Abstract
This study investigated the ecology, epidemiology and plasmid characteristics of extended-spectrum cephalosporin (ESC)-resistant E. coli in healthy pigs over a period of 4 years (2013–2016) following the withdrawal of ESCs. High carriage rates of ESC-resistant E. coli were demonstrated in 2013 (86.6%) and 2014 (83.3%), compared to 2015 (22%) and 2016 (8.5%). ESC resistance identified among E. coli isolates was attributed to the carriage of an IncI1 ST-3 plasmid (pCTXM1-MU2) encoding blaCTXM-1. Genomic characterisation of selected E. coli isolates (n = 61) identified plasmid movement into multiple commensal E. coli (n = 22 STs). Major STs included ST10, ST5440, ST453, ST2514 and ST23. A subset of the isolates belong to the atypical enteropathogenic E. coli (aEPEC) pathotype that harboured multiple LEE pathogenic islands. pCTXM1-MU2 was similar (99% nt identity) to IncI1-ST3 plasmids reported from Europe, encoded resistance to aminoglycosides, sulphonamides and trimethoprim, and carried colicin Ib. pCTXM1-MU2 appears to be highly stable and readily transferable. This study demonstrates that ESC resistance may persist for a protracted period following removal of direct selection pressure, resulting in the emergence of ESC-resistance in both commensal E. coli and aEPEC isolates of potential significance to human and animal health.
Similar content being viewed by others
Introduction
The World Health Organisation (WHO) has recently warned of a post-antibiotic era after discovering widespread occurrence of antimicrobial resistance (AMR) to critically important antimicrobials (CIAs), including extended-spectrum cephalosporins (ESC), fluoroquinolones (FQ), carbapenems and colistin in bacteria that cause serious infections in humans worldwide [1]. There are also concerns of CIA-resistant bacteria originating from food-producing animals through direct transfer of zoonotic pathogens such as multidrug-resistant (MDR) Salmonella spp. and Escherichia coli as well as indirect transfer of mobile AMR genes to commensal or pathogenic human bacteria through the food chain [2, 3].
Globally, there is much debate concerning antimicrobial usage in food-producing animals [4]. The development of AMR is complex, driven by both clonal expansion of MDR strains and horizontal transmission of mobile genetic elements carrying AMR genes, including plasmids, transposons and integrons, under antibiotic selection pressure [3]. In intensive livestock production systems, the emergence of CIA-resistance among commensal E. coli may lead to an increased resistance burden in the gut microbiota [5, 6].
Ceftiofur and cefquinome are ESCs registered worldwide primarily for the treatment of respiratory infections in food-producing animals. It has been argued that overuse or misuse of these ESCs has led to the emergence of a reservoir of ESC-resistant Enterobacteriaceae including E. coli and Salmonella enterica in livestock worldwide [4, 7, 8]. ESC resistance is predominantly plasmid-mediated and the most commonly identified ESC-resistance genes are blaCMY-2 and blaCTX-M variants [9, 10]. Since the first report of ESC-resistant E. coli and Salmonella serovars in food-producing animals was linked to the use of ESCs in livestock, there has been increasing public health concern due to the direct transfer of resistant bacteria or the potential for the transfer of plasmid-mediated resistance to humans [2, 11,12,13]. In addition to the risk of acquiring ESC-resistant plasmids, prolonged carriage and persistence of ESC-resistance after the use of ESCs in livestock is an important complication to be considered [14].
Recent studies in humans and animals have shown that once ESC-resistance emerges in the host’s microbiota, it takes a considerable time for resistance genes distributed among autochthonous Enterobacteriaceae to be lost, even when direct antibiotic selection pressure is removed. This demonstrates the long-term impact of ESC-resistance on both human and animal health [15, 16]. A recent study in Denmark has shown that CTX-M- producing E. coli can persist for 1 year in the gut of pigs in the absence of direct selection pressure [16]. Studies in humans have shown that ESC-resistant E. coli can persist in the human gastrointestinal tract for up to 3–5 years [15, 17]. The reasons for this persistence are unclear, however co-selection through use of other antimicrobials, strain fitness and plasmid stability have been proposed as selection foci for prolonged carriage of ESC-resistant E. coli in livestock [15, 16, 18, 19]. A number of international studies have evaluated the carriage of ESC-resistant E. coli in livestock systems. However, little is known about the longitudinal carriage of ESC-resistance among commensal E. coli in pig herds and the maintenance of ESC-resistance plasmids in the absence of direct selection pressure for prolonged periods.
In this study, we describe the ecology and epidemiology of ESC-resistant E. coli in healthy pigs at a single piggery in Australia over a period of 4 years following the voluntary withdrawal of ESC from use on farm. Data collected have been used to comprehensively describe transmission, evolution and persistence of ESC-resistant plasmids within host commensal E. coli clones.
Materials and Methods
Sampling and antimicrobial use history
All work involving animals was undertaken as part of a veterinary investigation by the veterinarian in charge or was undertaken with the approval of the University of Adelaide Animal Ethics Committee (approval number S-2013-182). Rectal swabs were obtained from randomly selected pigs from an Australian piggery with a history of off-label ceftiofur use on an individual animal basis over the preceding 4 years. This investigation was performed by a specialist pig veterinarian as part of the herd health management plan for the piggery, and samples were submitted to the University of Adelaide Veterinary Diagnostic Laboratory for culture. While ceftiofur use occurred in the first year of the study (2013) as the primary control agent for post-farrowing E.coli scours in the farrowing area, it was discontinued in 2014 in response to the initial findings from this study. None of the animals sampled in years 2014, 2015 and 2016 had exposure to ceftiofur. However, from 2014 amoxicillin (Amoxil) at 500ppm was used in the creep feed for Streptococcus suis management. In addition, lincomycin was used as required in the grower feed for ileitis management and suppression of a minor colitis associated with Brachyspira hyodysenteriae and kitasamycin was used in the finisher feed for ileitis and Erysipelas infections.
