Abstract
Marine microbial communities are critical for biogeochemical cycles and the productivity of ocean ecosystems. Primary productivity in the surface ocean is constrained by nutrients which are supplied, in part, by mixing with deeper water. Little is known about the time scales, frequency, or impact of mixing on microbial communities. We combined in situ sampling using the Environmental Sample Processor and a small-scale mixing experiment with lower euphotic zone water to determine how individual populations respond to mixing. Transcriptional responses were measured using the MicroTOOLs (Microbiological Targets for Ocean Observing Laboratories) microarray, which targets all three domains of life and viruses. The experiment showed that mixing substantially affects photosynthetic taxa as expected, but surprisingly also showed that populations respond differently to unfiltered deep water which contains particles (organisms and detritus) compared to filtered deep water that only contains nutrients and viruses, pointing to the impact of biological interactions associated with these events. Comparison between experimental and in situ population transcription patterns indicated that manipulated populations can serve as analogs for natural populations, and that natural populations may be frequently or continuously responding to nutrients from deeper waters. Finally, this study also shows that the microarray approach, which is complementary to metatranscriptomic sequencing, is useful for determining the physiological status of in situ microbial communities.
Similar content being viewed by others
Introduction
Marine microbial communities are the base of the oceanic food web. They catalyze diverse chemical transformations that are critical for primary and secondary production and for nutrient and trace element cycling [1,2,3]. Understanding the environmental controls on microbial productivity and community structure is key for predicting current and future marine ecosystem functions. This is challenging because of the complexities of both the environment and the microbial communities. Microbial assemblages comprise diverse species and strains that have different abundances and growth rates, and are controlled by different factors, including nutrient availability and grazing [4,5,6].
The environmental microbial community transcriptome, or metatranscriptome, provides a measure of physiological responses across microbial taxa, including nutrient status, photosynthesis, cell division, and many other processes [7,8,9,10]. Metatranscriptomic sequencing provides relatively unbiased cataloguing of transcripts since it does not rely on previously obtained gene sequences, but is most effective for abundant microorganisms [11,12,13,14]. Alternatively, one can employ a targeted microarray approach. The limitation of the microarray approach is that it can only detect known sequences, but it has the advantages that it can distinguish closely related taxa (by probe design), and that it can detect low abundance transcripts since it is not dependent upon sequencing depth [9]. The MicroTOOLs (Microbiological Targets for Ocean Observing Laboratories) microarray [15] was designed to measure the transcripts of key functional genes from diverse open ocean microorganisms at high taxonomic resolution and to detect transcripts from keystone, but less abundant taxa, such as dinitrogen (N2)-fixing microorganisms (diazotrophs). The MicroTOOLs array has 171,000 gene probes designed from marine environmental sequences, targeting all three domains of life as well as viruses, enabling the analysis of gene expression patterns from phylogenetically diverse plankton.
The North Pacific Subtropical Gyre (NPSG) is a relatively stable oligotrophic environment, with low surface concentrations of fixed inorganic nitrogen (N) and phosphorus (P). Much of our knowledge of the NSPG is based on 30 years of physical, chemical, and biological data collected as part of the Hawai‘i Ocean Time-series (HOT) program at the long-term monitoring site Station ALOHA (A Long-term Oligotrophic Habitat Assessment [16]). Station ALOHA, as a characteristic oligotrophic ocean environment, is inhabited by the cyanobacteria Prochlorococcus and Synechococcus, the photoheterotroph Pelagibacter ubique, and Archaea [16,17,18]. In such environments, an important source of nutrients is advective transport from depth, sometimes caused by mesoscale eddies. High-resolution autonomous profiling has shown that cyclonic eddies can transport nitrate + nitrite into the surface ocean and thereby stimulate primary production, but the delay until the increase in primary production can be detected makes it difficult to link ephemeral nutrient advection to responses [19, 20]. In addition to nutrients supplied to surface waters via vertical advection, diazotrophs also are important for supplying fixed N. Although it is known that there are links between nutrient availability and microbial community structure, it is not well-understood how the individual members of the community respond to shifts in nutrient availability. This study examined the individual responses from key members of the NPSG microbial community to nutrient influxes in two contexts, a shipboard mixing experiment and a suspected mixing event that may have occurred at the confluence of two mesoscale eddies. We hypothesized that experimentally mixed populations can serve as analogs to in situ populations for assessing physiological status in response to ephemeral nutrient advection.
Materials and methods
In situ sampling with the Environmental Sample Processor (ESP)
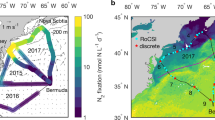
This study was conducted on the BioLINCS cruise from 6 to 21 September 2011 (described in [21]). In situ samples were collected and preserved by the ESP as described in Ottesen et al. [18]. Morning (05:00–06:00) and evening (17:00) ESP samples from September 14, 15, and 16 were analyzed (Fig. 1a).
BioLINCS transect and nutrient profiles. a Sea level anomaly is averaged from AVISO (Archiving, Validation and Interpretation of Satellite Oceanographic data) and MODIS (Moderate Resolution Imaging Spectroradiometer) Aqua, for 6–20 September 2011. The ESP drifter transit during the BioLINCS cruise is shown. The 14-16 September 2011 transit is depicted in white and shows that the ESP sampled between two anticylonic eddies on these dates. Station ALOHA is denoted by the black disc. (Figure kindly provided by John Ryan.) b Nitrite, chlorophyll and oxygen concentrations vs. depth for the entire ESP transit (7–18 September 2011) reveal an influx of nitrite from the nitrite maximum at 130 m, with decreasing concentration to 25 m depth at mid-day on 15 September. There was no clear stimulation of photosynthesis, using chlorophyll or oxygen as proxies, into the shallow mixed layer during this influx. The 24 m ESP depth is indicated by horizontal dashed lines. Locations at mid-day on 14, 15 and 16 September are indicated by vertical dashed lines, as is the location of samples collected on 12 September for mixing experiments. Please note the different scales
Mixing experiment
The mixing experiment simulated natural mixing of surface (25 m) and deep (130 m) waters in the NPSG (Fig. 2). We stress that “deep” is shorthand for “lower euphotic zone” and is shallower and lower in nutrients than deep water used in previous mixing experiments [13, 20, 22,23,24] in order to simulate mixing via small-scale eddy-induced nutrient advection [19]. From each depth, 40 L water samples were collected at 3:30 on 12 September (Fig. 1). Filtered surface water (FSW) and filtered deep water (FDW) were prepared with 0.2 μm Sterivex™ filters (Millipore, Billerica, MA, USA), which were changed every ~5 L to avoid organic nutrient contamination due to cell lysis on the filter. However, it is also possible that FDW contained organic matter that resulted from the breakage of cells during the filtration. Unfiltered water (Surf, Deep [communities] and UDW [unfiltered deep water treatments] in Fig. 2) was prepared with 64 μm mesh to remove large organisms and particles. Water samples were stored in the dark in a trace metal clean carboy prior to the mixing experiment. Mixed samples of each type had one replicate (n = 2). Each mixed sample had a total volume of 20 L with 80% and 20% of each water type as shown in Fig. 2. Mixed samples were incubated in Cubitainers® within deck sea water flow-through incubators for 24 h at 25–26 °C and 14% PAR to mimic surface conditions.
