytoplankton primary productivity in the North Pacific Subtropical Gyre. Mar Ecol Prog Ser. 2016;562:1–18.
Hartmann M, Gomez-Pereira P, Grob C, Ostrowski M, Scanlan DJ, Zubkov MV. Efficient CO2fixation by surface Prochlorococcusin the Atlantic Ocean. ISME J. 2014;8:2280–9.
Buitenhuis ET, Li WKW, Vaulot D, Lomas MW, Landry MR, Partensky F, et al. Picophytoplankton biomass distribution in the global ocean. Earth Syst. Sci Data. 2012;4:37–46.
Jardillier L, Zubkov MV, Pearman J, Scanlan DJ. Significant CO2 fixation by small prymnesiophytes in the subtropical and tropical northeast Atlantic Ocean. ISME J. 2010;4:1180–92.
Mackey KRM, Paytan A, Caldeira K, Grossman AR, Moran D, McIlvin M, et al. Effect of temperature on photosynthesis and growth in marine Synechococcus spp. Plant Physiol. 2013;163:815–29.
Johnson ZI, Zinser ER, Coe A, McNulty NP, Woodward EMS, Chisholm SW. Niche partitioning among Prochlorococcus ecotypes along ocean-scale environmental gradients. Science. 2006;311:1737–40.
Zinser ER, Johnson ZI, Coe A, Karaca E, Veneziano D, Chisholm SW. Influence of light and temperature on Prochlorococcus ecotype distributions in the Atlantic Ocean. Limnol Oceanogr. 2007;52:2205–20.
Moore LR, Goericke R, Chisholm SW. Comparative physiology of Synechococcusand Prochlorococcus: Influence of light and temperature on growth, pigments, fluorescence and absorptive properties. Mar Ecol Prog Ser. 1995;116:259–76.
Chen B, Liu H, Huang B, Wang J. Temperature effects on the growth rate of marine picoplankton. Mar Ecol Prog Ser. 2014;505:37–47.
Martiny AC, Pham CTA, Primeau FW, Vrugt JA, Moore JK, Levin SA, et al. Strong latitudinal patterns in the elemental ratios of marine plankton and organic matter. Nat Geosci. 2013 ;6:279–83.
Weber TS, Deutsch C. Ocean nutrient ratios governed by plankton biogeography. Nature . 2010;467:550–4.
Karl DM, Bidigare RR, Church MJ, Dore JE, Letelier RM, Mahaffey C et al. The nitrogen cycle in the North Pacific trades biome: an evolving paradigm. In: Nitrogen in the marine environment. 2008. New-York, USA: Academic press, pp. 705–69.
Fawcett SE, Lomas MW, Casey JR, Ward BB, Sigman DM. Assimilation of upwelled nitrate by small eukaryotes in the Sargasso Sea. Nat Geosci. 2011;4:717–22.
Martiny AC, Kathuria S, Berube PM. Widespread metabolic potential for nitrite and nitrate assimilation among Prochlorococcus ecotypes. Proc Natl Acad Sci Usa. 2009;106:10787–92.
Berube PM, Biller SJ, Kent AG, Berta-Thompson JW, Roggensack SE, Roache-Johnson KH, et al. Physiology and evolution of nitrate acquisition in Prochlorococcus. ISME J. 2014;9:1195–207.
Casey JR, Lomas MW, Mandecki J, Walker DE. Prochlorococcus contributes to new production in the Sargasso Sea deep chlorophyll maximum. Geophys Res Lett. 2007;34:L10604.
Kettler GC, Martiny AC, Huang K, Zucker J, Coleman ML, Rodrigue S, et al. Patterns and implications of gene gain and loss in the evolution of Prochlorococcus. PLoS Genet. 2007;3:2515–28.
Biller SJ, Berube PM, Berta-Thompson JW, Kelly L, Roggensack SE, Awad L, et al. Genomes of diverse isolates of the marine cyanobacterium Prochlorococcus. Sci Data. 2014;1:140034.
Kashtan N, Roggensack SE, Rodrigue S, Thompson JW, Biller SJ, Coe A, et al. Single-cell genomics reveals hundreds of coexisting subpopulations in wild Prochlorococcus. Science. 2014;344:416–20.