Sampling of the pigs was opportunistic, however it was stratified by age and production class. A total of 490 rectal swabs were collected over four consecutive years from healthy pigs. This included rectal swabs from 2013 (n = 90), 2014 (n = 60), 2015 (n = 140) and 2016 (n = 200) (Table 1).
Culture conditions and bacterial identification
Swabs were plated onto MacConkey agar (Thermofisher Scientific, Australia) supplemented with 4 µg/ml cefovecin (2013, Zoetis) or ceftiofur (2014–2016) (Sigma-Aldrich, Australia) and incubated overnight at 37 °C. Lactose fermenting colonies were identified, subcultured onto SBA (Thermofisher Scientific, Australia) and incubated at 37 °C overnight. E. coli was then identified from SBA plates by colony morphology, Gram stain and spot indole tests, and identifications were confirmed using MALDI-TOF (Bruker). One representative E. coli colony was subcultured on to SBA and preserved in brain heart infusion broth with 20% glycerol and stored frozen at −80 °C for later analysis. An additional E. coli isolate was selected from the selective agar if two morphologically distinct E. coli colony types were identified on the selective agar plate and on SBA.
Antimicrobial susceptibility testing
Antimicrobial susceptibility was evaluated using minimum inhibitory concentration (MIC) testing by the broth-microdilution method as recommended by the Clinical Laboratory Standards Institute (CLSI) in 96 well plates [20]. Susceptibility to eight antimicrobials was assessed (ampicillin, ceftiofur, ceftriaxone, ciprofloxacin, florfenicol, gentamicin, tetracycline and trimethoprim/sulfamethoxazole). E. coli ATCC 25922 was used as a quality control strain as recommended. MIC results were interpreted based on clinical breakpoints according to CLSI VET01S (ampicillin, ceftiofur, florfenicol, gentamicin, tetracycline, trimethoprim/sulphamethoxazole) [21]. CLSI M100 was used where no breakpoints were available in the VET01S manual (ceftriaxone, ciprofloxacin) [22].
DNA extraction for PCR assays
DNA extraction for preliminary screening was performed using 6% Chelex (Bio-Rad) as previously described with the exception that extractions were performed in 96 well plates [23].
Molecular characterisation
The detection of the blaCTX-M gene was performed using real-time PCR (RT-PCR) that amplifies all blaCTX-M variants as previously described with minor variations [24]. Briefly, primers and probe CTX-A_fwd. (CGGGCRATGGCGCARAC), CTX-A_rev. (TGCRCCGGTSGTATTGCC) and CTX-A_probe (6FAM-CCARCGGGCGCAGYTGGTGAC-BHQ1) were combined at final working concentrations of 400 μM for each primer and 120 μM for the probe, with 2 μl of DNA extract and TaqPath qPCR Master Mix (Life Technologies). Reactions were run on a Quantstudio 6 Flex (Life Technologies) in fast mode, with samples showing a sigmoidal curve with a Ct ≤ 40 considered positive for the presence of the blaCTX-M gene.
Genome sequencing
Whole genome sequencing was performed on a subset of ESC-resistant E. coli isolates (n = 61). The isolates were selected on the basis of antimicrobial susceptibility results and year of isolation. DNA extraction of E. coli isolates for whole genome sequencing was performed using MagMAX-96 DNA multi-sample kit (Thermo Fisher Scientific). Genome sequencing was performed on Illumina MiSeq and NextSeq platforms using Nextera XT DNA library preparation and MiSeq V3 2 × 300 or NextSeq High Output v2 (300 cycles) kits according to the manufacturer’s instructions. Reads were de novo assembled using CLC Genomics Workbench v9.5.4, and contig files uploaded to the Centre for Genomic Epidemiology (CGE) (http://www.genomicepidemiology.org/) for screening for AMR genes, MLST and plasmid replicon type. Clermont Phylogenetic group was inferred from whole genome sequence data [25].
Core genome single nucleotide polymorphisms (SNPs) were extracted using the Nullarbor pipeline on 60 E. coli isolates [26]. Gubbins was further used to identify regions under horizontal transfer [27]. Core genome SNPS within these regions were removed and SNP data was imported into MEGA6 to generate a maximum parsimony tree [28]. One representative isolate (SA152) was also subject to PacBio Sequencing. The sequence data was de novo assembled using PacBio software and Quiver, and annotation was performed using RAST 28, with annotation editing performed using Geneious v10.1.3. The presence of genomic islands within plasmids was predicted using IslandViewer 4 [29].
Intimin subtypes (eae) were identified by BLASTn analysis against previously deposited eae sequences in GenBank. Isolates carrying intimin genes were analysed for the presence of bundle forming pilli (bfp) genes by mapping reads against the E. coli adherence factor plasmid pB171 (Genbank accession number: AB024946). In addition, all the sequenced isolates were screened for the carriage of porcine A/E-associated gene (paa) gene [30].