Mixing experiment design. Shipboard experiments mixed filtered and unfiltered waters as indicated in the ratio 80:20 by volume and incubated them for 24 h at 14% PAR and 25–26 °C. Up- and down-regulated genes were identified by comparing samples as follows: Surf_FDW vs. Surf_Cnt (effects on surface community microbes due to nutrients); Surf_UDW vs. Surf_FDW (effects on surface community microbes due only to deep water microbes or particles); Deep_Cnt vs. Deep_FSW (effects on deep community microbes due to nutrient depletion and increased temperature and PAR; Supplementary Information). FSW = filtered surface water (25 m), FDW = filtered deep water (130 m); UDW = unfiltered deep water (equivalent to “Deep”). Labels above each sample graphic indicate which water was mixed
Nutrient concentrations were not measured from these incubations, but we assume that concentrations measured from the mid-day CTD cast at the same site (13:30 on 12 September) were similar. The nutrient concentrations from 25 m (σT = 23.18 vs. 23.17 during surface experimental water recovery) and 130 m (σT = 24.44 vs. 24.58 during deep experimental water recovery) depths on that CTD cast were measured as in Karl and Tien [25] and Dore and Karl [26] and used to estimate nutrient concentrations for the incubations (Table S1).
Acetylene reduction assays were performed on water collected from each treatment at 24 h and normalized to a 0.22 µm filtered seawater control incubation, as described previously [27]; Supplementary Information).
A volume of 4 L of seawater was collected for DNA and RNA from each carboy at 05:00, at the start of the experiment and 24 h later. Water was filtered using 0.2 μm Sterivex™ cartridges, and filters were immediately flash frozen in liquid nitrogen to preserve for analysis after the cruise. Samples were sent to UC Santa Cruz on dry ice and stored at −80 °C.
Nucleic acid extraction and preparation for analysis
DNA was extracted from filters using a modified Qiagen DNeasy Plant Kit (Valencia, California, USA) protocol as described in [28]. V3-V4 hypervariable region tag sequences were obtained using Illumina MiSeq sequencing of samples using a dual PCR approach [29] at the DNA Service Facility at the University of Chicago, Illinois. RNA was extracted using the Ambion RiboPure RNA purification kit (Life Technologies, Grand Island, NY, USA) with the addition of a bead-beating step during TRI Reagent extraction as described in Shilova et al. [15]. DNA was digested using the RNase-Free DNase Kit (Qiagen) according to the manufacturer’s protocol, and RNA quality and quantity was evaluated using the Agilent BioAnalyzer RNA Nano Kit (Agilent Technologies, Santa Clara, CA, USA) and Qiagen Qubit. All samples with an RNA Integrity Number greater than 9 were processed for microarray analyses (>7 RIN was recommended by the microarray facility). Microarray hybridization was performed at the Roy J. Carver Center for Genomics, The University of Iowa, USA.
16S rRNA gene diversity
Community composition in the mixing experiment samples was based on 16S rRNA V3-V4 hypervariable region tag sequences as described in Shilova et al. [32]. Operational taxonomic units (OTUs) were defined by clustering V3-V4 sequences at 97% nucleotide identity. The community composition was analyzed using the phyloseq R package [30] in R ([31]; www.R-project.org). 16S rRNA gene oligotyping for Prochlorococcus was done as described in Shilova et al. [32].
MicroTOOLs microarray
All microarray analyses were done using the MicroTOOLs R package (ver. 1.0; available at https://www.jzehrlab.com/microtools). The transcription values for each gene were obtained by robust multi-array average of hybridization values for all probes and quantile normalization across all samples [33, 34]. Within each sample, gene transcripts were detected if the gene had intensity z-scores > 3 s.d. above a background based on 16 non-marine negative control genes (Supplementary Information). On average 42% (~8744) of the genes were detected per sample. Differentially expressed (DE) genes were identified by comparing: Surf_Cnt versus Surf_FDW; Surf_FDW versus Surf_UDW; Deep_Cnt versus Deep_FSW (Fig. 2). A gene was DE if, in any of the comparisons, it had a >1.5-fold change and Benjamini–Hochberg adjusted p-value < 0.05, calculated relative to gene linear models created with the lmFit and eBayes functions in the limma R package (ver. 3.22.7; ref. [35]). The MicroTOOLs microarray data used in this study was deposited at NCBI GEO under accession GSE109218.
Results and discussion
In this study we used a floating Environmental Sample Processor (the ESP drifter) to collect and preserve microbes in situ. We also designed a deep water (130 m) addition experiment to measure microbial transcriptional responses as analogs to in situ populations, to see if samples collected from nature can inform us about aperiodic nutrient additions. We then compared the organisms and transcription patterns of in situ populations to those in experimental manipulations to determine the physiological status of in situ populations with respect to the effects of recent nutrient advection. For the experimental populations, we examined the separate effects of mixing deep water particles and the effects of dissolved nutrients. In the following sections, we first describe the physico-chemical environment, which includes evidence of a possible mixing event during in situ sampling (1.), then show that there were species-specific transcription responses to the event (2.). We then discuss the impact of mixing with deeper water on community composition (3.) and transcriptional patterns (4.), highlighting the differential effects of the particles vs. nutrients (and viruses) (5.). We examine the similarities between transcription patterns from experimental mixing relative to the in situ populations (6.). Finally, we discuss the performance of the microarray for assessing the status of natural microbial populations (7.).
1. Oceanographic conditions during ESP sampling
From 7 to 18 September 2011, the Environmental Sample Processor (ESP drifter) filtered and preserved cells in situ at 24 m depth every ~5 h as described in Ottesen et al. [18]. The ESP moved with a Lagrangian drift, first northward on the western edge of an anticyclonic eddy, then it slowed and drifted westward on 13 September toward the southern edge of a second anticyclonic eddy [21] (Fig. 1a). Water collected on 12 September for mixing experiments had surface nutrient concentrations typical for the NPSG in the late summer [21, 36] (Figs. 1b, 2). Nutrients increased sharply during the westward transit between the two eddies while ESP sampling occurred (05:00–06:00 and 17:00, 14–16 September), possibly due to mixing resulting from the vertical advection of deep water between 14 and 15 September (Fig. 1b, S1; Supplementary Information).
2. The in situ transcriptional response to mixing
Collectively, transcripts from a total of 12,745 genes (from ~21,000 total on the microarray) were detected in the in situ ESP samples. We categorized the genes with the most abundant transcripts (3923 genes total) by phylogeny, and observed distinct transcription patterns (Fig. 3a; Table S2). Haptophytes had the highest transcript levels in every sample, except for the evening samples collected on 15 and 16 September. This pattern was shared by other eukaryotic phytoplankton groups (eight groups, highlighted in Fig. 3a) which are represented on the array almost exclusively by RuBisCO large subunit genes (rbcL) [15]. Aside from the high transcription observed on the evening of 14 September, the rbcL transcription patterns from eukaryotic phytoplankton are consistent with diel transcription of RuBisCO by these groups [37, 38].
