Abstract
Phage–host interactions are critical to ecology, evolution, and biotechnology. Central to those is infection efficiency, which remains poorly understood, particularly in nature. Here we apply genome-wide transcriptomics and proteomics to investigate infection efficiency in nature’s own experiment: two nearly identical (genetically and physiologically) Bacteroidetes bacterial strains (host18 and host38) that are genetically intractable, but environmentally important, where phage infection efficiency varies. On host18, specialist phage phi18:3 infects efficiently, whereas generalist phi38:1 infects inefficiently. On host38, only phi38:1 infects, and efficiently. Overall, phi18:3 globally repressed host18’s transcriptome and proteome, expressed genes that likely evaded host restriction/modification (R/M) defenses and controlled its metabolism, and synchronized phage transcription with translation. In contrast, phi38:1 failed to repress host18’s transcriptome and proteome, did not evade host R/M defenses or express genes for metabolism control, did not synchronize transcripts with proteins and its protein abundances were likely targeted by host proteases. However, on host38, phi38:1 globally repressed host transcriptome and proteome, synchronized phage transcription with translation, and infected host38 efficiently. Together these findings reveal multiple infection inefficiencies. While this contrasts the single mechanisms often revealed in laboratory mutant studies, it likely better reflects the phage–host interaction dynamics that occur in nature.
Similar content being viewed by others
Introduction
Phage research has provided numerous fundamental biological discoveries over the last century. For example, in molecular biology, phage research helped to identify DNA as the hereditary material, elucidate how gene expression is regulated, and provide CRISPR-Cas genes that have revolutionized eukaryotic genome editing [1]. In ecology, phages are known to be abundant and to modulate microbial populations, ecosystem functioning, and evolution [2, 3]. Such importance has spurred recent sequencing and analytical advances to explore viral genomes across ecosystems ranging from soils, water, and plants to animals and humans [2, 3], which has also implicated phages in health and disease [4, 5].
Despite these advances, phage biology is hampered by several knowledge gaps, which prevent the incorporation of phages into predictive ecosystem models or their use in biotechnological applications. First, phage–host model systems exist for few phyla and environments [6,7,8,9,10], thus severely limiting our understanding of naturally occurring phage–host interactions. Second, inefficient infections in nature are woefully understudied, despite that phages might attach to any encountered host [11], not just one where infection is efficient. Such variable infection efficiency is especially relevant to broad host-range phages (generalists) given that, as predator–prey studies predict, they might infect more hosts than phages with narrow host ranges (specialists), but with the trade-off of reduced infection efficiency [2, 12, 13]. Third, common experimental approaches favor investigating efficient over inefficient lytic infections [14], the latter of which can be confused with lysogeny [15, 16]. Namely, phage–host range assays are measured more often as infection or not by, for example, observing phage-mediated cell clearing via spot tests [17, 18] than quantitatively. Yet, quantitative host ranges, i.e., efficiency of plating calculations from phage stock serial dilutions, enable evaluating hypotheses about ecological [19] and evolutionary [20] population units. As quantitative host ranges detect the gradient of infection efficiencies that occur in nature [14, 21] they provide, in turn, key data needed to parameterize predictive models [22, 23]. Additionally, knowledge of the molecular factors driving infection efficiency is constrained, as data largely derives from laboratory-induced mutations [24,25,26,27] and experiments that detect extreme phenotypes (e.g., failure to lyse) and assess single stages of the phage–host interaction. Such approaches are useful in the laboratory, but they are unlikely to capture naturally occurring subtleties in phage–host interaction variability, where many genetic changes would be accumulated within any given population over time.
Fortunately, ‘omics technologies (e.g., genomics, transcriptomics, proteomics, and metabolomics) can together provide simultaneous insight into all the stages of phage–host interactions and help uncover novel mechanisms—including drivers of infection efficiency—in environmentally diverse and less-well studied model systems. To date, population-wide transcriptomics studies have uncovered novel phage genes and regulatory RNAs [28,29,30,31], host-independent transcription of giant phages [28, 32], and universalities of phage transcription regardless of the host [28,29,30,31,32,33,34,35,36,37]. Additionally, metabolomics has illuminated how phage metabolic reprogramming can vary from phage to phage on the same host and be independent of transcriptional changes [29, 38]. However, similar quantitative, temporally resolved proteomics studies are lacking, and together with the scarcity of mechanistic knowledge of infection efficiency, numerous key questions remain unanswered: (1) What are the temporal dynamics of phage and host proteins during infection, how are they related to transcription, and how does this vary with infection efficiency?; (2) Do phages takeover the host proteome? If yes, how and when, and how might this vary across phage–host systems of varied infection efficiency?; (3) What are host defenses against phage proteins and how do they alter phage protein dynamics?
To address such knowledge gaps and specific research questions we chose to let “nature’s experiment” (i.e., phage infection across naturally occurring host genetic variation) guide our understanding of infection efficiency by assessing it an environmental model system—the Bacteroidetes bacterium Cellulophaga baltica and its phages [8]. Bacteroidetes bacteria are ubiquitous across diverse environments ranging from soils and water to humans; impacting nutrient turnover, and health and disease [8, 39, 40]. In the oceans, the Bacteroidetes are a dominant phylum and contain bacteria that associate with particles and constitute a nucleus for phage infection [39, 41]. To develop a model phage–host system, 12 novel phage genera were isolated on 21 C. baltica strains and then they were whole-genome sequenced, characterized in morphology, as well as in quantitative host range, in structural proteomics and in infection dynamics [8, 21, 42]. Among the phages, phi38:1 is a podovirus and belongs to one of the most abundant viral candidate genera in the global oceans [43] and is a generalist that infects 13 of 21 C. baltica strains tested. Another co-isolated phage, phi18:3, also a podovirus, represents a rare group in the global oceans [43] and is a specialist that infects its host of isolation [44], strain #18 (here “host18”) [21]. While both phages are virulent, infection efficiency for the generalist phage (phi38:1) drastically differs across two genetically nearly identical (i.e., 100% identical 16S rRNA [44]); >99.9% nucleotide identity across 93% of their genes [30] hosts that lack CRISPR-Cas genes [30]. Specifically, phi38:1 is more efficient (75-min latent period (LP)) on its original host of isolation (strain NN016038; here “host38”) than on alternative host18 (LP of 2.5 h detected via phageFISH or 11 h via plaque assays [15]). Previous experimental measurements and population-based, genome-wide transcriptomics revealed that host18 defenses contributed to this infection inefficiency by delaying phi38:1’s DNA replication and virion production [30], leading to the hypothesis that host18 impacts either phage protein production and dynamics or protein abundance.