Plasmid transfer by conjugation
Transferability of the resistance plasmid was performed by bacterial conjugation using E. coli J53AzR recipient strain as previously reported using two randomly selected ESC-resistant E. coli isolates (SA210 and SA148) were used for this experiment [31]. The selection of the trans-conjugants was made on MacConkey Agar containing sodium azide (150 mg/L) and ceftiofur (4 µg/mL).
Plasmid stability
Isolate SA152 was cultured onto sheep blood agar (SBA) (Edwards, Australia) and incubated overnight at 37 oC. A single colony of SA152 was inoculated into 10 mL of Luria Bertani broth. After overnight incubation, 0.1 mL of each sample was inoculated into 9.9 mL of the corresponding broth or broth/antibiotic. This was repeated across 9 days. On the 9th day inoculums were plated in serial dilutions onto MacConkey agar (non-selective) and MacConkey agar (selective agar) with 4 mg/L calculate the CFU/mL. This was completed using a Tecan 1.5 robot with Scirobotics hardware.
Results
Detection of ESC-resistant E. coli carriage
Evaluation of ESC-resistant E. coli using selective agar demonstrated the relatively high carriage rates in 2013 (86.6%) and 2014 (83.3%); compared to 2015 (22%) and 2016 (8.5%) (Table 1). Samples obtained from finisher pigs in 2015 and 2016 were particularly noteworthy for the very low frequency of samples yielding ESC-resistant E. coli (0 and 5%, respectively) compared to 2013 (85 %) [Table 1].
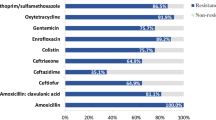
Phenotypic characterisation of AMR by MIC determination confirmed on that all presumptive ESC-resistant E. coli isolated (n = 219) were resistant to ESCs (ceftiofur MICs ≥ 64 µg/mL). The majority of ceftiofur-resistant isolates were also resistant to ampicillin (100%), ceftriaxone (99.1%) and trimethoprim/sulfamethoxazole (99.1%). A smaller proportion of isolates were resistant to tetracycline (47.0%), gentamicin (16.4%) and florfenicol (11.4%). None of the isolates was resistant to fluoroquinolones (ciprofloxacin). All the isolates were positive for blaCTX-M group 1 by RT-PCR, confirming the carriage of a blaCTX-M gene.
Molecular and genomic characterisation of ESC-resistant E. coli isolates
Phylogenetic characterisation of the isolates
A subset of representative isolates (n = 61) underwent whole genome sequencing to evaluate their full genomic characteristics. Genome sequencing confirmed the heterogeneous genetic make of the ESC-resistant E. coli isolates. Clermont phylogrouping revealed that the ESC-resistant E. coli isolates predominantly belonged to phylogenetic groups A (34.4%), C (29.5%) and B1 (24.6%) with remaining isolates classified as belonging to groups E (8.2%), D (1.6%) and B2 (1.6%). A total of 22 MLST types were identified among the ESC-resistant E. coli isolates, with ST10 (10/61), ST5440 (9/61), ST453 (6/61), ST2514 (6/61) and ST23 (4/61) being predominant. The CGE serotype predictor tool identified a number of serogroups including 18 O: types and 18 H: types (Table S1, supplemental material). The predominant serogroups identified among the ESC-resistant E. coli isolates were O8:H9 (7/61) and O8:H4 (7/61).
Phylogenetic characterisation using core genome SNPs also confirmed the polyclonal nature of the ESC-resistant E. coli population demonstrating a phylogenetically diverse ESC-resistant E. coli population in the piggery. The core-genome SNPs analysis mirrored the MLST analysis of the isolates (Fig. 1).
Dynamics and phylogenetic relationships of extended-spectrum cephalosporin resistant E. coli isolated from pigs over a period of four years from an Australian pig farm following the voluntary removal of ceftiofur. Transmission of IncI1-blaCTXM-1 plasmid (pCTXM1-MU2) in the pig farm among multiple E. coli lineages isolated from pigs of different age groups was demonstrated and a maximum parsimony tree using single nucleotide polymorphisms (SNPs) of extended-spectrum cephalosporin-resistant E. coli was generated. Year of isolation is marked with the colours green (2013), blue (2014), pink (2015) and brown (2016). MLST groupings for each cluster are also included. Isolates carrying the eae, ehx and paa gene are marked with an asterisk and written gene name
Antimicrobial resistance genes
Genome sequencing revealed that the gene responsible for ESC resistance was blaCTXM-1, a commonly identified ESC resistance gene among livestock E. coli. Additional AMR genes (aadA5, dfrA17 and sul2) were also identified among these isolates as detailed in Table S1, supplemental material, and as noted in Figs. 2 and 3. The floR resistance gene that encodes resistance to florfenicol (an important antimicrobial to treat various animal infections that is not used in human medicine) was present in three isolates.
Genomic map of the plasmid pCTXM1-MU2 carried by extended-spectrum cephalosporin-resistant E. coli isolates belonging to multiple E. coli lineages in an Australian pig farm following the voluntary removal of ceftiofur. Positions of gene classes are indicated by colour. a Expanded view of the putative genomic island consisting of aadA5 and dfrA17 on a Tn3 transposase. b Expanded view of the ISEcp1-CTX-M-1 region
Genomic map of the plasmid pIncF-MU4 obtained from isolate SA152. Positions of gene classes are indicated by colour. a Expanded view of the aerobactin locus. b Expanded view of the tetracycline resistance cassette associated with an IS2 transposase. c Expanded view of macrolide resistance genes associated with an IS3 transposase
Plasmid characterisation
Preliminary plasmid characterisation revealed that an IncI1 plasmid was present among all the ESC-resistant E. coli isolates and contig mapping revealed that an IncI1 plasmid carried a blaCTXM-1 gene. De novo construction of the full plasmid was not possible with Illumina sequencing.