ESP samples differed with respect to which genes were highly transcribed. A total of 3923 genes, with transcription levels in the top quartile of at least two samples (Table S2), were aggregated by the categories phylogroup (a) and metabolic or physiological process (b). Each heat map cell shows the mean of the gene normalized transcription intensities (Materials and methods) for a category (row) and sample without row or column scaling. Within each sample, the most highly transcribed categories are numbered 1 through 5. In a, the green brace denotes eukaryotic phytoplankton. In b, the gene categories in the cluster denoted with a * had large transcript level changes on 15 September likely after the hypothesized advection event
We also categorized the 3923 genes with abundant transcripts by metabolic and physiological processes (Fig. 3b; Table S2). In all samples, the first or second highest transcript levels came from energy metabolism genes, mostly photosynthesis genes from picocyanobacteria (1141 genes), viruses (326 genes) or diazotrophic cyanobacteria (32 genes). Nitrogen metabolism genes (417 total) were also highly transcribed. Among these, urtA, which encodes the urea-binding protein of the ABC-transporter, and amt, which encodes an ammonium transporter, had the first or second highest abundances in most samples (Fig. S2), attributable mainly to picocyanobacteria (144 urtA genes, 53 amt genes; Table S2).
Transcription patterns observed for Prochlorococcus and viruses suggested that a mixing event occurred between 14 and 15 September. The event, defined by a spike in nutrient concentrations (described in (1.)), coincided with decreases in transcript levels of a cluster of genes shown in Fig. 3b. This cluster has a high-to-low pattern comprised mainly of transcripts from high-light-adapted (HL) Prochlorococcus, in particular genes associated with carbon, iron, nitrogen, and phosphorus (Fig. 3b, S3; Supplementary Information). Among them was the phosphorus stress gene pstS, which encodes a high affinity phosphate-binding ABC transporter (Fig. S4; Table S2). Prochlorococcus pstS transcripts peaked in the morning and were reduced by the evening of 15 September (Fig. S4) in contrast to the diel pstS profile observed in cultures by Zinser et al. [39]. The reduced pstS transcripts indicate either abundance changes of Prochlorococcus cells expressing pstS or an input of phosphate, or both. Indeed, phosphate concentrations measured on 15 and 16 September (13:30) were high relative to historic summer phosphate concentrations at Station ALOHA, and the phosphate concentration on 16 September is the third highest seen near Station ALOHA during summer stratification since 1989 [21]. Increases in viral transcript abundances that occurred on 16 September (Figs. 3a, 4) are consistent with increased viral activity associated with increased nutrient availability [23]. The lag in the viral response after nutrient influx may correspond to a slow net growth of Prochlorococcus [40].
Hierarchical clustering of metatranscriptomic patterns reveals strong clustering among replicates and among treatments, supporting community composition analysis. Genes (rows) are categorized by function and genus, noted on the left. Three main gene clusters and several subclusters (shaded, in main cluster 1) are defined based on similarity of transcriptional patterns across samples, noted on the right. Heat map colors reflect the log2 of transcript levels from low (blue) to high (red), or are white if the organism associated with the gene was absent from a sample. FSW = filtered surface water, FDW = filtered deep water, UDW = unfiltered deep water, Cnt = control. Sample clusters have 100% bootstrap support unless otherwise indicated
Prochlorococcus and Synechococcus had distinct transcription changes following the advection event. HL Prochlorococcus peaked in the morning of 15 September but decreased by that evening as described above. In contrast, Synechococcus genes, mainly from RCC307 and JA-2-3B’a(2-13)-like strains, had their highest transcript abundances in the evening of 16 September (Figs. 4 and S5), as did energy metabolism genes from low-light-adapted (LL) Prochlorococcus (Fig. S3; Supplementary Information).
Responses from major N2-fixing (diazotrophic) cyanobacteria in the NPSG were measured by detecting nifH (which encodes the iron subunit of nitrogenase, the key enzyme for the fixation of dinitrogen to ammonia) transcripts. We detected nifH transcripts from the unicellular cyanobacteria Crocosphaera and Candidatus Atelocyanobacterium thalassa (UCYN-A, a symbiont of a haptophyte alga), as well as the filamentous, colony-forming cyanobacterium Trichodesmium (Figs. S2, S6) in all samples. For Crocosphaera and UCYN-A, nifH transcripts were high in the morning and evening samples collected on 14 September, before the hypothesized advection event that occurred between 14 and 15 September, but decreased after the event (Figs. S2, S6; Table S2).
3. Community composition after deep water mixing
The mixing experiment (Figs. 1, 2; Table S1; Supplementary Information) was designed to determine how surface mixed-layer microbial communities respond to the biological and chemical effects of vertical mixing with lower euphotic zone waters. After mixing with 20% “deep” (130 m) water (DW, Table S1), nutrient concentrations increased slightly, but the surface community never had nitrite + nitrate concentrations >87 nM in mixing experiments, or phosphate concentrations greater than 46 nM (increases of 1.3× and 1.1×, respectively). These increases are very small relative to concentrations from previous deep water mixing experiments in the region (using 700 m water that resulted in average increases of 353.7× and 5.9× background for nitrate and phosphate, respectively [20]).
In addition to the increases in nutrient concentrations, surface communities were likely exposed to different virus populations in FSW and FDW treatments. This is especially important to keep in mind given new evidence for light-induced activity and transcription of auxillary metabolic genes involved in photosynthesis from cyanophages [41, 42], as the mixing incubations were performed at the 25 m light level. Differences that might have been introduced as part of the filtration process also include increases in organic matter (in all controls and treatments with the exception of the Surf_UDW mixing experiment (Fig. 2)) and changes in dissolved gases. These parameters were unmeasured during this study but their potential effects were considered while interpreting the metatranscriptomic data.
Major phylogroups shifted in relative abundances after simulated deep water mixing. Using 16S rRNA gene sequencing, and clustering the sequences at >97% nucleotide identity, a total of 566 OTUs were identified with 100–200 unique OTUs per sample on average. At the start of the experiment (T0), the surface community control (Surf_Cnt) was dominated by Pelagibacteraceae and HL Prochlorococcus with relative abundances of 38% and 31%, respectively (Fig. 5a; Table S3; Supplementary Information). Synechococcus was rare (<0.3%), and plastids recovered from eukaryotic microorganisms comprised 4.8% of all sequences (0.8% stramenopiles and 4.0% haptophytes; Table S3). Twenty-four hours of incubation under low-nutrient control conditions (Surf_Cnt, T24) resulted in large decreases in relative abundances for Pelagibacteraceae and HL Prochlorococcus (22% and 1.4%, respectively; Table S3). For Prochlorococcus, incubation with filtered deep water (FDW) resulted in a much smaller decrease in relative abundance (25% of all sequences in Surf_FDW at T24; Table S3). Incubation with either filtered or unfiltered deep water (UDW) resulted in similar shifts in the surface microbial community composition by T24 (Fig. 5b), especially for Gamma- and Alphaproteobacteria and Cyanobacteria (Fig. 5a; Table S3).