Here we test this hypothesis and address the aforementioned questions to further assess mechanisms driving naturally occurring infection inefficiencies. We build upon our prior work [30] by adding (1) time-resolved, population-based, genome-wide proteomics data in both phi38:1 infections, and (2) a comparable transcriptomic–proteomic dataset of the efficient infection by phi18:3 on host18. Together, these efforts provide a window into the multistage mechanisms that drive infection efficiency in naturally occurring phage–host interactions, and thereby get beyond traditional, single-target mechanistic approaches to better reveal how phages infect their hosts in nature.
Materials and methods
Data availability
New RNA-Seq reads are available in the ArrayExpress database (https://www.ebi.ac.uk/arrayexpress/) under accession #E-MTAB-5448. Proteomics raw data have been deposited to the ProteomeXchange Consortium via the PRIDE [45] partner repository with the dataset identifier PXD005440 and 10.6019/PXD005440. Also available are the transcriptomics read counts and all analyses for transcriptomics and proteomics (Supplementary Dataset S1; http://datacommons.cyverse.org/browse/iplant/home/shared/iVirus/Cellulophaga_Omics/Howard-Varona_2018_ISMEJ_transcriptomics_and_proteomics), and the scripts used [30] (http://datacommons.cyverse.org/browse/iplant/home/shared/iVirus/Cellulophaga_Omics/Howard-Varona_2016_ISMEJ_transcriptomics).
Bacterial and phage genomes
Bacterial isolates C. baltica strains #18 (“host18”) and NN016038 (“host38”) and phages phi38:1 and phi18:3 used in this study were previously isolated and characterized [8, 15, 21, 44]. Host38 was isolated in the year 1994 [46] and the phages and host18 were isolated together in the year 2000 [44], all from surface waters of the Baltic Sea. The genomes are publicly available: C. baltica #18 (available at GenBank #CP009976 and refined at http://datacommons.cyverse.org/browse/iplant/home/shared/iVirus/Cellulophaga_Omics/Cellulophaga_baltica_strain_18/, which was here used), C. baltica NN016038 (GenBank #CP009887.1), phi18:3 (Genbank #NC_021794.1), and phi38:1 (available at GenBank #NC_021796.1 and refined at http://datacommons.cyverse.org/browse/iplant/home/shared/iVirus/Cellulophaga_Omics/141114_phi381_start33820bp.gb/ file “141114_phi381_start33820bp.gb”, which was here used). Improved annotations for all are in Supplementary Dataset S2 (http://datacommons.cyverse.org/browse/iplant/home/shared/iVirus/Cellulophaga_Omics/Howard-Varona_2018_ISMEJ_transcriptomics_and_proteomics). Phage promoter and terminator sequences were predicted using BProm [47] and TransTerm [48], respectively, and manually visualized and assessed within the Artemis genome browser [49]. Annotated genome maps were produced using EasyFig [50].
Phage–host manipulations, adsorption kinetics, and one-step growth curves
Cell growth and phage infections, including those to measure adsorption kinetics, were conducted as previously described [21, 30]. Briefly, cells were grown in Marine Luria Bertani medium to a density of 108 cells ml−1 in early logarithmic stage. To measure the adsorption of phi18:3 to C. baltica #18 (host18), cells were then infected with phage at a multiplicity of infection (MOI) of 0.1. Free phages were sampled right after phage addition (0 min) every 5 min for 30 min by 0.2 µm filtering out the cells and measuring phage concentration by plating serial dilutions. By 15 min, 63% of phi18:3 had adsorbed to host18 (Supplementary Figure S1). The adsorption kinetics of the other two infections were previously reported [21]. Namely, by 15 min, 55% of phi38:1 had adsorbed to host18 whereas 84% to C. baltica NN016038 (host38).
To measure the one-step growth curves, the exponentially growing cells were infected in 50 ml at a MOI of 3.7 with phi18:3 and tenfold diluted after a 15-min adsorption (this is time 0). Infections with phi18:3 were sampled at 0, 15, 30, 45, 60, and 120 min. Infections with phi38:1 were previously conducted and described [30]. The new proteomics samples here reported derived from the infection on host38 (0, 15, 30, 45, and 60 min) or host18 (0, 60, 120, 150, and 240 min). Samples were collected for proteomics (40 ml) and RNA (2 ml) as described below.
RNA extraction, sequencing, and read mapping
All RNA-Seq procedures were done as previously described [30]. Namely, samples were collected (2 ml) from minutes 0, 15, 30, 45, 60, and 120 (phi18:3 infecting C. baltica #18) and from minutes 0, 20, 40, 60, and 120 (phi38:1 infecting C. baltica #18), and spun down at 20,817 g for 5 min. The supernatant was then removed and tubes were flash-frozen in liquid nitrogen and stored at −80 °C until the extraction. RNA extractions were performed with the RNeasy Mini Kit (Qiagen cat. no. 74104), DNase treated with TURBO DNA-free (Ambion AM1907), and concentrated to 20 μl with the RNA Clean and Concentrator-5 kit (Zymo Research R1015). The RNA integrity number and concentration of the extracted RNA was determined via the Agilent Bioanalyzer 2100 (Agilent Technologies, USA).
Libraries of complementary DNA were prepared for SOLiD 5500 XL (Applied Biosystems) sequencing according to the manufacturer’s protocol (Life Technologies, USA). The 50 bp reads were mapped to the genomes of C. baltica #18 (GenBank # CP009976), phage phi38:1 (GenBank # NC_021796.1), or phage phi18:3 (GenBank # NC_021794.1) using Bowtie2 v.2.14. Coverage of the phage and host genomes was calculated as the number of base pairs mapped to a genome divided by the genome length (Supplementary Table S1, S2). All downstream analyses, including read count normalization, RPKM calculation, and differential expression were exactly as published [30].
Proteomics data collection and analysis
Samples from infections were collected, alongside noninfected controls, in biological triplicates and spun down at 10,000 × g for 15 min, washed by resuspending with 0.2 µm filtered 1X PBS and spun down for 10 min at 10,000 × g. Supernatant was discarded and pellets were flash-frozen and stored at −80 °C until processing for protein extraction.
Cell pellets were lysed, proteins denatured with buffered 8 M urea and 5 mM DTT, digested with trypsin, and cleaned-up by C18 solid phase extraction as described previously [51] for label-free analysis. For proteomics quantitation by a label-free method (for phi18:3 and phi38:1 infections on C. baltica #18), resulting peptides were analyzed by Liquid chromatography–mass spectrometry (MS)/MS analysis [51] with the resulting data analyzed using the accurate mass and time (AMT) tag approach [52,53,54,55,56].
