Abstract
High-throughput sequencing of ocean biomes has revealed vast eukaryotic microbial diversity, a significant proportion of which remains uncharacterized. Here we use a temporal approach to understanding eukaryotic diversity at the Scripps Pier, La Jolla, California, USA, via high-throughput amplicon sequencing of the 18S rRNA gene, the abundances of both Synechococcus and Synechococcus grazers, and traditional oceanographic parameters. We also exploit our ability to track operational taxonomic units (OTUs) temporally to evaluate the ability of 18S sequence-based OTU assignments to meaningfully reflect ecological dynamics. The eukaryotic community is highly dynamic in terms of both species richness and composition, although proportional representation of higher-order taxa remains fairly consistent over time. Synechococcus abundance fluctuates throughout the year. OTUs unique to dates of Synechococcus blooms and crashes or enriched in Synechococcus addition incubation experiments suggest that the prasinophyte Tetraselmis sp. and Gymnodinium-like dinoflagellates are likely Synechococcus grazers under certain conditions, and may play an important role in their population fluctuations.
Similar content being viewed by others
Introduction
Microbial communities form the foundation of marine ecosystems. Phytoplankton, including the cyanobacteria, are responsible for the majority of marine primary productivity [1, 2]. Heterotrophic and mixotrophic eukaryotes, commonly known as grazers, are primary consumers that comprise a crucial but once-overlooked trophic step [3]. Understanding basal trophic interactions relies on characterizing the long-underestimated diversity of marine microbial species. Two primary challenges have been the limitations of morphological species identification and the inability to identify most microbial species [4]. Environmental sequencing offers a potential solution: sequencing the 18S ribosomal RNA (rRNA) gene has revealed far greater species diversity among protists than that recorded using microscopy, including entirely novel clades [5,6,7,8,9,10].
Many sequencing-based studies of microbial diversity focus on spatial comparisons, implicitly assuming that a single time-point can represent the community at a sampling station. However, temporal studies have revealed considerable variability in community composition, especially at coastal sites [11,12,13,14]. Although some taxa remain consistent in their abundances, others exhibit sudden, transient blooms [15]. These temporal dynamics are not fully understood, but may reveal information about trophic interactions, as well as factors underlying harmful blooms.
Cyanobacteria, including members of the genus Synechococcus, play a prominent role in ocean primary productivity: they are estimated to comprise 8% of worldwide phytoplankton biomass and are responsible for up to 40% of marine carbon fixation [2, 16, 17]. Synechococcus species are globally abundant, especially in coastal environments [18, 19] such as the California Current Ecosystem [20].
The Scripps Institution of Oceanography pier, located in the Southern California Bight, has served as a long-term monitoring site since the early 1900s, when WE Allen recorded diatom and dinoflagellate abundances for over 10 years [21]. More recently, Synechococcus abundance has been monitored weekly or bi-weekly for at least 20 years ([20]; Palenik lab unpublished data), in addition to counts of harmful algal species and other physical and chemical metadata (SCCOOS, http://www.sccoos.org/). These historical data make the Scripps Pier an excellent site for a temporal study of the microbial community.
We have utilized environmental sequencing to describe the composition of the eukaryotic community at the Scripps Pier, and to explore the temporal dynamics of individual protist species and their potential roles as drivers of the onset or termination of Synechococcus blooms. Seasonality in Synechococcus cell density has previously been observed at the Scripps Pier and other coastal sites [20, 22,23,24,25]. The Synechococcus population at the Scripps Pier has historically bloomed once or twice every spring, typically followed by rapid declines of the population (days to weeks) back to basal levels ([20, 26]; Palenik lab unpublished data). The major processes responsible for loss of Synechococcus biomass are sedimentation and sinking via attachment to larger particles, viral lysis, and grazing—often by ciliates and nanoflagellates [27]. Although some Synechococcus grazers have been isolated, including Paraphysmonas sp., Pteridomonas sp., Gymnodinium sp., and various ciliates ([28,29,30]; Paz-Yepes, manuscript in preparation), most have not been validated as significant players in the ecology of Synechococcus. It is therefore largely unknown whether the same grazers that are readily maintained in culture are also important grazers of Synechococcus in the ocean.
The objectives of this study were to utilize the large amount of information provided by a temporal dataset of 18S rRNA amplicon sequences to (i) describe the composition and dynamics of the eukaryotic microbial community at the Scripps Pier site; (ii) identify putative grazers of Synechococcus; and (iii) evaluate the ecological significance of cultured species of grazers and other protists with respect to their role in shaping Synechococcus population dynamics at this site.
Materials and methods
Sample collection
Sampling was conducted in 2011 and 2012 as described in Tai and Palenik [20]. Briefly, surface seawater was collected weekly from the end of the Scripps Pier in La Jolla, CA (32°87′N, 117°26′W). Sampling dates were first selected for amplicon sequencing based on their correspondence with Synechococcus or grazer blooms or declines, with additional time-points included to increase overall temporal coverage. The resulting amplicon sequence data described 17 time-points covering diverse seasons over 2 years (Table 1).
DNA extraction and sequencing
For DNA extraction, 500 ml of seawater was filtered in triplicate onto 0.2 µm Supor filter discs (Pall) and stored at −80 °C. DNA was extracted with the DNEasy Blood and Tissue Kit (Qiagen, Valencia, CA, USA) and quantified by PicoGreen fluorescence (Quant-iT; Qiagen) as described in Tai and Palenik [20]. DNA from the 17 time-points was sent to Research & Testing Laboratories (Lubbock, TX, USA) for sequencing of the V4 region of the 18S gene on a Roche 454-FLX using the TAReuk454FWD1 (5′-CCAGCA(G/C)C(C/T)GCGGTAATTCC-3′) and TAReukREV3 (5′-ACTTTCGTTCTTGAT(C/T)(A/G)A-3′) primers [31]. For PCR conditions and complete methods, see S1.