Changes in microbial community composition in the surface and deep samples in response to mixing. Composition is based on 16S rRNA gene copy numbers. a Relative abundances of microbial phyla in each sample (color-coded). b Principal coordinate analysis on Bray–Curtis dissimilarity index showed clustering of samples by community origin and treatment. Relative abundance for each OTU was calculated as the number of nucleotide reads assigned to the OTU divided by the number of reads in the sample (e.g., Surf_FDW at T24). For each phylum, the mean of the relative abundances of the OTUs from the phylum was calculated
4. Transcriptional response to deep water mixing
The addition of filtered deep water resulted in taxon-specific transcription changes for genes associated with photosynthesis and responses to key nutrients (Surf_FDW vs. Surf_Cnt in Figs. 2 and 6a,c, S7; Table S2). Unless stated otherwise, results that follow indicate genes only if they were differentially expressed (DE, 1.5-fold change that was significant [p < 0.05]). Transcript levels in response to FDW addition increased at least 1.5-fold over the control for a total of 3846 genes, which were mainly from Prochlorococcus and associated with photosynthesis, transcription, pigment synthesis, and the metabolism of nitrogen, phosphorus, and iron (Fig. 6a,c; Table S2). Photosynthesis genes from phage and Synechococcus (especially RCC307) had increased transcript levels, as did the RuBisCO large subunit genes (rbcL) from eukaryotes and proteorhodopsin genes (bop) from Pelagibacter ubique. For all of these phylogroups (except phage), 16S rRNA relative abundances increased in the FDW treatment compared to the control at 24 h (Table S3), so community shifts likely explain some of the transcript increases, but not all. After normalizing for relative abundance changes, transcript level increases were still observed for Prochlorococcus and Synechococcus (mainly from photosynthesis genes), haptophytes (rbcL), and P. ubique (bop).
Metatranscriptomes from the surface communities were distinct in each treatment. a A volcano plot of Surf_FDW vs. Surf_Cnt shows that transcript levels changed in response to filtered deep water (FDW), i.e., due to changes in chemistry or viruses associated with deep water mixing. Transcript levels increased for Prochlorococcus photosystem (psa and psb) and urea transport (urtA) genes, and decreased for Synechococcus photosystem, transcription, carbon and nitrogen metabolism genes (psa, sigAII, rbcL, and glnA) in response to the addition of FDW. The phage photosystem genes also had increased transcript levels in response to FDW. Gray lines are at log2 1.5, the minimum fold change at which gene models were checked for differential expression (p < 0.05). b A volcano plot of Surf_UDW vs. Surf_FDW reveals transcript level changes due to biological interactions associated with mixing of unfiltered deep water (UDW) with the surface microbial community. Transcript levels among eukaryotic phytoplankton decreased due to biological interactions (depicted by rbcL genes associated with dominant eukaryotic groups), while Prochlorococcus transcript levels increased for P and N stress genes (pstS, phoH and urtA). c Metabolic processes of genes (in %, Y-axis) that changed at least 1.5-fold in response to FDW shown by phylogroup. For example, in response to FDW addition, nitrogen genes were 26% of the 133 diazotrophic cyanobacteria genes that decreased at least 1.5-fold. d As in c but shows the responses to UDW. Syn = Synechococcus, Pro = Prochlorococcus, euks = eukaryotes, diazos = diazotrophic (N2-fixing) cyanobacteria
Transcript levels decreased by at least 1.5-fold for 4972 genes as a result of FDW addition to the surface community (Surf_FDW vs. Surf_Cnt in Figs. 2 and 6a,c; Table S2). Transcripts with decreased levels were mainly from Synechococcus and Prochlorococcus and associated with the metabolism of carbon or key nutrients (nitrogen, phosphorus, iron), or with energy. Among the eukaryotes, decreases occurred mostly for diatoms and especially for nitrate reductase genes (NR). For each phylogroup, transcript level decreases occurred despite relative abundance increases in the FDW treatment compared to the control at 24 h (Table S3). Therefore, we believe these decreases are downregulation of genes.
The results show that changes in chemistry, and perhaps viruses, following FDW mixing elicit distinct responses from the surface microbial community by 24 h. Similar to a previous NPSG mixing experiment by Shi et al. [23], we observed increases in photosystem transcripts from Prochlorococcus (mostly HL clades) and cyanophages, but in our study it was in response to smaller nutrient increases and without deep water microbes or particles (Table S1). We also observed increased levels of Synechococcus transcripts for photosystem and carbon fixation genes and eukaryotic phytoplankton carbon fixation genes (rbcL were the most abundant genes on the array for this group; Figs. 4, 6). The significant changes in the transcription of photosystem and carbon fixation genes are expected responses to nutrient availability [15, 43].
In contrast to the responses from the above taxa, N2-fixing cyanobacteria had decreased transcript levels in response to FDW addition, in particular for N2 fixation genes. Significant nifH transcript abundance decreases were detected for UCYN-A and Trichodesmium, which is consistent with inhibition of N2 fixation by the presence of fixed N (as in Dekaezemacker and Bonnet [44]), or from a reduced competitive advantage compared to other organisms that can respond to the fixed nitrogen. For Trichodesmium, decreases also occurred for genes associated with energy (cox, ndh), photosynthesis, and the regulation of transcription (rpoA, sigD) and translation (rpsS16, rbsS11). Crocosphaera genes also had decreased transcript abundances in response to FDW addition, but only significantly for genes encoding a Zn2+-binding alkaline phosphatase (phoA) and a hypothetical protein homologous to Tery_2900 (which has a similar expression pattern to nifH in Trichodesmium IMS101 [unpublished]).
Our mixing experiment demonstrated that even relatively small nutrient increases (compared to previous mixing experiments i.e. [20, 23]; Table S1) can affect the surface microbial community structure and transcriptional patterns. Remarkably, different nutrient response patterns were apparent among the dominant phytoplankton taxa (Synechococcus vs. Prochlorococcus) as well as among the N2-fixing taxa (Figs. 4 and 6a,c, S7; Table S2). To our knowledge, this is the first documentation of these distinct ecological strategies in response to small-scale mixing.
5. Differential effects of particles and nutrients
The surface microbial community responded differently to unfiltered and filtered deep water additions (Surf_UDW vs. Surf_FDW in Figs. 2 and 6b,d, S7; Table S2). We observed 2809 genes with transcripts that increased at least 1.5-fold relative to levels seen with the FDW addition (Fig. 6b,d; Table S2). These increases were not likely due to differences in community composition between UDW and FDW treatments at 24 h, which were usually small (Fig. 5a; Table S3). Prochlorococcus genes associated with energy or the metabolism of carbon, nitrogen, phosphorus (pstS, phoH) and iron increased after the UDW addition (Fig. 6b,d; Table S2). For Synechococcus, few genes had transcript level increases (relative to FDW levels) in response to UDW (Fig. 6b,d). Some genes associated with nitrogen (urtA, nrtP) and iron (isiB) decreased 1.5-fold with FDW and increased 1.5-fold with UDW (Table S2), but the increases were not significant (DE).