For proteomics quantitation by iTRAQ methods (for phi38:1 on C. baltica NN016038 and #18), the label-free quantitation approach was followed except higher energy collisional dissociation was used for peptide fragmentation [57, 58]. All peptide database searches used the C. baltica and phage genomes described above.
Protein abundance was scaled via z-score of all infected and control samples for the host and phage separately, and across all replicates and time points. Average z-score values were used as standardized protein abundances in downstream analyses. All such analyses can be found in Supplementary Dataset S1 (http://datacommons.cyverse.org/browse/iplant/home/shared/iVirus/Cellulophaga_Omics/Howard-Varona_2018_ISMEJ_transcriptomics_and_proteomics).
Improving annotation of restriction/modification (R/M) genes in C. baltica #18
To better identify restriction-modification genes in silico, the NCBI-annotated bacterial genome used in this study (Supplementary Dataset S2, tab “C.baltica#18_genes”) was searched against KEGG, Uniref, Interproscan and Pfam with a published pipeline [102]; REBASE (Roberts et al. 2015) was searched for Type I and II R/M predictions; and genes were run against Protein Data Bank for structural homologs using Phyre2 (Kelley et al. 2015). A total of 38 endonucleases plus methyltransferases were obtained based on hits obtained from KEGG and Uniref (with e-value ≤ 2.3 × 10−13 and bit score ≥ 100) (Supplementary Dataset S2, tab “host18_Restr_Modif”), which included the Phyre homologs and REBASE predictions.
Results and discussion
Time-resolved, phage, and host genome-wide transcriptomics (RNA-Seq) and proteomics (tandem mass spectrometry) data were collected and analyzed from population-wide infections and compared to samples from uninfected controls, in biological triplicates (Supplementary Dataset S1 Tab. 1, Supplementary Table S1 and S2), and analyzed using available genomes and genome annotations (Supplementary Dataset S2). The infections included (Fig. 1; Supplementary Figure S1): (1) generalist phi38:1 (72.5 Kb dsDNA podophage) on its original host of isolation, host38 [44], (2) generalist phi38:1 on host18, and (3) specialist phi18:3 (71.4 Kb dsDNA podophage) on its original host of isolation, host18 [44]. Phages phi18:3 and phi38:1 do not share any homologous gene (Supplementary Figure S2) and belong to different genera, Cba183likevirus and Cba401likevirus, respectively, based on their genome similarity [8]. Phage phi18:3 does not infect host38 [21]. Critical new analyses from the previously reported phi38:1 transcriptomes [30] are used to both compare with significant new data—phi18:3 transcriptomes, and proteomes of all three infections—and address the hypotheses generated from prior work.
The C. baltica—phage model system investigated. Generalist phage phi38:1 infects more efficiently its original host of isolation (strain NN016038; “host38”) than alternative host strain #18 (“host18”). Specialist phage phi18:3 efficiently infects its original host of isolation, “host18,” and does not infect host38 [21]
Phage genomes and transcriptomes suggest drivers of host-range and infection efficiency
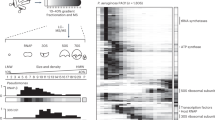
On host18, specialist phi18:3 had an efficient, 75-min infection (Fig. 2a) and a similar temporal transcriptome to that of phi38:1 infecting less efficiently this same host (Fig. 2b). Namely, both phages expressed all their genes by 60 min, and with early, middle, and late expression categories where transcript abundances peaked at 0, 15–20, and ≥40 min post infection, respectively (Fig. 2c, d; Supplementary Tables S3-S4 and Supplementary Dataset S1 tabs 9-10). Additionally, genes in each temporal category were functionally similar for both phages. Namely, expressed early were host takeover (i.e., those that begin phage gene expression and control host functions) genes, including a ribosyltransferase, serine/threonine phosphatase, or a lysozyme. Expressed middle were the DNA metabolism genes, and expressed late were the structural and lysis genes (Supplementary Figures 2, 3). Such transcriptional patterns are common among phages across diverse bacterial hosts and environments [30, 32, 33, 37, 59,60,61,62,63] and suggests that, regardless of their efficiency, phages that lack an RNA polymerase (RNAP) may successfully utilize the host’s RNAP for transcription, similar to well-known Enterophages (e.g., lambda or T4) [64,65,66]. Thus, the data at this point suggest that these universalities in phage transcription can be predicted via RNA-seq and are independent of host type, host range, and infection efficiency.
Infection dynamics and expression profile of phi18:3 and phi38:1 infecting C. baltica #18 (“host18”). One-step growth curve of the a efficient phi18:3 and b inefficient phi38:1 infections on host18. Represented are the average values of three biological replicates and their standard error, as well as the latent period (LP) and burst size (BS). Time 0 represents the dilution of the infection after a 15-min adsorption. Both phages express their genes in early, middle, and late categories, with highest expression at c 0, 15, and ≥30 min for phi18:3, and d 0, 20, and ≥40 min for phi38:1. The average log2RPKM values of the genes in each category and standard deviation are represented in the graphs. Numbers of genes in each temporal category are represented in parentheses
Beyond these overall similarities in phage transcriptomes, some expressed genes (Supplementary Tables S3-S4, Supplementary Dataset S2; Supplementary Figures S3, S4) suggested unique generalist versus specialist strategies from their predicted functions. For example, unique to generalist phi38:1 are 16 tRNAs (two transcribed early and 14 late) and DNA replication genes (e.g., DNA polymerase and primase, middle expressed, and late translated). Such genes might help a generalist phage utilize host translation and replicate phage DNA, respectively, across diverse hosts [67] given that phage genomes are known to code for genes that reprogram host metabolism [68, 69] and that they are not identified in a specialist phage (phi18:3). In contrast, expressed genes that were unique to phi18:3 included an anti-restriction gene and methylases (“anti-R/M” genes; transcribed and translated early), and the metabolic regulator MazG (middle transcribed and translated late). In Escherichiacoli, phages overcome host R/M defenses by, for example, expressing a suite of anti-restriction genes (e.g., T3, T7), or injecting such proteins into the cell (e.g., P1, T4) [70], and/or by self-methylating their DNA via methylases (e.g., T2, T4) [62]. Additionally, E. coli’s maz system is induced to prevent translation from stalling in response to diverse stresses, including phage infection [70, 71]. A variety of environmental phages encode for MazG, including cyanophages that transcribe it as a middle gene during their efficient infection [33]. Thus, these genes might enable phi18:3 to efficiently infect host18 by overcoming host R/M defenses through direct interaction with host’s endonucleases (anti restriction) [72] and self-DNA methylation (methylases) [70], and by preventing host metabolic shutdown during infection (MazG) [70, 71]. Other phages that lack such genes and still have efficient infections [28, 30,31,32] either protect their DNA through alternative modifications (e.g., glycosylation [70]) or, as in the case for phi38:1 infecting host38, do not trigger the host’s R/M system and might have adapted to utilize the host’s functions without the need for MazG [30].