PacBio sequencing of one representative isolate (SA152) was used to detect and determine the full length sequence of an IncI1 ST-3 plasmid that carried blaCTXM-1. Read mapping of the rest of the isolates confirmed the presence of this plasmid, designated pCTXM1-MU2 (GenBank accession number MF152729) in all sequenced isolates.
The genetic map of pCTXM1-MU2 is shown in Fig. 2. The plasmid was 110,255 nucleotides in length, had a GC content of 51% and carried 23 IncI1 associated transfer genes. The blaCTXM-1 gene of interest was present in a region spanning nucleotide positions 91,939 – 92,814 and the resistance genes dfrA17, aadA5, and sul2 were present in the regions 8856–8987, 9118 – 9906 and 14,021–14,836, respectively. Upon BLASTn search, pCTXM1-MU2 displayed 95–100% query coverage and 99% identity to three previously reported IncI1 E. coli plasmids originating from chickens pC60-108, pC59-112 and pC49-108 (accession numbers KJ484635.1, KJ484637.1 and KJ484638.1 respectively), with MAUVE alignment of the genomes demonstrating the presence of largely conserved collinear coding blocks. All plasmids contained the blaCTXM-1 gene in association with insertion sequence ISEcp1. In plasmid pCTXM1-MU2, ISEcp1 is flanked by 19 bp repeat sequences and is carried on a Tn3 transposon.
In addition to the IncI1 plasmid, the ESC-resistant E. coli (SA152) also carried two other plasmids, an IncH12 plasmid 258,112 nucleotides in length designated pIncHI2-MU3 (GenBank accession number MF174859) and a smaller IncF plasmid of 162,142 nucleotides designated pIncF-MU4 (GenBank accession number MF174860) (Fig. 3). Plasmid pIncHI2-MU3 contained the AMR genes blaTEM-12 and tetM on Tn3 transposons, along with lnu(F), aadA22, aac(3)-Iva and aph(4)-la. The plasmid also carried a number of metal resistance genes including copA, copB and proC copper resistance genes, cusA/B/C/F cation efflux genes and silE and silP silver resistance genes on a Tn3 transposase. No other resistance genes or virulence genes of significance were present. Plasmid pIncF-MU4 carried Colicin Ia and Colicin E1 genes. AMR genes tetA, tetC, tetD and tetR were present on an IS2 transposon, and the macrolide resistance genes macA and macB were present and associated with an IS3 transposase element.
Plasmid transferability and stability
The conjugation experiment revealed that the ESC-resistance-encoding pCTXM1-MU2 plasmid was easily transferred to E. coli J53. The efficiency of plasmid transfer from both donors (SA210 and SA148) were 3.4 × 10−2 and 2.7 × 10−2 respectively. Phenotypic antimicrobial susceptibility testing confirmed the resistance to ceftiofur and the transconjugates were positive for CTX-M by PCR.
Stability testing on SA152 in the absence of selection pressure with ESC revealed that the plasmid is stable within the host E. coli. No notable difference in CFU was observed after 9 overnight passages in LB broth plated on both MacConkey agar (1.9 × 10 11 CFU/mL) and MacConkey agar infused with ceftriaxone (1.5 × 10 11 CFU/mL), demonstrating the stability of the plasmid within the E. coli host without selection pressure.
To confirm the presence of pCTXM1-MU2 in isolates from multiple years, mapping of read files was performed against the curated plasmid sequence obtained from isolate SA152. Mapping of 12 isolates from 2013, 19 from 2014, 6 from 2015 and 8 from 2016 comprising 45 separate serotypes was performed. Coverage ranged from 97.7 to 100% (37 of 45 with 100% coverage) and pairwise alignment ranged from 95 to 98.5%. These results indicate the pCTXM1-MU2 plasmid was present in all isolates mapped, and that it was generally stable in genetic makeup over the four-year study period and across different E. coli serotypes.
Virulence characterisation
The virulence screening revealed that none of the isolates could be classified as porcine enterotoxigenic E. coli, demonstrated by the lack of key virulence factors such as enterotoxins STa, STb and LT. The majority of the sequenced E. coli isolates appeared to be porcine commensal E. coli and lacked specific virulence factors that are usually identified among E. coli pathotypes including extra-intestinal pathogenic E. coli [32, 33]. However, a small (8/61) proportion of isolates were positive for the eae gene (intimin), but negative for the bundle forming pilus gene bfpA and the virulence regulator gene perA, a genotype that classifies the isolates as atypical enteropathogenc E. coli (aEPEC) [Table S1, supplemental material] [34, 35]. The intimin gene subtypes present in these isolates varied by serotype and year of isolation, with eaeα1 present in three isolates from 2013 (two typed as ST794-O76:H7 and one as ST1040-O177:H45) and eaeε3 in three isolates from 2016 (three typed as ST301-O70:H2). The ST794-O76:H7 isolates also carried the porcine A/E-associated gene (paa) gene. A single isolate from 2013 carried eae-κ (typed as ST206-O49:H10). Another isolate from 2014 carried eaeθ (typed as ST327-H8) and carried the enterohaemorrhagic E. coli (EHEC) hemolysin gene ehxA and paa gene. None of the aEPEC carried Shiga toxin genes, bfp genes or other fimbrial genes recognised as having a particular role in attachment to the intestinal epithelium.