A total of 1748 genes had decreased transcript levels in the UDW addition relative to their levels in the FDW treatment by 24 h (Surf_UDW vs. Surf_FDW in Figs. 2 and 6b,d; Table S2). The similar community compositions among these samples (Fig. 5a; Table S3) suggest the decreases are mainly due to down-regulated genes. Decreases occurred for photosynthesis genes from Prochlorococcus, phage, and Synechococcus, and for proteorhodopsin genes from SAR11 strains (bop, not DE). For 301 of the 337 eukaryotic rbcL genes detected by the array, microbes or particles in UDW led to decreased transcripts relative to the FDW addition, most often for stramenopiles, haptophytes, and chrysophytes (e.g., Ochromonas in Fig. 6b; only DE for chrysophytes). Moreover, for 226 of these rbcL genes, the transcript levels in the UDW treatments were even lower than in the controls (Table S2), despite slightly higher relative abundances for stramenopiles and haptophytes in UDW compared to controls at 24 h (Table S3). For some genes associated with the metabolism of limiting nutrients, treatment with UDW resulted in larger decreases compared to treatment with FDW (i.e., a 1.5-fold decrease was observed both in the UDW vs. FDW treatments, and in FDW vs. the control). These included some Synechococcus nitrogen metabolism genes, Prochlorococcus iron metabolism genes, and diatom genes associated with nitrogen, iron, silica, and phosphorus. For N2-fixing cyanobacteria, few genes had decreased transcript abundances with UDW addition. These included transcript levels of UCYN-A photosystem genes which had increased with FDW addition.
Addition of deep water with associated microbes and particles (UDW) to the surface water community resulted in different effects on transcription than were observed with FDW addition alone, in particular decreases in picocyanobacteria photosystem genes and eukaryote RuBisCO (rbcL) (Fig. 6b,d, S7; Table S2). This may be partly explained by competition for nutrients by deep water microbial community microbes. Indeed, we detected transcripts from three genera of ammonia-oxidizing archaea only in the deep water samples, and previous data suggest they can out-compete oligotrophic diatoms for ammonium [45]. Competition could also have resulted from diatoms, prymnesiophytes, and pelagophytes, which can be abundant in deep euphotic water from the NPSG [46]. Diatoms and prymnesiophytes have also been observed to increase transcription of genes associated with growth when stimulated with deep water [24]. The addition of UDW resulted in significant (DE) increases for Prochlorococcus genes associated with light stress (pmm1359), phosphorus stress (phoH, pstS), and nitrogen or sulfur metabolism (metC). Genes associated with key nutrients (carbon, nitrogen, phosphorus) often increased with UDW, and often even more than they had with FDW (Fig. 6; Table S2). In contrast, Synechococcus nutrient-associated genes (carbon, nitrogen, phosphorus, iron) often decreased with FDW and even more so in the presence of UDW microbes or particles (Fig. 6; Table S2). Thus, biological interactions such as competition for nutrients between the deep and surface water microbial communities intensifies or weakens transcription of nutrient metabolism genes in Prochlorococcus and Synechococcus, respectively, depending on their genetics and physiology. These distinct nutrient utilization strategies among dominant open ocean cyanobacteria clades were unexpected. Different strategies among Synechococcus and Prochlorococcus will likely determine how changes associated with projected increases in stratification will impact the activities and composition of these photosynthetic communities.
N2-fixing cyanobacteria had distinct responses to deep water microbes and particles. For Trichodesmium and Crocosphaera, many transcripts from genes associated with energy metabolism, photosynthesis, or nitrogen metabolism decreased with the addition of FDW but increased in response to UDW (Surf_FDW vs Surf_UDW; Fig. S8; Table S2). Curiously, this reversal did not occur for UCYN-A or Richelia intracellularis, which are both symbionts with eukaryotic hosts (Fig. S8; Table S2). Trichodesmium nifH transcript levels in the UDW addition were greater than in the FDW addition, and intriguingly, rates of gross N2 fixation were 3–5 higher with UDW compared to FDW additions (Table S1). Thus, the higher rates of N2 fixation (Table S1) in the UDW additions compared to FDW were likely due to Trichodesmium. This suggests a net positive effect of microbial interactions or particles on N2 fixation, but further investigation of taxa-specific effects is needed.
The differential effect of unfiltered water relative to filtered water additions is intriguing, since it suggests several important mechanisms involved in shaping microbial communities and activities. The deeper water microorganisms may secrete substrates that have positive or negative effects on surface microbes [47,48,49,50], or may compete with surface microbes for available nutrients [45, 51]. Non-living particles, or detritus, may be sources of inorganic or organic nutrients, or may provide substrates for attachment. The effect of particles on surface microbial communities is an interesting result that deserves further study.
6. Comparison of in situ transcription to transcription in experiments
Transcription patterns from in situ samples were remarkably similar to those from the deep water mixing experiments. The samples were compared with respect to 2274 DE genes that responded to added deep water (DW, either FDW or UDW) or filtered surface water (FSW) by at least 1.5-fold (p < 0.05) relative to controls at 24 h (Materials and methods; Fig. 2; Table S2). Most (2218) of the DE genes were in response to FDW relative to the surface control (Surf_Cnt), 11 were in response to UDW relative to FDW, and 60 were in response to FSW relative to the deep control (Deep_Cnt vs. Deep_FSW in Supplementary Information).
The transcription patterns for the 2274 DE genes formed highly reproducible hierarchical clusters (Fig. 4). ESP samples (from 24 m depth) clustered first by time of day, likely due in part to diel changes in transcription, and then clustered with the surface samples to which DW was added (Fig. 4). Non-metric multidimensional scaling (NMDS) with all 13,638 detected genes corroborated the similarity of in situ and experimental surface water metatranscriptomes suggested by the 2274 DE genes alone (Figs. 7 and 4, respectively).
In situ and mixing experiment samples had similar metatranscriptomes when analyzed with non-metric multidimensional scaling (NMDS). In situ samples from 24 m were archived by the ESP at 5:00 and 17:00 on 14, 15, and 16 September. Surface community mixing experiment samples mixed 25 m water with 130 m deep water (FDW or UDW), all collected at 3:30 on 12 September. NMDS shows consistent clustering of ESP samples with surface community mixing experiment samples (Surf_FDW and Surf_UDW), and separate from the surface control (Surf_Cnt) and deep community samples (Deep_Cnt, Deep_FSW)
The gene transcription patterns (by row in Fig. 4) fell into three clusters that corresponded mainly to phylogenetic groups. For example, cluster 3 (312 genes) had mostly Synechococcus genes associated with nitrogen and phosphorus metabolism that had high transcript abundances in the control (Surf_Cnt) but low abundances in the DW treatments (Surf_FDW, Surf_UDW) and ESP samples. Several Trichodesmium genes associated with photosynthesis, transcription, translation, and responses to nutrients (nitrogen, iron, carbon) were also in this cluster (bottom rows) but remained at high levels following the DW addition.