Sequence analysis
Sequence reads were quality filtered and trimmed to 245 bp using the mothur software (version 1.35.1) with the pipeline described in the mothur 454 SOP ([32, 33], accessed 15 September 2015). Chimeras were removed using UCHIME [34] and the remaining sequences from each time-point were merged together for operational taxonomic unit (OTU) assignment. Sequences were clustered into OTUs at different similarity cutoffs ranging from 100 to 95%. Representatives from each OTU were classified based on the SILVA v123 taxonomy [35]. Sequences from each time-point were sub-sampled to the smallest sample size (1308 sequences) to prevent over-representing rare OTUs in more deeply sequenced samples. Certain sequences of interest were aligned on BLAST or SINA [36] for further taxonomic specificity. Unweighted Bray–Curtis analysis was conducted using SciPy [37]. The vegan package in R [38, 39] was used to conduct a canonical correspondence analysis of sample variation as explained by five variables: chlorophyll, alpha diversity, Synechococcus density, grazer density, and water temperature.
Data availability
Raw sequence reads and metadata were deposited into the NCBI Sequence Read Archive under accession number SRP132203 (https://www.ncbi.nlm.nih.gov/sra/SRP132203).
They are also available on the BCO-DMO page (https://www.bco-dmo.org).
Flow cytometry
Four replicate 1-ml seawater samples were preserved at a final concentration of 0.25% glutaraldehyde (Sigma-Aldrich, St Louis, MO, USA) for flow cytometry; upon addition of glutaraldehyde, samples were kept at room temperature for 10 min and then stored at −80 °C. Samples were processed as described in Collier and Palenik [40] (S2). Cell abundance counts were normalized using pre-counted 0.94 µm green fluorescent beads (Duke Scientific Corporation, Palo Alto, CA, USA) that were added to each sample. Events falling within a specific region were counted as Synechococcus cells (Fig. S3). Three-ml samples were frozen at −80 °C in 6.8% glycine betaine and sent to Bigelow Laboratory for Ocean Sciences, East Boothbay, Maine, for bulk flow sorting. Sorting gates were established for two different regions of the flow cytogram plots based on fluorescence, cell-size, and internal cell complexity (scatter). The objective was to capture putative mixotrophic grazers, which we expected to be larger in size than Synechococcus and emit both chlorophyll fluorescence (from having chloroplasts) and phycoerythrin fluorescence (from ingested Synechococcus). Bulk sorts of two specific regions of interest (R1 and R2, Fig. 4d) containing putative mixotrophic grazers were completed as described in S2, collected in UV-treated sterile 0.5 ml centrifuge tubes and stored immediately at −80 °C.
Clone libraries
Whole-genome amplifications were performed on cells from R1 and R2 (Fig. 4d). Cells were lysed in three rounds of heating (70 °C, 3 min)/freezing (−80 °C, 3 min) with vortex bead-beating (1 min, using ~50, 0.5 mm silica-zirconia beads per tube) in between the heating and freezing steps. The resulting crude cell-lysate was used in 20 µl Genomiphi V3 (GE Life Sciences, Marlborough, MA, USA) reactions that were primed with random hexamers. Amplification of DNA was confirmed by quantification on a Qubit fluorometer using the Broad Range Qubit Reagents (Thermo Fisher Scientific).
The clone libraries for R1 and R2 were made slightly differently but ultimately resulted in sequence data from the same region. For R1, a larger region of the 18S rRNA gene was initially amplified using the GoTaq HotStart Mastermix (Promega) with the Moon-A (5′-ACCTGGTTGATCCTGCCAG-3′) and Moon-B (5′-TGATCCTTCYGCAGGTTCAC-3′) primers [7]; the reaction was denatured at 94 °C for 2 min, then underwent 35 cycles of 30 s at 94 °C, 2 min at 52 °C, and 3 min at 72 °C, followed by a final extension for 10 min at 72 °C. The PCR product was ligated into pCR2.1-TOPO (Thermo Fisher Scientific) plasmid vectors and transformed into TOP10 chemically competent Escherichia coli cells following the manufacturer’s protocol (Invitrogen, Carlsbad, CA, USA). Plasmid DNA was extracted using the QIAprep Spin Miniprep Kit (Qiagen). The TAReuk454FWD1 and TAReukREV3 primers [31] were used as internal primers for Sanger-based DNA sequencing (Eton Biosciences, San Diego, CA, USA) of the cloned DNA templates to sequence the same V4 region of the 18S rRNA gene as the high-throughput sequencing. For R2, since the Moon-A–Moon-B amplification was deemed unnecessary, the initial amplification was conducted directly using the TAReuk primers (using the procedure from Stoeck et al. [31] and an annealing temperature of 45 °C for the last 25 cycles). PCR products were cloned as described for R1 and sequenced from the flanking M13F and M13R priming sites on the pCR2.1-TOPO vector.
Microscopy
On every sampling date, two 20 ml replicate seawater samples were preserved with glutaraldehyde (0.5%, final concentration), stained with 4,6-diamidino-2-phenylindole (DAPI) and vacuum filtered onto 2 µm polycarbonate filters (Spectrum). Filters were applied to a glass microscope slide with Immersol immersion oil (Zeiss, Oberkochen, Germany) and stored in the dark at −20 °C. Twenty pre-programmed random automated positions were counted on each slide using a Zeiss Axiovert 200 M inverted compound microscope. Grazers were identified as cells that demonstrated DAPI fluorescence within a defined nuclear region, a clear cell shape under Fluorescein Isothiocyanate filter settings, and localized phycoerythrin fluorescence (blue light, 488 nm) suggesting consumption of one or more Synechococcus cells.
Synechococcus enrichment experiments
On 29 July 2014, six flasks of 600 ml freshly collected seawater were placed in a rooftop incubator filled with running seawater at ambient temperature. The flasks were kept below the surface of the water using ring-weights and placed under shaded mesh to prevent photo-degradation of cells. Cultured representatives from four Synechococcus clades (clade I: CC9311, clade II: CC9605, clade III: WH9102, clade IV: CC9902) were spun down, re-suspended in F/4 culture media [41], and added to the seawater at a final concentration of 1 × 106 cells ml−1. After 5 days, 250 ml of seawater from each of these incubations was filtered, extracted for DNA, and 454-sequenced as described above.
Synechococcus feeding experiments
A Tetraselmis strain was isolated and cultured from a surface water sample collected at the Scripps Pier in January 2013. Mixtures of this Tetraselmis and Synechococcus cells were established and observed for ingestion and calculation of ingestion rates. More information on these feeding experiments is provided in S4.