Lomas MW, Bronk DA, van den Engh G. Use of flow cytometry to measure biogeochemical rates and processes in the ocean. Ann Rev Mar Sci. 2011;3:537–66.
Musat N, Foster R, Vagner T, Adam B, Kuypers MM. Detecting metabolic activities in single cells, with emphasis on nanoSIMS. FEMS Microbiol Rev. 2012;36:486–511.
Gao D, Huang X, Tao Y. A critical review of NanoSIMS in analysis of microbial metabolic activities at single-cell level. Crit Rev Biotechnol. 2016;36:884–90.
López-Lozano A, Diez J, El Alaoui S, Moreno-Vivián C, García-Fernández JM. Nitrate is reduced by heterotrophic bacteria but not transferred to Prochlorococcus in non-axenic cultures. FEMS Microbiol Ecol. 2002;41:151–60.
Duhamel S, Van Wambeke F, Lefevre D, Benavides M, Bonnet S. Mixotrophic metabolism by natural communities of unicellular cyanobacteria in the western tropical South Pacific Ocean. Environ Microbiol. 2018;20:2743-56.
Worden AZ, Nolan JK, Palenik B. Assessing the dynamics and ecology of marine picophytoplankton: The importance of the eukaryotic component. Limnol Oceanogr. 2004;49:168–79.
Baer SE, Lomas MW, Terpis KX, Mouginot C, Martiny AC. Stoichiometry of Prochlorococcus, Synechococcus, and small eukaryotic populations in the western North Atlantic Ocean. Environ Microbiol. 2017;19:1568–83.
Moutin T, Raimbault P, Poggiale J-C. Primary production in surface waters of the western mediterranean sea. Calculation of daily production. Comptes Rendus l’Académie Des Sciences. 1999;322:651–9.
Harrison WG, Harris LR, Irwin BD. The kinetics of nitrogen utilization in the oceanic mixed layer: Nitrate and ammonium interactions at nanomolar concentrations. Limnol Oceanogr. 1996;41:16–32.
Rees AP, Joint I, Donald KM. Early spring bloom phytoplankton-nutrient dynamics at the Celtic Sea shelf edge. Deep Res Part I Oceanogr Res Pap. 1999;46:483–510.
Ribalet F, Swalwell J, Clayton S, Jiménez V, Sudek S, Lin Y, et al. Light-driven synchrony of Prochlorococcus growth and mortality in the subtropical Pacific gyre. Proc Natl Acad Sci. 2015;112:8008–112.
L’Helguen S, Slawyk G, Le Corre P. Seasonal patterns of urea regeneration by size-fractionated microheterotrophs in well-mixed temperate coastal waters. J Plankton Res. 2005;27:263–70.
Clark DR, Rees AP, Joint I. Ammonium regeneration and nitrification rates in the oligotrophic Atlantic Ocean: implications for new production estimates. Limnol Oceanogr. 2008;53:52–62.
Price NM, Harrison PJ. Urea uptake by Sargasso Sea phytoplankton: saturated and in situ uptake rates. Deep Sea Res Part A Oceanogr Res Pap. 1988;35:1579–93.
Dugdale RC, Goering JJ. Uptake of new and regenerated forms of nitrogen in primary productivity. Limnol Oceanogr. 1967;12:196–206.
Raimbault P, Garcia N. Evidence for efficient regenerated production and dinitrogen fixation in nitrogen-deficient waters of the South Pacific Ocean: impact on new and export production. Biogeosciences. 2008;323–38.
Solomon C, Collier J, Berg G, Glibert P. Role of urea in microbial metabolism in aquatic systems: a biochemical and molecular review. Aquat Microb Ecol. 2010;59:67–88.
Fawcett SE, Lomas MW, Ward BB, Sigman DM. Global Biogeochemical Cycles production in the Sargasso Sea. Glob Biogeochem Cycles. 2014;28:86–102.
Berube PM, Coe A, Roggensack SE, Chisholm SW. Temporal dynamics of Prochlorococcus cells with the potential for nitrate assimilation in the subtropical Atlantic and Pacific oceans. Limnol Oceanogr. 2016;61:482–95.
Massana R. Eukaryotic Picoplankton in Surface Oceans. Annu Rev Microbiol. 2011;65:91–110.