Prochlorococcus genes in cluster 1 (1912 genes) had opposite patterns to Synechococcus genes in cluster 3. Cluster 1 included photosynthesis (energy) and N genes that had low transcript abundances in the control but high abundances in the DW and ESP samples (Fig. 4). Interestingly, most of the nitrogen and other nutrient-associated genes in cluster 1 fell within a subcluster of 456 genes (darker yellow), mainly from HL (66%) or unknown (31%) clades of Prochlorococcus. With respect to this subcluster, the ESP samples more closely resembled the UDW treatment (with deep microbes) than the FDW treatment (without; Fig. 4). The similarity was strongly corroborated by bootstrapping (Fig. S9). Altogether these observations show that DW nutrients induced HL Prochlorococcus to increase transcription of genes associated with photosynthesis and nutrient metabolism, but that nutrient metabolism gene increases were affected by the presence of deep water microbes or particles.
Phage photosystem genes had highest transcript levels in the DW treatments (green subclusters of cluster 1 in Fig. 4). They also had high transcript levels in the ESP sample taken 16 September at 17:00 after the hypothesized advection event, compared to all other ESP samples.
In cluster 2, Pelagibacter proteorhodopsin genes (bop) were low in deep controls (Deep_Cnt) but increased after FSW addition (Deep_FSW), in response to nutrient dilution, increased PAR, and/or increased temperature (Fig. 4).
Our results show that in situ metatranscriptomic patterns reflect responses to changes in nutrient availability. We observed similar patterns in the ESP samples and the mixing experiment samples to which FDW or UDW were added. The similarity was corroborated by sample clusters in both the NMDS analysis, which used all detected genes (Fig. 7), and in the differential expression analysis, which used only DE genes (Fig. 4). Remarkably, transcription patterns in the ESP samples more closely resembled those of experimental samples with added UDW than with FDW (Fig. 7). This observation is also supported by the subcluster of 456 Prochlorococcus nutrient genes (Figs. 4, S9). It is interesting to note that bootstrapping with just the 456 nutrient genes produced clusters that included the morning and evening ESP samples (Fig. S9). This suggests that environmental changes (not diel cycles) led to the differential expression of these 456 Prochlorococcus nutrient genes. Moreover, the similarities between the experimental and in situ transcriptional patterns (Figs. 4, 7) suggest that comparing experimental responses to FDW vs. UDW can help us gauge the extent of chemical vs. biological impacts of mixing of deep water with surface water in situ.
These results (1–6) show that the gene expression patterns in natural populations may reflect the recent history of nutrient entrainment by mixing. They also show that N2-fixing taxa and the dominant Prochlorococcus and Synechococcus taxa respond differently to nutrient availability and competition, indicative of niche partitioning, and that there is an effect of the particles (living and or dead) on gene expression of surface communities during mixing. Finally, the experiments show that gene expression in the natural communities sampled in situ are most similar to the experiments where deeper water was added, suggesting that natural communities in this region are frequently or continuously experiencing the effects of nutrient enrichment associated with mixing.
7. Performance of the microarray and interpretations relative to past studies
Natural communities collected in situ with the ESP and analyzed with the MicroTOOLs microarray were comprised of typical open ocean microbes found in surface waters: haptophytes, stramenopiles, Euryarchaeota, SAR11, Prochlorococcus (HLII clade mostly), and viruses. The high transcription of picocyanobacteria urtA detected in the ESP samples (Figs. S2, S3, S5) and in metatranscriptomic studies (e.g., Shi et al. [52]) supports the active utilization of urea by Prochlorococcus seen in previous studies [32]. Diel differences in transcription (reported in [18, 53]) may partly explain why the ESP samples clustered by morning or evening collection time (Fig. 4), as well as the evening transcription peaks of Synechococcus amt (Fig. S5) [39] and the morning transcription peaks of eukaryote RuBisCO (Fig. 3) [37, 38]. Notably, some of the diel patterns we observed were also seen using an approach called MAGC [9], which performed an in silico hybridization of next-generation sequencing reads from this same ESP deployment to MicroTOOLs probes. The high transcription of eukaryote RuBisCO on the evening of 14 September (Fig. 3) and of Prochlorococcus pstS on 15 September (Fig. S4) point to potential environmental responses that are distinct from diel transcriptional patterns. Heterotrophic microbes (e.g., Roseobacter, SAR116, SAR86, and SAR324 reported by Ottesen et al. [18]) were rarely, if at all, detected because they have few genes on the array. However, for the taxa and genes currently targeted, the transcription patterns observed with MicroTOOLs were generally similar to those seen in metatranscriptomic sequencing studies [23].
In addition to these findings, we found that the MicroTOOLs array detected environmental responses even from rare taxa, including transcripts from low-abundance organisms present in the ESP samples. Transcripts from Synechococcus (0.1–1.3% of community based on 16S rRNA gene relative abundances) derived from a mix of strains typically found in warm oligotrophic and transitional temperature waters [54]. Transcripts were also detected from strains for which the closest known reference sequences were from Clades I and IV (cold, high nutrient) and JA-2-3B’a(2–13) (hot-spring, but based on probes designed from sequences from Botany Bay, Australia). While JA-2-3B’a(2–13) was not recovered from 16S rRNA sequences in the experiments, transcripts were detected for multiple genes within these clades, lending confidence to the findings and underscoring the value of identifying novel strains of marine Synechococcus in situ (e.g., Farrant et al. [55]). N2-fixing microorganisms are critical components of NPSG microbial communities, but are often present at abundances several orders of magnitude lower than the dominant microbes, which include Prochlorococcus and SAR11. Gene expression from multiple N2-fixing taxa were detected using the array, and detected microbes were consistent with previous qPCR results from the same transect [21]. In contrast, a previous Illumina-based metatranscriptomic study from the same transect had too few reads from N2-fixers and Synechococcus for comparative analysis [18]. Thus, for the less abundant members of the NPSG, the sensitivity of the MicroTOOLs microarray enabled a quantitative comparison of transcription patterns that can be challenging with other approaches.