The eae gene in E. coli is typically found on a locus of enterocyte effacement (LEE) pathogenicity island. When reads from all eae positive strains were mapped to a pathogenicity island reference sequence (GenBank accession AF022236), a consensus sequence contig with a query coverage of 98% and an identity of 99% was produced. While Illumina sequencing could not definitively assemble the LEE pathogenicity island to the chromosome, read mapping to AF022236 displayed coverage of 40 × over the 5′ and 3′ termini of this sequence, which corresponds to chromosomal regions, further providing evidence this is likely to be a chromosomally inserted island, as previously reported [36, 37].
Discussion
In this study, we evaluated the carriage of ESC-resistant E. coli in a pig herd over a period of 4 years, and clarified the molecular characteristics of ESC-resistant E. coli and their associated plasmids responsible for ESC resistance. The major findings arising from this study are as follows: (i) ESC resistance identified among E. coli isolated in this study was attributed to the widespread dispersion and persistence of an IncI1 plasmid (pCTXM1-MU2) encoding the blaCTXM-1 gene. (ii) Genomic characterisation of ESC- resistant E. coli demonstrated that the ESC-resistant plasmid had moved into multiple commensal and several atypical enteropathogenic E. coli clonal lineages that harboured multiple LEE pathogenic island variants encoding the eae and/or ehx genes. (iii) Once ESC-resistant E. coli emerge in a livestock production system, it will take an extended period of time (at least 4 years) for significant reduction and/or elimination of ESC-resistant E. coli following withdrawal of the direct antimicrobial selection pressure. (iv) IncI1-blaCTXM-1 plasmids are globally disseminated, stable and highly transferable.
We have demonstrated that the dissemination of ESC-resistant E. coli and persistence of these clones in pig populations of different age categories is attributable to a highly transferable IncI1-blaCTXM-1 plasmid among different E. coli lineages. IncI1 plasmids carrying blaCTXM-1 are one of the most commonly identified plasmid-gene combinations imparting ESC resistance in pigs and poultry in Europe [38,38,40]. Genomic analysis of pCTXM1-MU2 demonstrated a close relationship with three IncI1-blaCTXM-1 plasmids isolated from poultry in Switzerland, with all blaCTXM-1 genes associated with an ISECP1 insertion sequence which is commonly described [41]. The close genomic similarity of the plasmids, conservation of blocks of coding regions and common occurrence in livestock suggest that plasmids with an IncI1 backbone carrying the blaCTXM-1 gene in this configuration are relatively stable, prompting inquiry into this plasmid’s origin and its dissemination into an Australia pig herd.
The first possible hypothesis is that this plasmid was carried by a Gram-negative bacterium in livestock or their environment and was mobilised and transferred to commensal E. coli in the presence of antibiotic selection pressure from the use of ceftiofur as an off-label treatment for scours. Following this, continued administration of routinely used antimicrobials such as amoxicillin for the control of Streptococcus suis may have contributed to the persistence of the plasmid (pCTXM1-MU2) in pigs of older age groups and the piggery environment via co-selection.
In Australia, a ban on the live importation of pigs has been in place for over 30 years, just before ceftiofur was registered for use in livestock globally in the late 1980s and extending into the1990s. As a result, ESC-resistant plasmids may have been present in the gut of pigs before the use of ceftiofur, for example with the establishment of Australia’s pig herd, or alternatively, Australian environmental bacteria may harbour IncI1-blaCTXM-1 plasmids. This seems unlikely given that the plasmid is 99% similar to previously described IncI1 plasmids isolated from poultry E. coli strains in Europe.
The close relationship of the IncI1-blaCTXM-1 plasmid identified in this study to others identified in European livestock isolates, along with the potential stability of this plasmid-gene combination implies a more recent common source or pathway of introduction. This in turn leads to the second hypothesis that the plasmid may have been introduced via reverse zoonotic transmission from pig workers who have had recent contact with livestock or contaminated food products from countries where the IncI1-blaCTXM-1 plasmid is endemic. Human-to-animal transmission of resistant bacteria has been demonstrated by the emergence and spread of MRSA ST398 in Australian piggeries [42, 43]. A study using whole genome sequencing of MRSA ST398 isolates from Australian pigs suggests multiple incursions of European-origin clonal lineages, most likely from human carriers visiting European piggeries [43].
A third hypothesis is that the plasmid was introduced into the piggery environment via wild birds, with transfer of the plasmid occurring via interactions between endemic and migratory bird species. Our recent study has demonstrated that the detection of FQ and ESC-resistant E. coli clone ST744 in Australian pigs may have been introduced into Australian pigs via humans or migratory birds, since ST744 is commonly detected among humans and birds in Asian countries [44, 45].
In this piggery, the IncI1-blaCTXM-1 plasmid was detected in multiple E. coli lineages including potentially pathogenic atypical EPEC clones, demonstrating high transferability of this plasmid without apparent clonal discrimination. The E. coli clones identified in this study have previously been reported in humans and animals in other parts of the world. For instance, ST10 is a broad host range E. coli type that has been detected in pigs, poultry and humans. This study also shows that the IncI1-blaCTXM-1 plasmid has moved into extraintestinal pathogenic clonal lineages such as ST93 (O5:H4) an avian pathogenic E. coli (APEC) and ST453 (O23:H16), another clone known to cause extraintestinal disease in humans (urinary tract infections) and metritis in cattle. ST10 and ST453 containing CTX-M genes have previously been isolated from humans and animals, including pigs and seagulls [46,46,48].