Vaulot D, Eikrem W, Viprey M, Moreau H. The diversity of small eukaryotic phytoplankton (≤3 μm) in marine ecosystems. Vol. 32, FEMS Microbiology Reviews. 2008. p. 795–820.
Ahlgren NA, Rocap G. Diversity and distribution of marine Synechococcus: Multiple gene phylogenies for consensus classification and development of qPCR assays for sensitive measurement of clades in the ocean. Front Microbiol. 2012;3:article 213.
Larkin AA, Blinebry SK, Howes C, Lin Y, Loftus SE, Schmaus CA, et al. Niche partitioning and biogeography of high light adapted Prochlorococcus across taxonomic ranks in the North Pacific. ISME J. 2016;10:1555–67.
Glibert PM, Wilkerson FP, Dugdale RC, Raven JA, Dupont CL, Leavitt PR, et al. Pluses and minuses of ammonium and nitrate uptake and assimilation by phytoplankton and implications for productivity and community composition, with emphasis on nitrogen-enriched conditions. Limnol Oceanogr. 2016;61:165–97.
Kent AG, Dupont CL, Yooseph S, Martiny AC. Global biogeography of Prochlorococcus genome diversity in the surface ocean. ISME J. 2016;10:1856-65.
Ohashi Y, Shi W, Takatani N, Aichi M, Maeda SI, Watanabe S, et al. Regulation of nitrate assimilation in cyanobacteria. J Exp Bot. 2011;62:1411–24.
Kang LK, Gong GC, Wu YH, Chang J. The expression of nitrate transporter genes reveals different nitrogen statuses of dominant diatom groups in the southern East China Sea. Mol Ecol. 2015;24:1374–86.
Song B, Ward BB. Molecular cloning and characterization of high-affinity nitrate transporters in marine phytoplankton. J Phycol. 2007;43:542–52.
Raven JA, Wollenweber B, Handley LL. A comparison of ammonium and nitrate as nitrogen sources for photolithotrophs. New Phytol. 1992;121:19–32.
Quesada A, Hidalgo J, Fernández E. Three Nrt2 genes are differentially regulated in Chlamydomonas reinhardtii. Mol Gen Genet. 1998;258:373–7.
L’Helguen S, Maguer JF, Caradec J. Inhibition kinetics of nitrate uptake by ammonium in size-fractionated oceanic phytoplankton communities: Implications for new production and f-ratio estimates. J Plankton Res. 2008;30:1179–88.
Sunagawa S, Coelho LP, Chaffron S, Kultima JR, Labadie K, Salazar G, et al. Structure and function of the global ocean microbiome. Science. 2015;348:1261359.
Fuhrman JA. Microbial community structure and its functional implications. Nature . 2009;459:193–9.
Ackermann M. A functional perspective on phenotypic heterogeneity in microorganisms. Nat Rev Microbiol. 2015;13:497–508.
Schreiber F, Littmann S, Lavik G, Escrig S, Meibom A, Kuypers MMM, et al. Phenotypic heterogeneity driven by nutrient limitation promotes growth in fluctuating environments. Nat Microbiol. 2016;1:16055.
Bódi Z, Farkas Z, Nevozhay D, Kalapis D, Lázár V, Csörgő B, et al. Phenotypic heterogeneity promotes adaptive evolution. PLOS Biol. 2017;1:e2000644.
Magdanova LA, Golyasnaya NV. Heterogeneity as an adaptive trait of microbial populations. Microbiology . 2013;82:1–10.
Hashimoto M, Nozoe T, Nakaoka H, Okura R, Akiyoshi S, Kaneko K, et al. Noise-driven growth rate gain in clonal cellular populations. Proc Natl Acad Sci. 2016;113:3251–6.
Mohr W, Vagner T, Kuypers MMM, Ackermann M, LaRoche J. Resolution of Conflicting signals at the single-cell level in the regulation of cyanobacterial photosynthesis and nitrogen fixation. PLOS ONE. 2013;8:e66060.
Kopf SH, McGlynn SE, Green-Saxena A, Guan Y, Newman DK, Orphan VJ. Heavy water and 15N labeling with NanoSIMS analysis reveals growth-rate dependent metabolic heterogeneity in chemostats. Environ Microbiol. 2015;17:2542–56.