Summary
Modification of gene transcription is the first response of organisms to environmental changes and thus can reveal taxa-specific sensitivities to conditions, including nutrient availability. This study revealed differential sensitivities among critical members of the NPSG surface microbial community—Prochlorococcus, Synechococcus, eukaryotic phytoplankton, N2-fixing cyanobacteria, and viruses—to deep water mixing, with distinct effects due to deep water nutrients versus deep water particles and organisms. Transcriptional patterns in the mixing experiments were repeatable and, for abundant taxa, were consistent with patterns seen in previous studies. For example, the addition of deep water (filtered or not) stimulated transcription of viral photosynthetic genes. However, the sensitivity of MicroTOOLs also enabled us to detect transcriptional responses from less abundant, but key, members. A surprising example was the differential responses among N2-fixers to nutrient increases (an overall decrease in transcription) versus in the presence of deep water particles and organisms (when Trichodesmium and Crocosphaera transcription returned to nearly the control levels). It is well recognized that biological interactions impact biogeochemistry in the open ocean [56,57,58,59,60], and our mixing experiment points to the impact of environmental perturbations (chemistry, biology) in dictating which organisms thrive. Most importantly, the metatranscriptomic patterns from samples collected in situ show that, though undetectable using biogeochemical proxies for phytoplankton responses (Fig. 1), small-scale mixing events can stimulate the photosynthetic community at the periphery of eddies during the stratified summer months in the NPSG.
References
DeLong EF, Karl DM. Genomic perspectives in microbial oceanography. Nature. 2005;437:336–42.
Azam F, Malfatti F. Microbial structuring of marine ecosystems. Nat Rev Microbiol. 2007;5:782–91.
Falkowski PG, Fenchel T, DeLong EF. The microbial engines that drive Earth’s biogeochemical cycles. Science. 2008;320:1034–9.
Coleman ML, Sullivan MB, Martiny AC, Steglich C, Barry K, DeLong EF, et al. Genomic islands and the ecology and evolution of Prochlorococcus. Science. 2006;311:1768–70.
Ivars-Martinez E, Martin-Cuadrado A-B, D’Auria G, Mira A, Ferriera S, Johnson J, et al. Comparative genomics of two ecotypes of the marine planktonic copiotroph Alteromonas macleodii suggests alternative lifestyles associated with different kinds of particulate organic matter. ISME J. 2008;2:1194–212.
Johnson ZI, Zinser ER, Coe A, McNulty NP, Woodward EMS, Chisholm SW. Niche partitioning among Prochlorococcus ecotypes along ocean-scale environmental gradients. Science. 2006;311:1737–40.
Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, et al. The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007;5:e77.
de Vargas C, Audic S, Henry N, Decelle J, Mahé F, Logares R, et al. Eukaryotic plankton diversity in the sunlit ocean. Science. 2015;348:1261605.
Shilova IN, Robidart JC, DeLong EF, Zehr JP. Genetic diversity affects the daily transcriptional oscillations of marine microbial populations. PLoS ONE. 2016;11:e0146706.
Sunagawa S, Coelho LP, Chaffron S, Kultima JR, Labadie K, Salazar G, et al. Ocean plankton. Structure and function of the global ocean microbiome. Science. 2015;348:1261359.
Hewson I, Poretsky RS, Dyhrman ST, Zielinski B, White AE, Tripp HJ, et al. Microbial community gene expression within colonies of the diazotroph, Trichodesmium, from the Southwest Pacific Ocean. ISME J. 2009;3:1286–300.
Poretsky RS, Hewson I, Sun S, Allen AE, Zehr JP, Moran MA. Comparative day/night metatranscriptomic analysis of microbial communities in the North Pacific Subtropical Gyre. Environ Microbiol. 2009;11:1358–75.
McCarren J, Becker JW, Repeta DJ, Shi Y, Young CR, Malmstrom RR, et al. Microbial community transcriptomes reveal microbes and metabolic pathways associated with dissolved organic matter turnover in the sea. Proc Natl Acad Sci USA. 2010;107:16420–7.
Ottesen EA, Marin R, Preston CM, Young CR, Ryan JP, Scholin CA, et al. Metatranscriptomic analysis of autonomously collected and preserved marine bacterioplankton. ISME J. 2011;5:1881–95.
Shilova IN, Robidart JC, Tripp HJ, Turk-Kubo K, Wawrik B, Post AF, et al. A microarray for assessing transcription from pelagic marine microbial taxa. ISME J. 2014;8:1476–91.
Karl DM, Church MJ. Ecosystem structure and dynamics in the North Pacific Subtropical Gyre: new views of an old ocean. Ecosystems. 2017;20:433–57.
Karl DM. A sea of change: biogeochemical variability in the North Pacific Subtropical Gyre. Ecosystems. 1999;2:181–214.
Ottesen EA, Young CR, Gifford SM, Eppley JM, Marin R, Schuster SC, et al. Multispecies diel transcriptional oscillations in open ocean heterotrophic bacterial assemblages. Science. 2014;345:207–12.
Johnson KS, Riser SC, Karl DM. Nitrate supply from deep to near-surface waters of the North Pacific Subtropical Gyre. Nature. 2010;465:1062–5.
Mahaffey C, Björkman KM, Karl DM. Phytoplankton response to deep seawater nutrient addition in the North Pacific Subtropical Gyre. Mar Ecol Prog Ser. 2012;460:13–34.
Robidart JC, Church MJ, Ryan JP, Ascani F, Wilson ST, Bombar D, et al. Ecogenomic sensor reveals controls on N2-fixing microorganisms in the North Pacific Ocean. ISME J. 2014;8:1175–85.
McAndrew PM, Björkman KM, Church MJ, Morris PJ, Jachowski N, Williams PJlB, et al. Metabolic response of oligotrophic plankton communities to deep water nutrient enrichment. Mar Ecol Prog Ser. 2007;332:63–75.
Shi Y, McCarren J, DeLong EF. Transcriptional responses of surface water marine microbial assemblages to deep-sea water amendment. Environ Microbiol. 2012;14:191–206.
Alexander H, Rouco M, Haley ST, Wilson ST, Karl DM, Dyhrman ST. Functional group-specific traits drive phytoplankton dynamics in the oligotrophic ocean. Proc Natl Acad Sci USA. 2015;112:E5972–9.
Karl DM, Tien G. MAGIC: a sensitive and precise method for measuring dissolved phosphorus in aquatic environments. Limnol Oceanogr. 1992;37:105–16.
Dore JE, Karl DM. Nitrification in the euphotic zone as a source for nitrite, nitrate, and nitrous oxide at Station ALOHA. Limnol Oceanogr. 1996;41:1619–28.
Wilson ST, Böttjer D, Church MJ, Karl DM. Comparative assessment of nitrogen fixation methodologies, conducted in the oligotrophic North Pacific Ocean. Appl Environ Microbiol. 2012;78:6516–23.
Turk-Kubo KA, Achilles KM, Serros TR, Ochiai M, Montoya JP, Zehr JP. Nitrogenase (nifH) gene expression in diazotrophic cyanobacteria in the Tropical North Atlantic in response to nutrient amendments. Front Microbiol. 2012;3:73–103.
Green SJ, Venkatramanan R, Naqib A. Deconstructing the polymerase chain reaction: understanding and correcting bias associated with primer degeneracies and primer-template mismatches. PLoS ONE. 2015;10:e0128122.
McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 2013;8:e61217.
The R Core Team. R: a Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna, 2017).