Absence of transcriptional–translational synchronization in inefficient infections
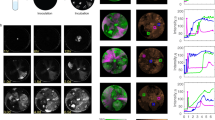
The presence of early proteins in phi18:3 (Fig. 3a, b, Supplementary Figure S3) suggested that this phage was able to rapidly utilize the host’s translational machinery to translate very early (i.e., within 15 min of infection). Notably, among these early proteins was the anti-restriction protein, which would be one of the proteins needed immediately upon infection to evade host R/M systems. To test the hypothesis that phi18:3 utilizes host translation early we measured phage protein dynamics (Fig. 3) and observed that, indeed, most (88%) of phi18:3’s early transcripts produced early proteins (Supplementary Figure S3) and that overall protein abundances paralleled those of the transcripts, with early, middle and late classifications readily apparent (Fig. 3a–c). One exception was a DNA replication protein that peaked early despite having a middle transcript (Fig. 3b). Presumably this protein was detected prior to its transcript because it was contained in the virion and introduced upon infection, even though it was not previously detected in a preliminary virion screen with other phages [8]. Other phages, including cyanophage Syn9 [33], T4 [73], and other Enterophages [70] also carry proteins in the virion that function in host takeover and early phage expression as T4’s Alt protein [73]. Notably, the observed 45-min peak in protein abundance (Fig. 3a–c) was not an artifact as interpreted from multiple proteomics controls, and yet is difficult to ascribe to particular biology. One possibility would be that while our population-wide averaged measurements captured the overall signal in these phage–host interactions, the 45-min anomaly might result from cell-to-cell variability in phage–host interaction initiation and infection dynamics. This could perhaps result from some asynchronous infections that are even coupled with variation caused by phenotypic heterogeneity in the underlying clonal population of cells [74]—phenomena that might be evaluated with single-cell ‘omics technologies as they emerge for prokaryotic phage–host systems.
Temporal transcriptional–translational dynamics of phi18:3 and phi38:1. For each infection, three vertical graphs represent the early, middle, or late phage transcripts. In those, averaged transcript expression is graphed with the thickest line, with “n” representing how many genes were averaged. The standardized, relative protein abundances of those transcripts are individually graphed in thinner lines, and they are early, middle, late, or a variation of those. The number in each category is also represented by an “n.” Time 0 represents the dilution of the infection after a 15-min adsorption. a–c Proteins are early (35%), middle (2%), middle late (5%), or late (58%) and they largely appear after their respective a early, b middle, and c late transcripts when phi18:3 infects host18. d–f Some (14%) phage proteins are early but most (86%) are late regardless of d early, e middle, or f late transcription of their genes when phi38:1 infects host18. g–i Phage proteins are early (4%), middle (4%), middle late (51%), late (39%), or constant (1%; protein abundance does not significantly change over time; see Supplementary Dataset S1, Tab. 10) and closely follow the g early, h middle, or i late transcription of their genes when phi38:1 infects host38. Calculations are in Supplementary Dataset S1
In contrast, the abundances of 86% of phi38:1’s proteins peaked later than the “late” transcriptional group (Supplementary Figure S4a-b) and largely plateaued after 60 min (Fig. 3d–f). These findings suggest either a mixed protein dynamics signal deriving from new rounds of infection happening after 150 min (when 4% of the cells lyse based on phageFISH data [15])—which could be resolved with better single-cell technologies—or slower protein dynamics and delayed translation signal deriving from population-wide data. If the latter, such biological signal could explain the previously observed delayed particle production [30] and even lysis, as follows. Namely, among phi38:1’s genes with non-delayed translation was a peptidase (gp021) that was transcribed and translated early (Supplementary Table S3; Supplementary Figure S4a-b). Given that a variety of peptidases function as endolysins and accumulate in the cell throughout the infection until holins are made [75, 76], one hypothesis is that the holin (which is unknown in phi38:1’s genome) is delayed or not produced in the inefficient infection. Regardless, these expression data together suggest that only specialist phi18:3 synchronizes host-derived translation with transcription to synthesize proteins in a timed and regulated manner.
To determine whether phi38:1 could have better translation in another host, we measured its protein dynamics while infecting its original host, host38, and compared them to our prior transcriptome dynamics [30]. This revealed that phage proteins were no longer delayed as when infecting host18 and instead were produced in early, middle, and late categories, with over half of them (55%) being middle/middle late (Fig. 3g–i; Supplementary Figure S4c-d). Notably, upregulation of host38’s translation genes was also middle/middle late [30], thus suggesting that phage phi38:1 translation in this host is mainly timed with the activation of host translation as occurs with phi18:3 on host18. Given that phage protein translation is expected to closely follow transcription based on lessons from phages infecting (efficiently) well-known systems (e.g., Bacillus phage phi29 synthesizes >60% of its proteins before middle transcription [77]), the inability of phi38:1 to synchronize translation with transcription in host18 may contribute to this inefficient infection.
Hosts’ transcriptomes and proteomes reveal causes for inefficient phage infections
Given that phage proteome analyses suggested differences in utilizing host functions, we next examined host18’s transcriptome and proteome during phi18:3 and phi38:1 infections and compared them to host38’s responses. Globally, host18’s transcription was repressed throughout infection by phi18:3, but not by phi38:1 (Fig. 4a), as evidenced by the relative decrease in host transcript abundance, which was not derived from a sequencing skew toward phage transcripts (Supplementary Figure S5). This suggested that, like phi38:1 on its original host38 [30], phi18:3 successfully shutdown transcription in its own original host (Fig. 4a), whereas phi38:1 did not in an alternative host [30]. Phage induced, host-transcriptional repression as a sign of host takeover has been described across phage–host model systems [28,29,30, 33, 35,36,37]. Mirroring the transcriptional response, the host’s proteome was differentially impacted by phage infection. Namely, host18’s protein abundances significantly decreased with phi18:3 (t-test, p ≤ 0.05; Fig. 4b) and increased with phi38:1 (Fig. 4c). In contrast, when phi38:1 infected host38 host protein abundances no longer significantly increased (t-test, p > 0.05; Supplementary Figure S6b). Thus, these proteomic data suggest that variably efficient infections differentially impact the host proteome such that only the former stalls host protein synthesis, which is likely phage mediated, whereas the latter does not impact the host proteome. Here, the increase in host18 protein abundances at 4 h during the infection with phi38:1 (Fig. 4c) suggests that this host continues translating, perhaps as a defense mechanism against the phage.