Results
Eukaryotic community diversity
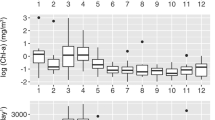
Eukaryotic diversity was assessed at 17 time-points during a 2-year span (Table 1). A total of 67,295 DNA sequences were obtained after quality trimming and alignment. Although multiple approaches are possible for interpreting the ecological implications of environmental sequence data [42, 43], we employed average neighbor clustering methods to analyze the taxa (OTUs) in our samples. The 17 samples together contained 6945 OTUs (before rarefaction) when using a 97% sequence identity cutoff to assign approximate “species-level” OTUs. Hereafter, “OTUs” will refer to those assigned at 97% cutoff unless otherwise specified, and the number in the name of an OTU reflects its overall abundance (OTU 1 refers to the most abundant OTU and so on). See Supplementary Table (S8). Rarefaction curves of each sample indicate that few samples were “saturated” at the level of sub-sampling, suggesting that greater sequencing depth would be required to represent the entire microbial community for most dates (Fig. 1a). In total, approximately 0.6% of OTUs were “abundant” (comprising 1% or more of total sequences), whereas 68% were “rare” (fewer than 0.01% of total sequences). A wide range of taxonomic diversity was still present in the rare sequences, although a higher proportion were “unclassified” (Fig. S5). After sub-sampling to the least common number of sequences (n = 1308) at each time-point, the total number of OTUs across all time-points was 2110. The estimated species richness on each date (using the Chao1 indicator) was highly variable, ranging from 80 to 961 OTUs (Fig. 1b).
a Rarefaction curves for each of the samples (with OTUs assigned at a 97% similarity cutoff). Vertical line represents level of sub-sampling that was used for subsequent analysis. b Estimated number of OTUs, based on the Chao1 richness indicator, present at each time-point. c Canonical correspondence analysis of communities and selected environmental variables. d Dendrogram clustering communities at each sampling point based on Bray–Curtis dissimilarity
Relating sequences to eukaryotic taxa requires classification of OTUs; we did this using mothur [32] and the Silva v123 database [35]. The eukaryotic community composition was temporally dynamic, with alveolates consistently comprising the largest proportion of the classified sequences (Fig. 2a). Alveolates are known to have high copy numbers of the 18S amplicon [44], so it is unclear whether their prominence in environmental sequence data can be related to their abundance. We therefore focus on changes in the relative (rather than the absolute) abundance of OTUs in our data. Although representatives from all other major protistan lineages were present at nearly every time-point, their relative abundances varied throughout the sampling period. A large portion of the sequences and OTUs were unclassified, even at the highest taxonomic levels, suggesting a large amount of novel eukaryotic diversity. The sampling dates that were most similar based upon five variables (chlorophyll, Chao1 diversity, temperature, grazer density, and Synechococcus density), as determined by canonical correspondence analysis (Fig. 1c), did not necessarily cluster together based on the similarities of their eukaryotic communities (Fig. 1d). The microbial eukaryotic communities that were sampled during Synechococcus blooms and peaks in grazer abundance were slightly more similar to one another than they were to the communities at other time-points, as demonstrated by Bray–Curtis dissimilarity (Fig. 1d).
Alveolates also dominated the phylogenetic diversity, represented as the proportion of total OTUs (not sequences) within each taxonomic group (Fig. 2b). Some other groups had a relatively low abundance of overall sequence numbers, but high phylogenetic diversity within the group. For example, on average picomonads were represented by 3% of total OTUs but only 0.2% of the total number of sequences. The relative richness of OTUs (number of OTUs per higher-level taxonomic group) was temporally stable compared with the proportional representation of sequence-types to the total sequence count. For example, although the relative proportion of alveolate sequences ranged widely between time-points (Fig. 2a), the number of alveolate OTUs was consistently around a quarter of total OTUs (Fig. 2b). The most abundant alveolates included dinoflagellates that have been previously found be ecologically important at the Scripps Pier, including Prorocentrum, Lingulodinium, and Akashiwo (SCCOOS, http://www.sccoos.org/; [15]).
Individual OTUs varied greatly in their temporal distributions. Of the 20 most abundant OTUs overall, 9 were present on at least 15 sampling dates. The dinoflagellates Heterocapsa rotundata (OTU 3) and Scrippsiella sp. (OTU 5) were consistently among the most abundant OTUs. Other genera that were both abundant and consistently present included Gyrodinium (OTUs 13 and 16), Tetraselmis (OTU 19), a diatom (Mediophyceae, OTU 20), and two prasinophytes (Ostreococcus, OTU 6, and Micromonas, OTU 12). These may comprise a “core community” of phytoplankton species, ranging over several major taxa. Other highly abundant OTUs were present more sporadically among the different time-points. Some, such as Akashiwo sanguinea (OTU 1) were likely bloom formers, observed abundantly on only one or several dates.
Choice of a cutoff for species-level identification
The clustering methods used in this article require pre-determined sequence similarity cutoffs. The cutoff value chosen for approximate species-level distinctions had an important effect on the resulting community characterization; at lower cutoffs, sequences from multiple OTUs clustered together, “absorbing” some of the rarer sequences that were assigned to discrete OTUs at higher cutoff values. We examined the clustering of sequences into the most abundant OTUs at different cutoffs to see whether they maintained consistent temporal patterns and found examples of three cases: some OTUs were indeed consistent regardless of cutoff (Fig. 3a). In a second case, a group of copepod sequences clustered together as OTU 2 at 97%, but separated into at least three OTUs with distinct abundance patterns at 99% (Fig. 3b). In a third case, sequences that had similar classifications and abundance patterns were assigned to different OTUs at 99%, such as OTUs 18 and 28, both classified as Neoceratium (Fig. 3c). Additional temporal sampling may provide evidence for separating these taxa. The interspecific 18S variation between copepod species (following the reproductive species concept) has been found to range from 1 to 6% [45]. As dynamics of three distinct copepod groups (Fig. 3b) were seen at 99% but not at 97%, examining temporal patterns at different cutoffs may be a useful method for determining potential species within taxa—and may be warranted especially when the focus of the study is a single taxonomic group. Within the top 25 OTUs defined by at a similarity of 99%, five OTUs with distinct temporal patterns (three copepod and two rhizarian) appeared to be inappropriately joined into two OTUs at 97% (Figure S6).