Shilova IN, Mills MM, Robidart JC, Turk-Kubo KA, Björkman KM, Kolber Z, et al. Differential effects of nitrate, ammonium, and urea as N sources for microbial communities in the North Pacific Ocean. Limnol Oceanogr. 2017;62:2550–74.
Bolstad BM, Irizarry RA, Åstrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–93.
Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64.
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47.
Dore JE, Letelier RM, Church MJ, Lukas R, Karl DM. Summer phytoplankton blooms in the oligotrophic North Pacific Subtropical Gyre: historical perspective and recent observations. Prog Oceanogr. 2008;76:2–38.
Pichard SL, Campbell L, Kang JB, Tabita FR, Paul JH. Regulation of ribulose bisphosphate carboxylase gene expression in natural phytoplankton communities. I. Diel Rhythms. Mar Ecol Prog Ser. 1996;139:257–65.
John DE, Wang ZA, Liu X, Byrne RH, Corredor JE, López JM, et al. Phytoplankton carbon fixation gene (RuBisCO) transcripts and air-sea CO2 flux in the Mississippi River plume. ISME J. 2007;1:517–31.
Zinser ER, Lindell D, Johnson ZI, Futschik ME, Steglich C, Coleman ML, et al. Choreography of the transcriptome, photophysiology, and cell cycle of a minimal photoautotroph, Prochlorococcus. PLoS ONE. 2009;4:e5135.
Liu H, Nolla HA, Campbell L. Prochlorococcus growth rate and contribution to primary production in the equatorial and subtropical North Pacific Ocean. Aquat Microb Ecol. 1997;12:39–47.
Thompson LR, Zeng Q, Chisholm SW. Gene expression patterns during light and dark infection of Prochlorococcus by cyanophage. PLoS ONE. 2016;11:e0165375.
Puxty RJ, Evans DJ, Millard AD, Scanlan DJ. Energy limitation of cyanophage development: implications for marine carbon cycling. ISME J. 2018;12:1273–86.
Tolonen AC, Aach J, Lindell D, Johnson ZI, Rector T, Steen R, et al. Global gene expression of Prochlorococcus ecotypes in response to changes in nitrogen availability. Mol Syst Biol. 2006;2:53.
Dekaezemacker J, Bonnet S. Sensitivity of N2 fixation to combined nitrogen forms (NO3 - and NH4 +) in two strains of the marine diazotroph Crocosphaera watsonii (Cyanobacteria). Mar Ecol Prog Ser. 2011;438:33–46.
Martens-Habbena W, Berube PM, Urakawa H, de la Torre JR, Stahl DA. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature. 2009;461:976–9.
Li B, Karl DM, Letelier RM, Bidigare RR, Church MJ. Variability of chromophytic phytoplankton in the North Pacific Subtropical Gyre. Deep Sea Res II. 2013;93:84–95.
Decho A. Microbial exopolymer secretions in ocean environments: their role(s) in food webs and marine processes. Oceanogr Mar Biol Annu Rev. 1990;28:73–153.
Hassler CS, Norman L, Mancuso Nichols CA, Clementson LA, Robinson C, Schoemann V, et al. Iron associated with exopolymeric substances is highly bioavailable to oceanic phytoplankton. Mar Chem. 2015;173:136–47.
Amin SA, Hmelo LR, van Tol HM, Durham BP, Carlson LT, Heal KR, et al. Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature. 2015;522:98–101.
Landa M, Burns AS, Roth SJ, Moran MA. Bacterial transcriptome remodeling during sequential co-culture with a marine dinoflagellate and diatom. ISME J. 2017;11:2677–90.
Ouverney CC, Fuhrman JA. Marine planktonic Archaea take up amino acids. Appl Environ Microbiol. 2000;66:4829–33.
Shi Y, Tyson GW, Eppley JM, DeLong EF. Integrated metatranscriptomic and metagenomic analyses of stratified microbial assemblages in the open ocean. ISME J. 2011;5:999–1013.
Aylward FO, Eppley JM, Smith JM, Chavez FP, Scholin CA, DeLong EF. Microbial community transcriptional networks are conserved in three domains at ocean basin scales. Proc Natl Acad Sci USA. 2015;112:5443–8.
Sohm JA, Ahlgren NA, Thomson ZJ, Williams C, Moffett JW, Saito MA, et al. Co-occurring Synechococcus ecotypes occupy four major oceanic regimes defined by temperature, macronutrients and iron. ISME J. 2016;10:333–45.
Farrant GK, Dore H, Cornejo-Castillo FM, Partensky F, Ratin M, Ostrowski M, et al. Delineating ecologically significant taxonomic units from global patterns of marine picocyanobacteria. Proc Natl Acad Sci USA. 2016;113:E3365–74.
Amin SA, Parker MS, Armbrust EV. Interactions between diatoms and bacteria. Microbiol Mol Biol Rev. 2012;76:667–84.
Krupke A, Hmelo LR, Ossolinski JE, Mincer TJ, Van Mooy BA. Quorum sensing plays a complex role in regulating the enzyme hydrolysis activity of microbes associated with sinking particles in the ocean. Front Mar Sci. 2016;3:55.
Thompson AW, Foster RA, Krupke A, Carter BJ, Musat N, Vaulot D, et al. Unicellular cyanobacterium symbiotic with a single-celled eukaryotic alga. Science. 2012;337:1546–50.
Thompson AW, Zehr JP. Cellular interactions: lessons from the nitrogen‐fixing cyanobacteria. J Phycol. 2013;49:1024–35.
Van Mooy BA, Hmelo LR, Sofen LE, Campagna SR, May AL, Dyhrman ST, et al. Quorum sensing control of phosphorus acquisition in Trichodesmium consortia. ISME J. 2012;6:422–9.
Acknowledgements
The authors would like to thank the Center for Microbial Oceanography, Research and Education (C-MORE grant number EF0424599, J.Z. and D.M.K.), the Simons Collaboration on Ocean Processes and Ecology (a grant from the Simons Foundation – SCOPE Award ID 329108, J.Z. and D.M.K.), the David and Lucile Packard Foundation, the Gordon and Betty Moore Foundation MEGAMER Facilities Grant (number 1761 to J.Z.) and Investigator Award (number 3794 to D.M.K.) and National Science Foundation Dimensions of Biodiversity Program (grant number 1241221, J.Z.) for funding and resources. The expertise of the international MicroTOOLs Team was crucial in the development of a successful environmental microarray. The MicroTOOLs array hybridization was performed at the Roy J. Carver Center for Genomics, The University of Iowa, USA. Steve Poulos and Lance Fujieki kindly provided Seaglider density data and images. We also thank Ariel Rabines, Roman Marin III, John Ryan, Gene Massion, Blake Watkins, Mariona Segura-Noguera, Susan Curless and the captain and crew of the R/V Kilo Moana.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Robidart, J.C., Magasin, J.D., Shilova, I.N. et al. Effects of nutrient enrichment on surface microbial community gene expression in the oligotrophic North Pacific Subtropical Gyre. ISME J 13, 374–387 (2019). https://doi.org/10.1038/s41396-018-0280-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-018-0280-0