Global transcriptome and proteome during C. baltica #18 (“host18”) infection with phi18:3 and phi38:1. OE over expressed, UE under expressed. a Host genes are globally UE throughout the infection with phi18:3, and OE throughout the infection with phi38:1. Represented is the fold change of gene expression between infected and uninfected cells, normalized by the fraction of infected cells. b, c Host protein abundances b significantly (t-test between 0 and 60 min, p ≤ 0.05) decrease by the end of the infection with phi18:3, and c significantly (t-test against 0 min, p ≤ 0.05) increase throughout the infection (≥120 min) with phi38:1. None of the uninfected host protein abundances significantly increase (t-test against 0 min, p > 0.05) over time. Represented in b and c is the average values of three biological replicates and standard error. Statistical analyses can be found in Supplementary Dataset S1 (Tab. 12). Time 0 represents the dilution of the infection after a 15-min adsorption
With global host responses suggesting differences in host takeover in the variably efficient infections, we next examined phage impacts on specific host functions. First, we evaluated the response of host18’s R/M system for three reasons: (1) both C. baltica hosts lack the common bacterial CRISPR-Cas defenses [78], (2) bacteria commonly restrict incoming phage DNA [70, 79, 80], and (3) early expression of 22 early proteins by phi18:3 suggested a phage counter defense [70]. Those early proteins included both the anti-R/M genes and potentially the 14 proteins with unknown functions that surrounded those anti-R/M genes and which are predicted to be transcribed under the same promoter (Supplementary Table S3; Supplementary Figure S3). Indeed, host18 R/M genes were differentially over expressed during infection with phi18:3, but not with phi38:1 (Fig. 5a, Supplementary Table S5). Proteomics suggested that overall protein abundances significantly (t-test, p ≤ 0.05) decreased during phi18:3 infection, but increased with phi38:1 (Supplementary Figure S7). Notably, our annotation of these genes as R/M derives from in silico predictions of restriction endonucleases and methyltransferases (see Methods section), and awaits experimental evaluation. Additionally, as R/M genes are known to have ubiquitous roles [81], beyond defense to phage the genes shown here might also function in SOS response or DNA replication. Should they be involved in defense against phage DNA, data here presented suggest that host18 transcriptionally activated its R/M system against phi18:3, but its proteins likely encountered phi18:3’s early translated genes involved in anti-R/M counter defenses and were inactivated, as occurs in other phage–host systems [70]. Such transcriptional activation, is not commonly reported in phage–host interactions as it is in, for example, chlorella virus [81]. While this remains an open question, the observed over-expression of R/M genes (Fig. 5a) could be an example of inducible R/M systems against not just phage phi18:3 DNA entry, but also replication. In contrast, the lack of these genes in phi38:1 would prevent such inactivation and likely allow host18 R/M proteins to accumulate from the genes’ basal expression and contribute to the previously observed host targeting of phi38:1 DNA [15, 30].
Expression of host18’s restriction/modification (R/M) and translation genes in response to phi18:3 and phi38:1. OE over expressed; UE under expressed. Represented in a stacked bar graph is the fold change of expression of infected relative to uninfected cells, normalized by the fraction of infected cells. Solid bars represent the differential expression values whereas dotted bars represent the non-differentially expressed genes. a The R/M system is clearly differentially expressed in response to phi18:3; mostly OE throughout the infection; and UE occurs during middle infection with phi38:1. Host18 displays basal expression (non-differentially expressed genes) of the R/M system regardless of phage infection. b Host18 genes involved in protein translation are OE 4.5-fold (early), 1.7-fold (middle), and 1.4-fold (late) higher in response to phi18:3 than to phi38:1. The values of expression in host18 infected with phi38:1 do not significantly increase over time (t-test, p-value > 0.05; Supplementary Dataset S1 Tab. 7). Host18 displays basal expression (non-differentially expressed genes) of the translation genes regardless of phage infection. Time 0 represents the dilution of the infection after a 15-min adsorption
Second, we examined host translation genes because phi18:3 synthesized early proteins (Fig. 3a–c) and because previous transcriptome data suggested that differential transcription of host translation genes in host38 versus host18 contributed to phi38:1’s virion formation delay [30]. Here, host18 transcriptomes (Fig. 5b) and proteomes (Supplementary Figure S8) suggested that phi18:3 activated the host’s translational machinery for its reproduction better than phi38:1 did. Namely, from the beginning of the infection (early) host18’s genes had higher differential expression with phi18:3 (e.g., 6.5-fold) than with phi38:1 (1.4-fold; Fig. 5b). Additionally, such difference in translation activation at the early stage was reflected in the proteome: host18 infected with phi18:3 had produced 93% of those translation proteins, whereas only 2% when infected with phi38:1 (Supplementary Figure S8), thus suggesting that phi18:3 was better poised to utilize host18’s translation than phi38:1. The difference between this efficient translation in phi18:3-infected host18 and that observed in phi38:1 infected host38—the other, more efficient, original host infection—was that phi18:3-infected host18 cells expressed their translation genes early—4.5-fold higher than when infected with phi38:1 (Fig. 5b)—whereas over-expression began during mid infection in host38 (Fig. 5c in ref. [30]). We posit that host translational activation enabled better phage transcriptional–translational synchronization and thus efficient infections on the original hosts, and that early activation of translation in host18 enabled phi18:3 to translate early its host-takeover proteins.
Third, although the majority (86%) of phi38:1’s proteins were produced late when infecting host18, some (14%) were produced early (Fig. 3d–f; Supplementary Table S4), which suggests that phi38:1 proteins may be encountering host18’s proteases and contributing to disrupting transcriptional–translational synchronization. Notably, alternative explanations might exist for the delay in phi38:1 protein abundances as previously mentioned, but here we considered the highly conserved protease ClpP because it can target several phage (e.g., lambda, P1) proteins and also be inhibited by phage counter defenses [82, 83]. This host18 gene was differentially under expressed (−0.67-fold) in response to phi18:3, but not to phi38:1 (Fig. 6a), where the gene was not even differentially expressed. Protein abundances significantly (t-test, p ≤ 0.05) decreased (~3×) from start to end of the phi18:3 infection (Fig. 6b, c). These findings suggest that ClpP was inactivated only by phi18:3 and not phi38:1, which would in turn lead to ClpP accumulation to possibly target degradation of phi38:1 proteins. Such dynamics would be consistent with delays both in virion synthesis previously observed [30] and protein synthesis observed here (Fig. 3d–f). While ClpP effects are well-studied in phage lambda [84], this and other conserved proteases might constitute underexplored bacterial strategies that modulate phage infection efficiency.