Examples of OTU abundances throughout the time-series, with OTUs at 99% (left) and 97% (right) that presumably represent the same organism. Leftmost graphs show an OTU or multiple OTUs at 99%, whereas rightmost graphs overlay the sum of the OTU abundances from the left graph (blue) with a corresponding OTU at 97% (green). a A case where an OTU remains consistent at both cutoffs. b A case where three separate OTUs with distinct temporal patterns are assigned at 99%, but the sum of their abundances is roughly equal to the abundance pattern of a similarly classified OTU at 97%. In this case, we assume the OTUs assigned at 99% represent different species due to their very different abundance patterns, and use of a 97% cutoff masks this diversity. c A case where two separate OTUs with very similar temporal patterns are assigned separately at 99% but the sum of their abundances closely matches a single OTU as assigned at 97%. In this case it appears that use of a 99% cutoff creates “false diversity.” The discrepancy in abundance between OTU 8 and the sum of OTUs 18 and 28 is due to many other sequences being “absorbed” into OTU 8 when clustered at 97% (color figure online)
Synechococcus spp. and grazer abundance at the Scripps Pier
The abundances of Synechococcus and their grazers at the Scripps Pier were temporally dynamic and exhibited blooms and crashes that were detectable within the weekly measurements (Fig. 4a). Although the Synechococcus population generally remained below 2 × 105 cells ml−1, their abundance exceeded this density on nine dates (2011: 7 April, 14 April, 21 April, 19 May, and 7 July; 2012: 19 January, 10 May, 21 May, and 2 August). Of these, three were dates when the abundance of Synechococcus more than doubled within a week, and three were dates when a bloom decreased by more than half within a week. The fluctuations in abundance of Synechococcus did not correlate significantly with temperature (2011: Pearson’s r = 0.23, p-value = 0.087; 2012: Pearson’s r = 0.27, p-value = 0.05) or chlorophyll (2011: Pearson’s r = 0.10, p-value = 0.483; 2012: Pearson’s r = −0.01, p-value = 0.957). Synechococcus grazers (as detected with intracellular prey by fluorescence microscopy) were maximally abundant on eight dates (2011: 10 May, 7 July, 21 July, 25 August, and 15 September; 2012: 29 May, 21 June, and 26 July). The grazer peaks on 7 July 2011, 25 August 2011, and 2 August 2012 corresponded with peaks in Synechococcus abundance. Dates identified as crashes of Synechococcus blooms (Figs. 1, 4a) were those in which Synechococcus abundance declined by approximately fourfold (or greater) within the previous 2 weeks. Synechococcus abundance only had a weak significant correlation with grazer abundances (Pearson’s r = 0.26, p < 0.05).
a Synechococcus and Synechococcus grazer density in cells/ml in 2011 and 2012. Synechococcus cells were counted using flow cytometry; grazers were counted using epifluorescence microscopy as described in methods. Marked time-points (blue circles) show dates where 18S environmental sequences were analyzed. Chlorophyll (green triangles) and temperature (red dots) are shown along with Synechococcus abundances. Synechococcus abundance exceeded 2 × 105 cells ml−1 on nine dates (2011: 7 April, 14 April, 21 April, 19 May, 7 July; 2012: 19 Jan, 10 May, 21 May, and 2 August). Stars denote dates that have both Synechococcus and grazer blooms. b Epifluorescence microscopy image of a single Tetraselmis cell isolated from the SIO pier. Scale bar = 5 μm. c Image of a Tetraselmis cell that has been fed Synechococcus. Arrow points to Synechococcus cells that have been ingested by the Tetraselmis cells. d, e Examples of flow cytometry plots from a date with novel cell populations (R1 and R2, circled), 2 August 2012 (d), compared with a more “typical” date, 29 May 2012 (e). The blue region marks the control beads, red region denotes putative cryptophytes, and the orange region represents the Synechococcus population (color figure online)
Identifying putative grazers of Synechococcus
Some of the increases in grazer populations that were identified by microscopy corresponded with a novel autofluorescent population observed in flow cytometric plots on several dates, most prominently 25 August 2011 and 2 August 2012 (Fig. 4d, e). This population appeared near the photosynthetic picoeukaryotic algae gating region but had elevated phycoerythrin (relative orange fluorescence) signal. We hypothesized that this indicated consumption of Synechococcus by a photosynthetic species, and thus possible mixotrophy. To identify these populations from 2 August 2012, we made 18S rRNA gene clone libraries from small collections of cells that were flow sorted from gating three regions R1 and R2 (Fig. 4d; S3). Region R1 contained mostly dinoflagellate sequences, particularly Gymnodinium sp., which matched a highly abundant Gymnodinium OTU from the dataset (OTU 23). This OTU was present at the Scripps Pier throughout the time-series (Table 2), although its highest abundance was during the week after the Synechococcus bloom. Region R2 contained prasinophyte cells, mainly of the genus Tetraselmis, most of which matched the sequence from OTU 37 (Table 2). This OTU was only present on 25 August 2011 and 2 August 2012, in high abundances, indicating it may be a summer bloom former.
A Tetraselmis species isolated from the Scripps Pier, which corresponded with OTU 19 (distinct from OTU 37 from region R2 above), was observed to ingest CC9311 Synechococcus cells when they were provided as a food source (Figs. 4b, c). Ingestion rates of Synechococcus by Tetraselmis increased with increasing Synechococcus concentration (Fig. S7). Fitted to a Michaelis–Menten equation (S4, Eq. 2), the maximum ingestion rate (MIR) and KIR (the prey concentration sustaining 1/2 MIR) of Tetraselmis on Synechococcus were 0.18 Synechococcus cells grazer−1 h−1 and 1.5 × 107 cells ml−1, respectively (S4, Eq. 2). The maximum clearance rate was 0.13 nl grzaer−1 h−1. A strain of Gymnodinium was isolated from the Scripps Pier that clustered with OTU 23 and was observed to consume Synechococcus cells (Yeong Du Yoo, personal communication). Given the multiple lines of evidence (cell sorting, sequencing and culture work) it is possible that strains of both Gymnodinium and Tetraselmis could be mixotrophic grazers of Synechococcus at this sampling station, presumably under very specific environmental conditions.