Finzi-Hart JA, Pett-Ridge J, Weber PK, Popa R, Fallon SJ, Gunderson T, et al. Fixation and fate of C and N in the cyanobacterium Trichodesmium using nanometer-scale secondary ion mass spectrometry. PNAS . 2009;106:6345–50.
Ploug H, Musat N, Adam B, Moraru CL, Lavik G, Vagner T, et al. Carbon and nitrogen fluxes associated with the cyanobacterium Aphanizomenon sp. in the Baltic Sea. ISME J. 2010;4:1215–23.
Foster RA, Sztejrenszus S, Kuypers MM. Measuring carbon and N2 fixation in field populations of colonial and free-living unicellular cyanobacteria using nanometer-scale secondary ion mass spectrometry. Raven J, editor. J Phycol. 2013;49:502–16.
Flores E, Herrero A. Nitrogen assimilation and nitrogen control in cyanobacteria. Biochem Soc Trans. 2005;33:164–7.
Chisholm SW. Prochlorococcus. Curr Biol. 2017;27:R447–8.
Bec B, Husseini-Ratrema J, Collos Y, Souchu P, Vaquer A. Phytoplankton seasonal dynamics in a Mediterranean coastal lagoon: Emphasis on the picoeukaryote community. J Plankton Res. 2005;27:881–94.
Shilova IN, Mills MM, Robidart JC, Turk-Kubo KA, Björkman KM, Kolber Z et al. Differential effects of nitrate, ammonium, and urea as N sources for microbial communities in the North Pacific Ocean. Limnology and Oceanography. 2017;62:2550-74.
Van Mooy BAS, Devol AH. Assessing nutrient limitation of Prochlorococcus in the North Pacific subtropical gyre by using an RNA capture method. Limnol Oceanogr. 2008;53:78–88.
Mahaffey C, Björkman KM, Karl DM. Phytoplankton response to deep seawater nutrient addition in the North Pacific Subtropical Gyre. Mar Ecol Prog Ser. 2012;460:13–34.
Rii YM, Bidigare RR, Church MJ. Differential responses of eukaryotic phytoplankton to nitrogenous nutrients in the North Pacific Subtropical Gyre. Front Mar Sci. 2018;5:article 92.
Berube PM, Biller SJ, Kent AG, Berta-Thompson JW, Roggensack SE, Roache-Johnson KH, et al. Physiology and evolution of nitrate acquisition in Prochlorococcus. ISME J. 2015;9:1195–207.
Acknowledgements
We would like to thank the Schmidt Ocean Institute for providing the vessel to conduct this research and the captain and crew of the R/V Falkor for their help during the cruise. We are grateful to Aimee Neeley (NASA) for providing us with the Chl a data. We also thank Smail Mostefaoui for his assistance with nanoSIMS analyses at the French National Ion MicroProbe Facility hosted by the Muséum National d’Histoire Naturelle (Paris). N. C. and H. B. were supported by the “Laboratoire d’Excellence” LabexMER (ANR-10-LABX-19) and co-funded by a grant from the French government under the program “Investissements d’Avenir”. SD was funded by the National Science Foundation (OCE-1434916 and OCE-1458070). IC was funded through Schmidt Ocean Institute and NASA's PACE mission.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Berthelot, H., Duhamel, S., L’Helguen, S. et al. NanoSIMS single cell analyses reveal the contrasting nitrogen sources for small phytoplankton. ISME J 13, 651–662 (2019). https://doi.org/10.1038/s41396-018-0285-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-018-0285-8
This article is cited by
-
Tracking nitrogen allocation to proteome biosynthesis in a marine microbial community
Nature Microbiology (2023)
-
Single-cell view of deep-sea microbial activity and intracommunity heterogeneity
The ISME Journal (2023)
-
Synechococcus nitrogen gene loss in iron-limited ocean regions
ISME Communications (2023)
-
Tracing carbon and nitrogen microbial assimilation in suspended particles in freshwaters
Biogeochemistry (2023)
-
Effects of cyanate enrichment on growth of natural phytoplankton populations in the subtropical Pacific
Journal of Oceanography (2023)