Gene expression and protein abundance of “host18” protease ClpP during infection with phi18:3 and phi38:1. a The gene is only differentially expressed, and under expressed (UE), upon phi18:3 infection. Protein abundance over time during b phi18:3 infection decreases significantly (t-test, p ≤ 0.05), but during c phi38:1 infection increases significantly (t-test, p ≤ 0.05). The abundances in the controls b, c are not statistically significantly different from time 0 (t-test, p > 0.05), unlike in the infected treatment. Represented in b, c are the average of three biological replicates and their standard error. The t-tests can be found in Supplementary Dataset S1 (Tab. 7). Time 0 represents the dilution of the infection after a 15-min adsorption
A mechanistic model for drivers of variable efficient phage–host interactions
These data provide support for recent mechanistic work in environmental phage–host model systems, while also bringing new perspective on the mechanisms driving infection efficiency as follows. First, host takeover in efficient infections seems to involve global repression of host transcription across systems [28,29,30, 33, 35,36,37]. Our findings with a specialist phage are consistent with these studies. Additionally, they show that host takeover in efficient infections might also stall the host proteome. Second, here efficient infections also seem to activate the host’s translational machinery to translate phage genes to proteins. Unlike giant phages that can code for translation genes [85] and complement the host’s translation machinery, phages are host-dependent for synthesizing proteins. Third, we also show that the timing of such activation either occurs early, if the phage requires producing proteins to counteract host defenses against phage DNA (e.g., in specialist phi18:3 to fight host18 R/M), or later, perhaps reflecting that phages have alternative DNA protection strategies (e.g., glycosylation [70]), can localize to the cell poles [81], or the hosts lack, or fail to activate R/M defenses (e.g., in generalist phi38:1 infecting host38 [30]). In contrast, phage protein dynamics during inefficient infections are not only delayed but they likely lose temporal synchronization with their transcriptional counterparts. We suggest that host proteases, for example, ClpP, likely reduce phage protein abundances in inefficient infections. As these proteases are highly conserved [86] and phages can have inhibitors for them [84], proteases might be a widespread, underexplored, and critical mechanism driving phage infection efficiency. Finally, here we have provided insight into only some of the possibly additional mechanisms contributing to variation in infection efficiency. Specifically, additional mechanisms could be found in the ~260 unique bacterial genes (published in Supplementary Dataset S1 in ref. [30]), likely beyond the best described ones that are not present in these hosts, such as CRISPR-Cas, and the conserved toxin/antitoxin and abortive systems. While mechanistic knowledge of phage–host interactions in nature is arguably in its infancy, this and other work [28,29,30, 33, 35,36,37] establish a baseline with which to investigate infection efficiency and phage–host interactions in more phylogenetically diverse lytic phage–host systems. While such work is largely lytic cycle focused, recent ‘omics-enabled measurements in lysogenic infections are revealing both the complexity of these systems ([59, 87,88,89, 103]) and more fully helping establish paradigms for naturally occurring phage–host interaction biology.
Defining and better characterizing infection efficiency—a discussion just beginning
The work presented here invites discussing infection efficiency, as efficiency is a term with relative uses in the literature. Briefly, in phage studies, the term “efficient” is used to describe either infections or phages that lyse the most cells the fastest [28, 29], have the shortest LP/best growth rate [104], display better intracellular capabilities (e.g., in DNA replication [105] or gene transcription [37], or are proficient at hijacking host RNAP [37]). In contrast, the eukaryotic viral literature evaluates whether each step of the complex viral infection, from attachment to dispersion, is efficient [106,106,107,108,109,110,111,112,113].
Against this vast eukaryotic virus literature, we sought to classify each of the three infections presented here as efficient or inefficient. The infection of phi18:3 on host18 has both the shortest LP and largest burst size (BS), thus it can safely be classified as the most efficient infection of the three. However, phi38:1’s infections on host18 and host38 are more difficult to classify since the former has a LP of > 2.5 h and a BS of ~39, and the latter has a LP of 75 min and BS of nearly 10. To better clarify the longer term implications of such infection dynamics, we performed new experiments where we infected host18 and host38 separately with phi38:1 for 28 h and measured cell density and phage production (Supplementary Figure S9). The results showed that (A) infected host18 cells maintained lower OD than infected host38 cells, and that (B) more phage progeny was produced, and earlier, in host38 than in host18.
Given all knowledge of phi38:1’s infection, we assessed the relative efficiencies for each stage of infection as is often evaluated in the eukaryotic virus literature. This revealed that phi38:1 on host18 is, compared to the infection of either phi38:1 on host38 or phi18:3 on host18: (1) less efficient at forming plaques [21]; (2) less efficient at infecting cells [15, 21, 30]; (3) less efficient at overcoming host defenses (this work); (4) equally efficient at transcribing phage genes [30]; (5) less efficient at repressing host transcription [30]; (6) less efficient at repressing and taking over host translation [30]; (7) less efficient at replicating DNA [15, 30]; (8) less efficient at producing and/or assembling phage particles [30]; (9) less efficient at lysing cells [15]; and (10) less efficient at producing progeny throughout 28 h (this work). Given that phi38:1 on host18 is less efficient for nine out of ten steps, such infection is the least efficient of the three studied here.
Pragmatically, such inefficient infections are challenging to study using population-wide measurements. Specifically, synchronizing infections is not simple, as even our best efforts were still prey to a low fraction of cells initially infected and cell lysis spanning long time periods (4% of cells initially lysed at 2.5 h as compared to the bulk by 11 h [15]). To compensate for this, we assumed that the transcriptome and proteome of uninfected cells would be similar to uninfected control cells. While we see this as a reasonable assumption due to the lack of evidence for uninfected to infected cell communication mechanisms and the strong signal expected for infected cells (driven by the here observed higher RNA in infected versus uninfected samples; Supplementary Dataset S1 Tab. 1), the ability to apply single-cell ‘omics technologies to these systems would be invaluable for experimentally evaluating such assumption.
Conclusions
While sampling, sequencing, and informatic advances better unveil and classify the diversity of naturally occurring phage genes and genomes [3, 43, 90,91,92], their biology often remains underexplored due to (1) knowledge mainly deriving from targeted studies in laboratory model systems that underrepresent natural diversity of infection efficiencies and of phage–host interactions, and (2) the experimental challenges of studying environmental phage–host systems, most of which lack genetic tools. Consequently, inefficient infections are mechanistically underexplored, and knowledge of phage success is mostly limited to host defenses either blocking phage adsorption and DNA survival, or causing transcription and lysis delays [79, 80].