To further investigate putative grazers, we enriched a natural plankton community from the Scripps Pier with representatives of four Synechococcus clades and sequenced the community after 5 days. This resulted in a different shift in the community composition of microbial eukaryotes in each of the samples (compared with a control that was not enriched with Synechococcus). We are aware that “surface” effects alone may cause significant shifts in the diversity of the protistan assemblage [46]. The number of OTUs found within each community ranged from 158 to 237 with only 44 OTUs common to all the samples. Although our experimental treatments consisted of single enrichments for each of the Synechococcus clades (except for a duplicate CC9311 treatment) we observed several OTUs that experienced noticeable and consistent (including between replicates) shifts in their relative abundance compared with the un-enriched community. There was an increase in the abundance of several dinoflagellates (members of Gymnodiniphyceae and Syndiniales) and one stramenopile (Fig. 5). Notably, the Gymnodinium OTU with the greatest increase addition also matched with OTU 23, the OTU that was identified in the bulk sorts—providing another line of evidence that this species may be an important Synechococcus grazer at this sampling station. The relative abundance of the only abundant Tetraselmis OTU, which did correspond with the bloom-former OTU 37, declined in all treatments.
Changes in sequence abundance of selected OTUs in seawater enriched with four different Synechococcus species with respect to un-enriched control. OTUs shown had the largest overall increases or decreases in abundance compared with the control. Starred Gymnodiniphycidae OTU corresponds with the Gymnodinium OTU found in the R2 flow cytometry region from 2 August 2012. SAR Superkingdom of Stramenopiles, Alveolates, Rhizarians. Patterns represent the Synechococcus strain added to the enrichment
We further examined Gymnodinium-like species diversity due to its presence during Synechococcus blooms and because it has been demonstrated previously to exhibit mixotrophy in culture [28, 47, 48]. Phylogenetic trees of pier sequences that classified within Gymnodiniphycidae included a number of OTUs, which clustered with the 18S sequences from two Scripps Pier Gymnodinium-like species isolates (Fig. 6). These isolates contributed to the clade Suessiaceae and may be putative grazers of Synechococcus, given that members of this clade appear to be abundant on the two August 2012 dates during which the R2 population appears and disappears (Fig. 4d). Several other Gymnodiniphycidae clades, including Gyrodinium sp., did not cluster with any known mixotroph sequences within the tree, although this does not rule them out as possible Synechococcus grazers.
Consensus maximum-likelihood tree of Gymnodiniphycidae sequences found at pier, as well as sequences of dinoflagellate lab isolates. Colors represent four taxonomic groups as described in the SILVA database, Suessiaceae (pink), Gyrodinium (yellow), and two other separately designated clades (green and blue). Arrows point to most abundant OTUs (starred) and show their abundance patterns over time. Dates 1–17 correspond with dates from other figures: in order, 14 Apr 2011; 5 May 2011; 7 Jul 2011; 21 Jul 2011; 25 Aug 2011; 15 Sept 2011; 17 Nov 2011; 29 Dec 2011; 26 Jan 2012; 1 Mar 2012; 10 May 2012; 29 May 2012; 14 Jun 2012; 21 Jun 2012; 2 Aug 2012; 11 Aug 2012; 25 Oct 2012 (color figure online)
Environmental relevance of cultured protists
We evaluated the ecological relevance of protistan strains isolated as part of other projects from the Scripps Pier by aligning the 18S rRNA gene sequences that were previously obtained from these strains (Yeong Du Yoo, Brian Palenik, Javier Paz-Yepes, unpublished data) to those from our pier time-series (Table 2). Nine strains clustered with sequences within the 100 most abundant OTUs, whereas two were extremely rare. The two poorly represented OTUs were both known heterotrophic nanoflagellate grazers (Pteridomonas and Paraphysomonas). In contrast, a cercozoan isolate from the Scripps Pier that grazes on Synechococcus (Paz-Yepes, manuscript in preparation) was more common, corresponding to OTU 262. The nine isolated strains that were common OTUs represented dinoflagellates, raphidophytes, and chlorophytes. Two species were isolated during blooms (OTU 1, Akashiwo sanguinea, and OTU 37, a Tetraselmis species) were indeed bloom formers, present only at certain sampling times but in high abundances (Table 2).
Discussion
We have characterized the dynamics of the eukaryotic microbial community at the Scripps Pier to explore overall diversity and look into the factors driving Synechococcus ecology, which are not well understood at this site. Our sequencing-based approach uncovered eukaryotic species richness at the Scripps Pier at least 10-fold greater than that described decades ago using microscopy [21]. This corresponds with a similar theme worldwide; the 2015 TARA Oceans expedition conducted high-throughput sequencing at 47 sites [5] and estimated that there are approximately 150,000 eukaryotic species in the marine photic zone globally. Like the TARA data, we found that a large proportion of our sequences remained phylogenetically unclassified; the fourth most abundant OTU at the Scripps pier, present only during grazer blooms (7 July 2011, 21 July 2011, 25 August 2011, 21 June 2012), was unclassifiable beyond Eukaryota. Because microbial eukaryotes are not particularly well-represented in comparative DNA databases, it is unclear how these unclassified OTUs contribute to the coastal ecology and will require further sequencing efforts or probing with directed molecular techniques.
Despite the large range of species richness between time-points, the relative phylogenetic community structure within higher-level taxonomic designations was fairly stable, a trend that was also found between different sites in Taib et al. [49]. There does appear to be a “core community” of phytoplankton OTUs (dinoflagellates, diatoms, and prasinophytes) that are present abundantly most of the time, and many of these OTUs correspond to species recorded in past work [15, 21, 50]. However, there are many OTUs that appear to be bloom formers: sparse or absent at most time-points but highly abundant at a few. External drivers, including seasonal changes, nutrient inputs, and water transport, are likely to play a role in the varied levels of species diversity on different dates, but the causes for most blooms remain elusive. For example, although Akashiwo sanguinea’s dominance in the sequence data on 29 Dec 2011 was corroborated by cell counts and high chlorophyll measurements through SCCOOS, it did not correspond with any abnormal temperature, salinity, or nutrient concentration, nor did it happen during a time of year when upwelling typically occurs. Many OTUs appeared only once or twice throughout the time-series. These “conditionally rare taxa” may have important but yet-unknown functional roles in the community [51,52,53].
These results highlight the importance of temporal data in a field dominated by comparisons of spatially distinct communities sampled at a single time-point. Only three OTUs from the Scripps Pier were present throughout all time-points, but even these were not consistent in their relative temporal abundances. Thus, even if the goal is characterizing only the most abundant species, our data suggest that a single time-point is not sufficiently representative. Previous work has demonstrated rapid and large-scale changes in the composition of protistan communities during short-term experiments [46] and it is important for such shifts to be documented for an established time-series study at high temporal resolution. One instance of a dramatic shift in the eukaryotic community was observed between 2 August 2012 and 11 August 2012. The former was one of the most species-rich dates (>800 estimated OTUs) and had a Synechococcus bloom concurrent with an increase in grazers (observed by microscopy). By 11 August 2012, both Synechococcus and grazer abundances declined more than fivefold and the number of OTUs had decreased by an order of magnitude. These large changes in diversity and community composition within nine days highlight the need to explore time-series data at greater temporal resolution to better characterize boom-and-bust cycles. Additionally, it is presumed that there would be a time-lag between a bloom of a given species and a detectable response in a trophically-related species, but this might only be detected with higher-frequency sampling.