A small number of the E. coli lineages that harboured IncI1-blaCTXM-1 also carried virulence factors associated with aEPEC. Atypical EPEC are categorised as E. coli that possess the LEE pathogenicity island and lack bundle-forming pili (BFP) and Shiga-toxin genes. A number of studies have associated aEPEC with paediatric diarrhoea, however, defining aEPEC as true pathogens is contentious due to the detection of these isolates in both diarrhoeic patients and control subjects [35, 49, 50]. In Australia, aEPEC have been isolated from clinical cases of diarrhoea in humans as and can be carried by healthy cattle and sheep [51]. In the present study, we identified eight aEPEC isolates belonging to different MLST groups and serotypes, confirming the presence of a heterogeneous group of ESC-resistant aEPEC strains in a single piggery. The carriage of intimin gene variants and ehx genes associated with EHEC pathotypes may indicate that these isolates are potential zoonotic pathogens. However, definitive testing in animal models would be required to confirm this as only a small number of studies have evaluated the full molecular characteristics, virulence potential and phylogeny of aEPEC [34, 35, 49, 50]. Further investigation is therefore warranted to fully characterise the virulence potential of these ESC-resistant aEPEC isolates and evaluate their ability to cause clinical disease in both humans and animals.
In this study, we have shown that once emerged, ESC-resistant E. coli is readily transmissible between pigs during different stages of production, resulting in an initial high carriage rate and persistence within the piggery over the 4-year study period. Genomic characterisation showed that the IncI1-blaCTXM-1 plasmid has remained highly stable over this period. However, despite continued detection of ESC-resistant E. coli over the course of the study, we demonstrated that withdrawal of ceftiofur had a significant impact on reducing ESC-resistant E. coli carriage in pigs in all age groups sampled, from 86.6% of pigs sampled in the first year following ceftiofur withdrawal, to 22.1 and 17.5% in the third and fourth years, respectively. This reduction was particularly apparent in finisher pigs, which may be related to the withholding period prior to slaughter when antimicrobial selection pressure is no longer a factor.
In conclusion, this study demonstrates the ecological dynamics of a highly transmissible, successful ESC-resistance plasmid in a pig herd and clarifies the genomic characteristics of ESC-resistant commensal E. coli harbouring the plasmid. This study shows that ceftiofur resistance may persist for a protracted period following removal of selection pressure, resulting in the emergence of multidrug resistance in both commensal E. coli and potentially virulent atypical EPEC isolates of significance to both human and animal health. This study also reveals that the IncI1-blaCTXM-1 plasmid pCTXM1-MU2 is a stable, highly transmissible plasmid that may confer additional fitness advantages to multiple E. coli lineages. Based on these results, a review of the current use of ESCs in livestock would be prudent, and ongoing surveillance combined with genomic characterisation of CIA-resistant bacteria is vital to mitigate the spread of antimicrobial resistance in livestock and its potential impact to human health.
References
WHO. Critically important antimicrobials for human medicine. 5th revision. Geneva: World Health Organization; 2017.
Marshall BM, Levy SB. Food animals and antimicrobials: impacts on human health. Clin Microbiol Rev. 2011;24:718–33.
Mukerji S, O’Dea M, Barton M, Kirkwood R, Lee T, Abraham S. Development and transmission of antimicrobial resistance among Gram-negative bacteria in animals and their public health impact. Essays Biochem. 2017;61:23–25.
Collignon P, Aarestrup FM. Extended-spectrum β-lactamases, food, and cephalosporin use in food animals. Clin Infect Dis. 2007;44:1391–2.
Cavaco LM, Abatih E, Aarestrup FM, Guardabassi L. Selection and persistence of CTX-M-producing Escherichia coli in the intestinal flora of pigs treated with amoxicillin, ceftiofur or cefquinome. Antimicrob Agents Chemother. 2008;52:3612–6.
Hansen KH, Bortolaia V, Damborg P, Guardabassi L. Strain diversity of CTX-M-producing enterobacteriaceae in individual pigs: insights into the dynamics of shedding during the production cycle. Appl Environ Microbiol. 2014;80:6620–6.
Agersø Y, Aarestrup FM, Pedersen K, Seyfarth AM, Struve T, Hasman H. Prevalence of extended-spectrum cephalosporinase (ESC)-producing Escherichia coli in Danish slaughter pigs and retail meat identified by selective enrichment and association with cephalosporin usage. J Antimicrob Chemother. 2012;67:582–8.
Agersø Y, Aarestrup FM. Voluntary ban on cephalosporin use in Danish pig production has effectively reduced extended-spectrum cephalosporinase-producing Escherichia coli in slaughter pigs. J Antimicrob Chemother. 2013;68:569–72.
Ewers C, Bethe A, Semmler T, Guenther S, Wieler LH. Extended-spectrum β-lactamase-producing and AmpC-producing Escherichia coli from livestock and companion animals, and their putative impact on public health: a global perspective. Clin Microbiol Infect. 2012;18:646–55.
Jahanbakhsh S, Smith MG, Kohan-Ghadr H-R, Letellier A, Abraham S, Trott DJ, et al. Dynamics of extended-spectrum cephalosporin resistance in pathogenic Escherichia coli isolated from diseased pigs in Quebec, Canada. Int J Antimicrob Agents. 2016;48:194–202.