In spite of a century of studying phages, novel phage biology continues to be uncovered. For example, phages are now known to employ complex communication systems [93, 94], and to play roles in health and disease that can lead to non-host-derived immunity in humans [5, 95,96,97]. As these knowledge gaps are filled in, it becomes critical to incorporate phages into ecosystem models [98] that will ultimately help advance phage-based biotechnological [99] and therapeutic applications [100]. Fundamental to this, however, is the need to understand strategies driving phage–host interaction biology and infection efficiency. With combinatorial genome-wide ‘omics applied to two environmental bacterial host strains and two phages with different infection efficiency on a shared host, we reveal here that multiple mechanisms alter phage infection efficiency across nearly identical hosts. Such findings in naturally occurring systems may better mimic phage infection in nature than the induced genetic mutations from traditional laboratory experiments. If so, this data, if generalizable, contribute to better understanding infection efficiency and, in turn, of how phages can be engineered [101] to better “edit” the microbial communities that dictate ecosystems’ and human health.
References
Salmond GP, Fineran PC. A century of the phage: past, present and future. Nat Rev Microbiol. 2015;13:777–86.
Diaz-Munoz SL, Koskella B. Bacteria-phage interactions in natural environments. Adv Appl Microbiol. 2014;89:135–83.
Paez-Espino D, Eloe-Fadrosh EA, Pavlopoulos GA, Thomas AD, Huntemann M, Mikhailova N, et al. Uncovering Earth’s virome. Nature. 2016;536:425–30.
De Paepe M, Leclerc M, Tinsley CR, Petit MA. Bacteriophages: an underestimated role in human and animal health? Front Cell Infect Microbiol. 2014;4:39.
Manrique P, Bolduc B, Walk ST, Van Der Oost J, De Vos WM, Young MJ. Healthy human gut phageome. Proc Natl Acad Sci USA. 2016;113:10400–5.
Bruder K, Malki K, Cooper A, Sible E, Shapiro JW, Watkins SC, et al. Freshwater metaviromics and bacteriophages: a current assessment of the state of the art in relation to bioinformatic challenges. Evol Bioinform Online. 2016;12:25–33.
Gillis A, Mahillon J. Phages preying on Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis: past, present and future. Viruses. 2014;6:2623–72.
Holmfeldt K, Solonenko N, Shah M, Corrier K, Riemann L, Verberkmoes NC, et al. Twelve previously unknown phage genera are ubiquitous in global oceans. Proc Natl Acad Sci USA. 2013;110:12798–803.
Mahony J, van Sinderen D. Current taxonomy of phages infecting lactic acid bacteria. Front Microbiol. 2014;5:7.
Perez Sepulveda B, Redgwell T, Rihtman B, Pitt F, Scanlan DJ, Millard A. Marine phage genomics: the tip of the iceberg. FEMS Microbiol Lett. 2016;363:15.
Murray AG, Jackson GA. Viral dynamics: a model of the effects size, shape, motion and abundance of single-celled planktonic organisms and other particles. Mar Ecol Progress Ser Oldendorf. 1992;89:103–16.
Campbell A. Conditions for the existence of bacteriophage. Evolution. 1961;15:153–65.
Koskella B, Meaden S. Understanding bacteriophage specificity in natural microbial communities. Viruses. 2013;5:806–23.
Dang VT, Sullivan MB. Emerging methods to study bacteriophage infection at the single-cell level. Front Microbiol. 2014;5:724.
Dang VT, Howard-Varona C, Schwenck S, Sullivan MB. Variably lytic infection dynamics of large Bacteroidetes podovirus phi38: 1 against two Cellulophaga baltica host strains. Environ Microbiol. 2015;17:4659–71.
Howard-Varona C, Hargreaves KR, Abedon ST, Sullivan MB. Lysogeny in nature: mechanisms, impact and ecology of temperate phages. ISME J. 2017;11:1511–20.
Hyman P, Abedon ST. Bacteriophage host range and bacterial resistance. Adv Appl Microbiol. 2010;70:217–48.
Ross A, Ward S, Hyman P. More is better: selecting for broad host range bacteriophages. Front Microbiol. 2016;7:1352.
Gregory AC, Solonenko SA, Ignacio-Espinoza JC, Labutti K, Copeland A, Sudek S, et al. Genomic differentiation among wild cyanophages despite widespread horizontal gene transfer. BMC Genom. 2016;17:930.
Duhaime MB, Solonenko N, Roux S, Verberkmoes NC, Wichels A, Sullivan MB. Comparative omics and trait analyses of particle-associated Pseudoalteromonas phages advance the phage OTU concept. Front Microbiol. 2017;8:1241.
Holmfeldt K, Howard-Varona C, Solonenko N, Sullivan MB. Contrasting genomic patterns and infection strategies of two co-existing Bacteroidetes podovirus genera. Environ Microbiol. 2014;16:2501–13.
Edwards RA, McNair K, Faust K, Raes J, Dutilh BE. (2016). Computational approaches to predict bacteriophage–host relationships. FEMS Microbiol Rev 40:258–272.
Weitz JS, Poisot T, Meyer JR, Flores CO, Valverde S, Sullivan MB, et al. Phage–bacteria infection networks. Trends Microbiol. 2013;21:82–91.
Cheetham GM, Jeruzalmi D, Steitz TA. Transcription regulation, initiation, and “DNA scrunching” by T7 RNA polymerase. Cold Spring Harb Symp Quant Biol. 1998;63:263–7.
Heineman RH, Molineux IJ, Bull JJ. Evolutionary robustness of an optimal phenotype: re-evolution of lysis in a bacteriophage deleted for its lysin gene. J Mol Evol. 2005;61:181–91.
Bertozzi Silva J, Storms Z, Sauvageau D. Host receptors for bacteriophage adsorption. FEMS Microbiol Lett. 2016;363:4.
Roychoudhury P, Shrestha N, Wiss VR, Krone SM. Fitness benefits of low infectivity in a spatially structured population of bacteriophages. Proc Biol Sci. 2014;281:20132563.
Ceyssens PJ, Minakhin L, Van Den Bossche A, Yakunina M, Klimuk E, Blasdel B, et al. Development of giant bacteriophage varphiKZ is independent of the host transcription apparatus. J Virol. 2014;88:10501–10.
Chevallereau A, Blasdel BG, De Smet J, Monot M, Zimmermann M, Kogadeeva M, et al. Next-generation “-omics” approaches reveal a massive alteration of host RNA metabolism during bacteriophage infection of Pseudomonas aeruginosa. PLoS Genet. 2016;12:e1006134.
Howard-Varona C, Roux S, Dore H, Solonenko NE, Holmfeldt K, Markillie LM, et al. Regulation of infection efficiency in a globally abundant marine Bacteriodetes virus. ISME J. 2016;11:284–295.
Mojardín L, Salas M.Global transcriptional analysis of virus-host interactions between phage φ29 and Bacillus subtilis. J Virol. 2016;90:9293–9304.