The OTUs described in this study are based upon a specific method of clustering sequences (average neighbor joining with a 97% similarity cutoff). Numerous other methods have been developed to accomplish the goal of converting sequence data into ecologically meaningful information about species dynamics [34, 43]. Previous studies have proposed sequence similarity cutoffs ranging from 95 to 99.5%, and it appears increasingly likely that the “correct” species-level cutoff might differ between higher-level taxa [6, 12, 54, 55]. We found that examining temporal patterns of OTUs at different cutoffs may be useful in evaluating how well that cutoff represents a specific population. We assumed that if two OTUs with similar taxonomic classifications also had highly similar temporal abundance patterns, the cutoff at which they clustered together might be appropriate for identifying species or ecotypes, and did find this to be the case for some OTUs at 97%. Moving forward, temporal information may aid in the definition of meaningful units of taxonomic classification by providing significantly greater coverage for any single OTU and thus justifying when taxa are split into multiple OTUs.
Another objective of this work was to closely track the Synechococcus population and search for factors underlying Synechococcus blooms and declines, with a focus on identifying grazers using both sequence data and other lines of evidence. At other locations, patterns of temperature, stratification, and nutrient concentrations are more reliably coincident with Synechococcus abundance [23]. However, at the Scripps Pier no significant correlations with temperature or chlorophyll (a proxy for overall phytoplankton biomass) were observed in 2011–2012, and the Synechococcus population did not show a strong seasonal pattern as is observed at many other sites [22, 56, 57]. This might be because of the relatively small temperature range at this site (13–23 °C) compared with others; additionally, Synechococcus has been found to only reliably correlate with temperature below 14 °C [58]. It is also possible that our higher sampling frequency captures variation that is present but undetected at other sites.
Oceanographic features and climate events surely impact the community in many ways, but the available measurements do not explain all large community shifts. Longer-term forcing factors in the Southern California Bight include the El Nino Southern Oscillation, the Pacific Decadal Oscillation, and the North Pacific Gyre Oscillation [59]. The Scripps Pier is influenced by the equator-ward flow of the California Current and the pole-ward flow of the California Undercurrent, among others. The latter weakens in the spring and leads to seasonal variation in salinity, stratification, and nutrient availability [60]. Upwelling events typically occur during the spring and summer [61], but previous work at this site has not implicated coastal upwelling in explaining chlorophyll variations [62]. Much of the water movement at the Scripps Pier is attributed to wind-driven currents and this may be explanatory for some of the variations in microbial diversity. Given the lack of strong connections between Synechococcus abundances and oceanographic factors, we sought to explore the role of biotic factors such as grazing.
We detected a high incidence of Gymnodinium sequences during a Synechococcus grazer bloom (in both the amplicon data and clone libraries made from the “R2” flow cytometry sort region). The abundant Gymnodinium OTU sequence clustered with that of a Gymnodinium species known to ingest Synechococcus cells, and [28, 47] have suggested that feeding might be a driving force for bloom formation in some dinoflagellates. Gymnodinium appears to have a grazing threshold for Synechococcus; significant ingestion is only observed when prey density exceeds 20 µg C l−1 [48]. Assuming 0.294 pg C Synechococcus cell−1 [48], the Synechococcus concentration on 2 August 2012 was 67.7 µg C l−1, a condition that could induce grazing.
We also found an unexpected association between Tetraselmis and Synechococcus during a bloom period. Tetraselmis OTU 37 was found in an unusual region of our flow cytograms (“R1”) on dates of a widely observed “Green Bloom,” and its 18S sequence matched that of a Tetraselmis isolate collected during one of these blooms. Other dates had different Tetraselmis OTUs present but lacked the novel population in the flow cytograms. Given that at least one Tetraselmis species isolated at the Scripps Pier (OTU 19) has been observed to ingest Synechococcus when it is presented at high density, it is possible that some Tetraselmis species do exhibit conditional mixotrophy at our site. However, we did not see an increase in Tetraselmis OTU sequences in an experiment where seawater was enriched with Synechococcus strains, despite OTU 37 being present, as might be expected if Synechococcus was being used as a food source. However, it is impossible to replicate the exact nutrient, temperature, and microbial community conditions that were present on the dates of the “Green Blooms” so this does not eliminate the possibility of Tetraselmis mixotrophy. We recognize that the unconventional “R1” flow cytometry population could also have been a result of physical attachment between Tetraselmis cells and Synechococcus as has been occasionally observed when they are co-cultured (Palenik lab, unpublished data), but further work is necessary to confirm or reject either hypothesis. Regardless, mixotrophy is not always considered in traditional food web schemes, but the Gymnodinium and Tetraselmis findings suggest it may play a role in consumption of Synechococcus at this sampling site.
Another unexpected finding was that, although the 18S sequences of many cultured dinoflagellates and green algae (isolated from the Scripps Pier) clustered with abundant OTUs, the well-characterized Synechococcus grazers Pteridomonas and Paraphysmonas did not, even at the relatively low sequence similarity threshold of 95%. This could indicate that efforts to isolate heterotrophic grazers select for taxa that are not dominant environmentally. It is also possible that their sequence signal was drowned out by that of organisms with higher rRNA gene copy numbers or some other DNA amplification bias [63]. In reviewing various public sequence databases, Del Campo and Massana [64] observed that chrysophytes such as Paraphysmonas, which are observed under the microscope frequently, were not found as commonly in sequence data. This is an important consideration for future work that uses amplicon sequencing to explore these types of trophic interactions.
We have described the changing microbial community at the Scripps Pier over 2 years and found it to be diverse and highly dynamic, with only a few species consistently present. Amplicon sequencing captured blooms of certain microbial eukaryotes. Our results clearly emphasize the importance of temporal studies in describing microbial communities.
References
Pomeroy LR. The ocean’s food web, a changing paradigm. Bioscience. 1974;24:499–504.