Bradford PA, Petersen PJ, Fingerman IM, White DG. Characterization of expanded-spectrum cephalosporin resistance in E. coli isolates associated with bovine calf diarrhoeal disease. J Antimicrob Chemother. 1999;44:607–10.
Vieira AR, Collignon P, Aarestrup FM, McEwen SA, Hendriksen RS, Hald T, et al. Association between antimicrobial resistance in Escherichia coli isolates from food animals and blood stream isolates from humans in europe: an ecological study. Foodborne Pathog Dis. 2011;8:1295–301.
Allen KJ, Poppe C. Occurrence and characterization of resistance to extended-spectrum cephalosporins mediated by β-lactamase CMY-2 in Salmonella isolated from food-producing animals in Canada. Can J Vet Res. 2002;66:137–44.
Jørgensen CJ, Cavaco LM, Hasman H, Emborg H-D, Guardabassi L. Occurrence of CTX-M-1-producing Escherichia coli in pigs treated with ceftiofur. J Antimicrob Chemother. 2007;59:1040–2.
Brolund A. Overview of ESBL-producing Enterobacteriaceae from a Nordic perspective. Infect Ecol Epidemiol. 2014; 4:24555 https://doi.org/10.3402/iee.v3404.
Hansen KH, Damborg P, Andreasen M, Nielsen SS, Guardabassi L. Carriage and fecal counts of cefotaxime M-producing Escherichia coli in pigs: a longitudinal study. Appl Environ Microbiol. 2013;79:794–8.
Alsterlund R, Axelsson C, Olsson-Liljequist B. Long-term carriage of extended-spectrum beta-lactamase-producing Escherichia coli. Scand J Infect Dis. 2012;44:51–4.
Hossain A, Reisbug MD, Hanson ND. Plasmid-encoded functions compensate for the biological cost of AmpC overexpression in a clinical isolate of Salmonella typhimurium. J Antimicrob Chemother. 2004;53:964–70.
Titelman E, Hasan CM, Iversen A, Nauclér P, Kais M, Kalin M, et al. Faecal carriage of extended-spectrum β-lactamase-producing Enterobacteriaceae is common 12 months after infection and is related to strain factors. Clin Microbiol Infect. 2014;20:O508–15.
Clinical and Laboratories Standards Institute Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals; Second informational supplement, CLSI document VET01-S4. Wayne, PA, USA: CLSI, 2013; 2013. 2013
CLSI. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals (VET01-S3). 3rd ed. Wayne, PA: Clinical and Laboratory Standards Institute; 2015a.
CLSI. Performance standards for antimicrobial susceptibility testing; (M100-S25) twenty fifth informational supplement. Wayne, PA: Clinical and Laboratory Standards Institute; 2015b.
Abraham S, Chin J, Brouwers HJM, Zhang R, Chapman TA. Molecular serogrouping of porcine enterotoxigenic Escherichia coli from Australia. J Microbiol Methods. 2012;88:73–76.
Roschanski N, Fischer J, Guerra B, Roesler U. Development of a multiplex real-time PCR for the rapid detection of the predominant beta-lactamase genes CTX-M, SHV, TEM and CIT-Type AmpCs in Enterobacteriaceae. PLOS One. 2014;9:e100956.
Clermont O, Christenson JK, Denamur E, Gordon DM. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep. 2013;5:58–65.
Seemann T GdSA, Bulach DM, Schultz MB, Kwong JC, Howden BP. Nullarbor Github https://github.com/tseemann/nullarbor.
Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43:e15–e15.
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–9.
Bertelli C, Laird MR, Williams KP, Lau BY, Hoad G, Winsor GL, et al. IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017;45:W30–35.
Batisson I, Guimond MP, Girard F, An H, Zhu C, Oswald E, Fairbrother JM, et al. Characterization of the novel factor paa involved in the early steps of the adhesion mechanism of attaching and effacing Escherichia coli. Infect Immun. 2003;71:4516–25.
Sidjabat HE, Townsend KM, Hasnon ND, Bell JM, Stokes HW, Gobius KS, Moss SM, Trott DJ. Identification of blaCMY-7 and associated plasmid-mediated resistance genes in multidrug-resistant Escherichia coli isolated from dogs at a veterinary teaching hospital in Australia. J Antimicrob Chemother. 2006;57:840–8.
Mitchell NM, Johnson JR, Johnston B, Curtiss R, Mellata M. Zoonotic potential of Escherichia coli isolates from retail chicken meat products and eggs. Appl Environ Microbiol. 2015;81:1177–87.
Bekal S, Brousseau R, Masson L, Prefontaine G, Fairbrother J, Harel J. Rapid identification of Escherichia coli pathotypes by virulence gene detection with DNA microarrays. J Clin Microbiol. 2003;41:2113–25.
Ingle DJ, Tauschek M, Edwards DJ, Hocking DM, Pickard DJ, Azzopardi KI, et al. Evolution of atypical enteropathogenic E. coli by repeated acquisition of LEE pathogenicity island variants. Nat Microbiol. 2016;1:15010.
Tennant SM, Tauschek M, Azzopardi K, Bigham A, Bennett-Wood V, Hartland EL, et al. Characterisation of atypical enteropathogenic E. coli strains of clinical origin. BMC Microbiol. 2009;9:117.
Perna NT, Mayhew GF, Pósfai G, Elliott S, Donnenberg MS, Kaper JB, et al. Molecular evolution of a pathogenicity island from enterohemorrhagic Escherichia coli O157:H7. Infect Immun. 1998;66:3810–7.