Waterbury JB, Watson SW, Guillard RR, Brand LE. Widespread occurrence of a unicellular, marine, planktonic, cyanobacterium. Nature. 1979;277:293–4.
Fenchel T. Ecology of heterotrophic microflagellates. IV. Quantitative occurrence and importance as bacterial consumers. Mar Ecol Prog Ser. 1982;9:35–42.
Potter D, Lajeunesse T, Saunders G, Anderson R. Convergent evolution masks extensive biodiversity among marine coccoid picoplankton. Biodivers Conserv. 1997;107:99–108.
De Vargas C, Audic S, Henry N, Decelle J, Mahé F, Logares R, et al. Eukaryotic plankton diversity in the sunlit ocean. Science. 2015;348:1261605.
Massana R, del Campo J, Sieracki ME, Audic S, Logares R. Exploring the uncultured microeukaryote majority in the oceans: reevaluation of ribogroups within stramenopiles. ISME J. 2014;8:854–66.
Moon-van der Staay SY, De Wachter R, Vaulot D. Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature. 2001;409:607–10.
Not F, Latasa M, Scharek R, Viprey M, Karleskind P, Balagué V, et al. Protistan assemblages across the Indian Ocean, with a specific emphasis on the picoeukaryotes. Deep-Sea Res Pt I. 2008;55:1456–73.
Seenivasan R, Sausen N, Medlin LK, Melkonian M. Picomonas judraskeda Gen. Et Sp. Nov.: the first identified member of the Picozoa Phylum Nov., a widespread group of Picoeukaryotes, formerly known as “Picobiliphytes”. PLoS ONE. 2013;8:e59565.
Cheung MK, Au CH, Chu KH, Kwan HS, Wong CK. Composition and genetic diversity of picoeukaryotes in subtropical coastal waters as revealed by 454 pyrosequencing. ISME J. 2010;4:1053–9.
Cleary AC, Durbin EG. Unexpected prevalence of parasite 18S rDNA sequences in winter among Antarctic marine protists. J Plankton Res. 2016;38:401–17.
Collado-Fabbri S, Vaulot D, Ulloa O. Structure and seasonal dynamics of the eukaryotic picophytoplankton community in a wind-driven coastal upwelling ecosystem. Limnol Oceanogr. 2011;56:2334–46.
Countway PD, Vigil PD, Schnetzer A, Moorthi SD, Caron DA. Seasonal analysis of protistan community structure and diversity at the USC microbial observatory (San Pedro Channel, North Pacific Ocean). Limnol Oceanogr. 2010;55:2381–96.
Massana R, Balagué V, Guillou L, Pedros-Alio C. Picoeukaryotic diversity in an oligotrophic coastal site studied by molecular and culturing approaches. FEMS Microbiol Ecol. 2004;50:gp231–243.
Goericke R. The structure of marine phytoplankton communities- patterns, rules and mechanisms. Cal Coop Ocean Fish. 2011;52:182–97.
Agawin NSR, Duarte CM, Agustí S. Nutrient and temperature control of the contribution of picoplankton to phytoplankton biomass and production. Limnol Oceanogr. 2000;45:591–600.
Flombaum P, Gallegos JL, Gordillo RA, Rincón J, Zabala LL, Jiao N, et al. Present and future global distributions of the marine Cyanobacteria Prochlorococcus and Synechococcus. PNAS. 2013;110:9824–9.
Olson RJ, Chisholm SW, Zettler ER, Armbrust E. Pigments, size, and distributions of Synechococcus in the North Atlantic and Pacific Oceans. Limnol Oceanogr. 1990;35:45–58.
Partensky F, Blanchot J, Vaulot D. Differential distribution and ecology of Prochlorococcus and Synechococcus in oceanic waters: a review. Bull Inst Oceanogr. 1999;19:457–75.
Tai V, Palenik B. Temporal variation of Synechococcus clades at a coastal Pacific Ocean monitoring site. ISME J. 2009;3:903–15.
Allen WE. Occurrence of marine plankton diatoms in a ten-year series of daily catches in southern California. Am J Bot. 1936;23:60–3.
Agawin NSR, Duarte CM, Agustí S. Growth and abundance of Synechococcus sp. in a Mediterranean Bay: seasonality and relationship with temperature. Mar Ecol Prog Ser. 1998;170:45–53.
Moisan TA, Blattner KL, Makinen CP. Influences of temperature and nutrients on Synechococcus abundance and biomass in the southern Mid-Atlantic Bight. Cont Shelf Res. 2010;30:1275–82.
Taylor AG, Landry MR, Selph KE, Wokuluk JJ. Temporal and spatial patterns of microbial community biomass and composition in the Southern California Current Ecosystem. Deep-Sea Res PT II. 2014;112:117–28.
Worden AZ, Nolan JK, Palenik B. Assessing the dynamics and ecology of marine picophytoplankton: the importance of the eukaryotic component. Limnol Oceanogr. 2004;49:168–79.
Palenik BP. Picophytoplankton seasonal cycle at the SIO pier, La Jolla, California. J Phycol. 2000;36(s3):53–53.
Christaki U, Courties C, Karayanni H, Giannakourou A, Maravelias C, Kormas KA, et al. Dynamic characteristics of Prochlorococcus and Synechococcus consumption by bacterivorous nanoflagellates. Microb Ecol. 2002;43:341–52.
Jeong HJ, Park JY, Nho JH, Park MO, Ha JH, Seong KA, et al. Feeding by red-tide dinoflagellates on the cyanobacterium Synechococcus. Aquat Microb Ecol. 2005;41:131–43.
Apple JK, Strom SL, Palenik B, Brahamsha B. Variability in protist grazing and growth on different marine Synechococcus isolates. Appl Environ Microbiol. 2011;77:3074–84.
Zwirglmaier K, Spence E, Zubkov MV, Scanlan DJ, Mann NH. Differential grazing of two heterotrophic nanoflagellates on marine Synechococcus strains. Environmen Microbiol. 2009;11:1767–76.
Stoeck T, Bass D, Nebel M, Christen R, Jones MD, Breiner HW, et al. Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol Ecol. 2010;19(s1):21–31.
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–41.
Schloss PD, Gevers D, Westcott SL. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS ONE. 2011;6:e27310.
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200.
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2012;41:D590–6. gks1219
Pruesse E, Peplies J, Glöckner FO. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics. 2012;28:1823–9.
Jones E, Oliphant E, Peterson P, et al. SciPy: open source scientific tools for Python. 2001. http://www.scipy.org/ [online] Accessed 08 Aug 2017.
Oksanen J, Blanchet F, Kindt R, Legendre P, Minchin P, O’Hara R et al. Vegan: community Ecology Package. R package version 2.0-10. R Package version 1 (http://cran.r-project.org). 2013.
R Core Team. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL http://www.R-project.org/. 2013.
Collier JL, Palenik B. Phycoerythrin-containing picoplankton in the Southern California Bight. Deep-Sea Res PT II. 2003;50:2405–22.
Guillard RR. Culture of phytoplankton for feeding marine invertebrates. Culture of marine invertebrate animals. Springer Boston, MA, US;1975. p. 29–60.
Eren AM, Maignien L, Sul WJ, Murphy LG, Grim SL, Morrison HG, Sogin ML. Oligotyping: differentiating between closely related microbial taxa using 16S rRNA gene data. Methods Ecol Evol. 2013;4:1111–9.
Mahé F, Rognes T, Quince C, de Vargas C, Dunthorn M. Swarm: robust and fast clustering method for amplicon-based studies. PeerJ. 2014;2:e593.
Prokopowich CD, Gregory TR, Crease TJ. The correlation between rDNA copy number and genome size in eukaryotes. Genome. 2003;46:48–50.
Bucklin A, Frost B, Bradford-Grieve J, Allen L, Copley N. Molecular systematic and phylogenetic assessment of 34 calanoid copepod species of the Calanidae and Clausocalanidae. Mar Biol. 2003;142:333–43.
Countway PD, Gast RJ, Savai P, Caron DA. Protistan diversity estimates based on 18S rDNA from seawater incubations in the western North Atlantic. J Eukaryot Microbiol. 2005;52:95–106.
Jeong HJ, Du Yoo Y, Lee KH, Kim TH, Seong KA, Kang NS, et al. Red tides in Masan Bay, Korea in 2004-2005: I. Daily variations in the abundance of red-tide organisms and environmental factors. Harmful Algae. 2013;30:S75–S88.
Strom SL. Growth and grazing rates of the herbivorous dinoflagellate Gymnodinium sp. from the open subarctic Pacific Ocean. MEPS. 1991;78:103–13.
Taib N, Mangot JF, Domaizon I, Bronner G, Debroas D. Phylogenetic affiliation of SSU rRNA genes generated by massively parallel sequencing: new insights into the freshwater protist diversity. PLoS One. 2013;8:e58950.
Reid FM, Lange CB, White MM. Microplankton species assemblages at the Scripps Pier from March to November 1983 during the 1982-1984 El Nino event. Bot Mar. 1985;28:443–52.
Logares R, Mangot JF, Massana R. Rarity in aquatic microbes: placing protists on the map. Res Microbiol. 2015;166:831–41.
Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR, et al. Microbial diversity in the deep sea and the underexplored “rare biosphere”. PNAS. 2006;103:12115–20.
Caron DA, Countway PD. Hypotheses on the role of the protistan rare biosphere in a changing world. Aquat Microb Ecol. 2009;57:227–38.
Wolf C, Frickenhaus S, Kilias ES, Peeken I, Metfies K. Protist community composition in the Pacific sector of the Southern Ocean during austral summer 2010. Polar Biol. 2014;37:375–89.
Caron DA, Countway PD, Savai P, Gast RJ, Schnetzer A, Moorthi SD, et al. Defining DNA-based operational taxonomic units for microbial-eukaryote ecology. Appl Environ Microbiol. 2009;75:5797–808.
Hunter-Cevera KR, Neubert MG, Olson RJ, Solow AR, Shalapyonok A, Sosik HM. Physiological and ecological drivers of early spring blooms of a coastal phytoplankter. Science. 2016;354:326–9.
Shapiro LP, Haugen EM. Seasonal distribution and temperature tolerance of Synechococcus in Boothbay Harbor, Maine. Estuar, Coast Shelf Sci. 1988;26:517–25.
Li WK. Annual average abundance of heterotrophic bacteria and Synechococcus in surface ocean waters. Limnol Oceanogr. 1998; 43:1746–53.
Di Lorenzo E, Schneider N, Cobb KM, Franks PJS, Chhak K, Miller AJ et al. North Pacific Gyre Oscillation links ocean climate and ecosystem change. Geophys Res Lett. 2008;35:Article Number L08607.
Lynn RJ, Simpson JJ. The California current system: the seasonal variability of its physical characteristics. J Geophys Res: Oceans. 1987;92:12947–66.
Dorman CE, Palmer DP. Southern California summer coastal upwelling. Coastal Upwelling. American Geophysical Union Washington, DC, US;1981. p. 44–56.
Kim HJ, Miller AJ, McGowan J, Carter ML. Coastal phytoplankton blooms in the Southern California Bight. Prog Oceanogr. 2009;82:137–47.
Lim EL, Dennett MR, Caron DA. The ecology of Paraphysomonas imperforata based on studies employing oligonucleotide probe identification in coastal water samples and enrichment cultures. Limnol Oceanogr. 1999;44:37–51.
Del Campo J, Massana R. Emerging diversity within chrysophytes, choanoflagellates and bicosoecids based on molecular surveys. Protist. 2011;162:435–48.
Acknowledgements
Funding for this project was provided through a National Science Foundation grant (DEB-1233085) to BP, as well as National Research Foundation of Korea (NRF) grants funded by the Korea government (MSIP) (NRF-2015M1A5A1041808) award to YDY. We thank M Carter and M Hilbern for collecting access to the Scripps Pier Chlorophyll Program dataset, funded by NOAA and COTS through the Southern California Coastal Ocean Observing System (SCCOOS). We thank Mike Landry (SIO) for use of his automated microscopy system used for obtaining images for Synechococcus grazer counts.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Nagarkar, M., Countway, P.D., Du Yoo, Y. et al. Temporal dynamics of eukaryotic microbial diversity at a coastal Pacific site. ISME J 12, 2278–2291 (2018). https://doi.org/10.1038/s41396-018-0172-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-018-0172-3
This article is cited by
-
Dynamics of the Eukaryotic Microbial Community at the Breeding Sites of the Large Yellow Croaker Pseudosciaena crocea in the Southern East China Sea
Journal of Ocean University of China (2022)