Sekiya K, Ohishi M, Ogino T, Tamano K, Sasakawa C, Abe A. Supermolecular structure of the enteropathogenic Escherichia coli type III secretion system and its direct interaction with the EspA-sheath-like structure. Proc Natl Acad Sci USA. 2001; 98: 11638-11643.
Borjesson S, Bengtsson B, Jernberg C, Englund S. Spread of extended-spectrum beta-lactamase producing Escherichia coli isolates in Swedish broilers mediated by an incI plasmid carrying blaCTX-M-1. Acta Vet Scand. 2013;55:3.
Dahmen S, Haenni M, Madec J-Y. IncI1/ST3 plasmids contribute to the dissemination of the blaCTX-M-1 gene in Escherichia coli from several animal species in France. J Antimicrob Chemother. 2012;67:3011–2.
Zurfluh K, Wang J, Klumpp J, Nüesch-Inderbinen M, Fanning S, Stephan R. Vertical transmission of highly similar blaCTX-M-1-harbouring IncI1 plasmids in Escherichia coli with different MLST types in the poultry production pyramid. Front Microbiol. 2014; 5:519.
Wang J, Stephan R, Power K, Yan Q, Hächler H, Fanning S. Nucleotide sequences of 16 transmissible plasmids identified in nine multidrug-resistant Escherichia coli isolates expressing an ESBL phenotype isolated from food-producing animals and healthy humans. J Antimicrob Chemother. 2014;69:2658–68.
Groves MD, O’Sullivan MVN, Brouwers HJM, Chapman TA, Abraham S, Trott DJ, et al. Staphylococcus aureus ST398 detected in pigs in Australia. J Antimicrob Chemother. 2014;69:1426–8.
Sahibzada S, Abraham S, Coombs GW, Pang S, Hernández-Jover M, Jordan D, et al. Transmission of highly virulent community-associated MRSA ST93 and livestock-associated MRSA ST398 between humans and pigs in Australia. Sci Rep. 2017;7:5273.
Abraham S, Jordan D, Wong HS, Johnson JR, Toleman MA, Wakeham DL, et al. First detection of extended-spectrum cephalosporin- and fluoroquinolone-resistant Escherichia coli in Australian food-producing animals. J Glob Antimicrob Resist. 2015;3:273–7.
Hasan B, Sandegren L, Melhus A, Drobni M, Hernandez J, Waldenstrom J, et al. Antimicrobial drug-resistant Escherichia coli in wild birds and free-range poultry, Bangladesh. Emerg Infect Dis. 2012;18:2055–8.
Gerhold G, Schulze MH, Gross U, Bohne W. Multilocus sequence typing and CTX-M characterization of ESBL-producing E. coli: a prospective single-centre study in Lower Saxony, Germany. Epidemiol Infect. 2016;144:3300–4.
Schink A-K, Kadlec K, Kaspar H, Mankertz J, Schwarz S. Analysis of extended-spectrum-β-lactamase-producing Escherichia coli isolates collected in the GERM-Vet monitoring programme. J Antimicrob Chemother. 2013;68:1741–9.
Simões RR, Poirel L, Da Costa PM, Nordmann P. Seagulls and beaches as reservoirs for multidrug-resistant Escherichia coli. Emerg Infect Dis. 2010;16:110–2.
Afset JE, Bevanger L, Romundstad P, Bergh K. Association of atypical enteropathogenic Escherichia coli (EPEC) with prolonged diarrhoea. J Med Microbiol. 2004;53:1137–44.
Nguyen RN, Taylor LS, Tauschek M, Robins-Browne RM. Atypical enteropathogenic Escherichia coli infection and prolonged diarrhea in children. Emerg Infect Dis. 2006;12:597–603.
Hornitzky MA, Mercieca K, Bettelheim KA, Djordjevic SP. Bovine feces from animals with gastrointestinal infections are a source of serologically diverse atypical enteropathogenic Escherichia coli and Shiga toxin-producing E-coli strains that commonly possess intimin. Appl Environ Microbiol. 2005;71:3405–12.
Acknowledgements
We would like to thank University of Adelaide Veterinary Diagnostic laboratory for their assistance in microbiology. This study was funded by the DVM clinical research programme, University of Adelaide and Small Grant Scheme of School of Veterinary Life Sciences, Murdoch University. We thank Dr Glen Carter, Ms Sarah Baines, Prof. Benjamin Howden from the Doherty Applied Microbial Genomics, The Doherty Institute for Infection & Immunity, University of Melbourne for their assistance in performing PacBio sequencing.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Abraham, S., Kirkwood, R.N., Laird, T. et al. Dissemination and persistence of extended-spectrum cephalosporin-resistance encoding IncI1-blaCTXM-1 plasmid among Escherichia coli in pigs. ISME J 12, 2352–2362 (2018). https://doi.org/10.1038/s41396-018-0200-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-018-0200-3
This article is cited by
-
Comparison of antimicrobial resistant Escherichia coli isolated from Irish commercial pig farms with and without zinc oxide and antimicrobial usage
Gut Pathogens (2023)
-
The “Cins” of Our Fathers: Rejuvenated Interest in Colicins to Combat Drug Resistance
Journal of Microbiology (2023)
-
Phenotypic and genotypic characteristics of Escherichia coli with non-susceptibility to quinolones isolated from environmental samples on pig farms
Porcine Health Management (2019)