Wagemans J, Blasdel BG, Van Den Bossche A, Uytterhoeven B, De Smet J, Paeshuyse J, et al. Functional elucidation of antibacterial phage ORFans targeting Pseudomonas aeruginosa. Cell Microbiol. 2014;16:1822–1835.
Doron S, Fedida A, Hernandez-Prieto MA, Sabehi G, Karunker I, Stazic D, et al. (2016). Transcriptome dynamics of a broad host-range cyanophage and its hosts. ISME J 10:1437–1455.
Duplessis M, Russell WM, Romero DA, Moineau S. Global gene expression analysis of two Streptococcus thermophilus bacteriophages using DNA microarray. Virology. 2005;2:192–208.
Fallico V, Ross RP, Fitzgerald GF, Mcauliffe O. Genetic response to bacteriophage infection in Lactococcus lactis reveals a four-strand approach involving induction of membrane stress proteins, D-alanylation of the cell wall, maintenance of proton motive force, and energy conservation. J Virol. 2011;85:12032–42.
Lindell D, Jaffe JD, Coleman ML, Futschik ME, Axmann IM, Rector T, et al. Genome-wide expression dynamics of a marine virus and host reveal features of co-evolution. Nature. 2007;449:83–6.
Blasdel BG, Chevallereau A, Monot M, Lavigne R, Debarbieux L. Comparative transcriptomics analyses reveal the conservation of an ancestral infectious strategy in two bacteriophage genera. ISME J. 2017;11:1988–1996.
De Smet J, Zimmermann M, Kogadeeva M, Ceyssens P-J, Vermaelen W, Blasdel B, et al. High coverage metabolomics analysis reveals phage-specific alterations to Pseudomonas aeruginosa physiology during infection. ISME J. 2016;10:1823–1835.
Thomas F, Hehemann JH, Rebuffet E, Czjzek M, Michel G. Environmental and gut bacteroidetes: the food connection. Front Microbiol. 2011;2:93.
Wexler AG, Goodman AL. An insider’s perspective: bacteroides as a window into the microbiome. Nat Microbiol. 2017;2:17026.
Dang H, Lovell CR. Microbial surface colonization and biofilm development in marine environments. Microbiol Mol Biol Rev. 2016;80:91–138.
Holmfeldt K, Solonenko N, Howard-Varona C, Moreno M, Malmstrom RR, Blow MJ, et al. Large-scale maps of variable infection efficiencies in aquatic bacteroidetes phage–host model systems. Environ Microbiol. 2016;18:3949–61.
Roux S, Brum JR, Dutilh BE, Sunagawa S, Duhaime MB, Loy A, et al. Ecogenomics and potential biogeochemical impacts of globally abundant ocean viruses. Nature. 2016;537:689–93.
Holmfeldt K, Middelboe M, Nybroe O, Riemann L. Large variabilities in host strain susceptibility and phage host range govern interactions between lytic marine phages and their Flavobacterium hosts. Appl Environ Microbiol. 2007;73:6730–9.
Vizcaino JA, Csordas A, Del-Toro N, Dianes JA, Griss J, Lavidas I, et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44:D447–56.
Johansen JE, Nielsen P, Sjoholm C. Description of Cellulophaga baltica gen. nov., sp. nov. and Cellulophaga fucicola gen. nov., sp. nov. and reclassification of [Cytophaga] lytica to Cellulophaga lytica gen. nov., comb. nov. Int J Syst Bacteriol. 1999;49:1231–40.
Solovyev V, Salamov A. Automatic annotation of microbial genomes and metagenomic sequences. In: Li RW, editor. Metagenomics and its applications in agriculture, biomedicine and environmental studies. New York, NY: Nova Science Publishers; 2011. p. 61–78.
Kingsford CL, Ayanbule K, Salzberg SL. Rapid, accurate, computational discovery of Rho-independent transcription terminators illuminates their relationship to DNA uptake. Genome Biol. 2007;8:1.
Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16(10):944–5.
Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27:1009–10.
Elfenbein JR, Knodler LA, Nakayasu ES, Ansong C, Brewer HM, Bogomolnaya L, et al. Multicopy single-stranded DNA directs intestinal colonization of enteric pathogens. PLoS Genet. 2015;11:e1005472.
Ansong C, Schrimpe-Rutledge AC, Mitchell H, Chauhan S, Jones MB, Kim Y-M, et al. A mutli-omic systems approach to elucidating Yersinia virulence mechanisms. Mol Biosyst. 2013;9:44–54.
Jaitly N, Monroe ME, Petyuk VA, Clauss TR, Adkins JN, Smith RD. Robust algorithm for alignment of liquid chromatography–mass spectrometry analyses in an accurate mass and time tag data analysis pipeline. Anal Chem. 2006;78:7397–409.
Monroe ME, Shaw JL, Daly DS, Adkins JN, Smith RD. MASIC: a software program for fast quantitation and flexible visualization of chromatographic profiles from detected LC-MS(/MS) features. Comput Biol Chem. 2008;32:215–7.
Monroe ME, Tolic N, Jaitly N, Shaw JL, Adkins JN, Smith RD. VIPER: an advanced software package to support high-throughput LC-MS peptide identification. Bioinformatics. 2007;23:2021–3.
Petritis K, Kangas LJ, Yan B, Monroe ME, Strittmatter EF, Qian WJ, et al. Improved peptide elution time prediction for reversed-phase liquid chromatography-MS by incorporating peptide sequence information. Anal Chem. 2006;78:5026–39.
Conlon BP, Nakayasu ES, Fleck LE, Lafleur MD, Isabella VM, Coleman K, et al. Activated ClpP kills persisters and eradicates a chronic biofilm infection. Nature. 2013;503:365–70.
Nakayasu ES, Tempel R, Cambronne XA, Petyuk VA, Jones MB, Gritsenko MA, et al. Comparative phosphoproteomics reveals components of host cell invasion and post-transcriptional regulation during francisella infection. Mol Cell Proteomics. 2013;12:3297–309.
Zhao X, Chen C, Shen W, Huang G, Le S, Lu S, et al. Global transcriptomic analysis of interactions between Pseudomonas aeruginosa and Bacteriophage PaP3. Sci Rep. 2016;6:19237.
Lavigne R, Lecoutere E, Wagemans J, Cenens W, Aertsen A, Schoofs L, et al. A multifaceted study of Pseudomonas aeruginosa shutdown by virulent podovirus LUZ19. MBio. 2013;4:2.
Lin X, Ding H, Zeng Q. (2016). Transcriptomic response during phage infection of a marine cyanobacterium under phosphorus-limited conditions. Environ Microbiol 18:450–60.
Miller ES, Kutter E, Mosig G, Arisaka F, Kunisawa T, Ruger W. Bacteriophage T4 genome. Microbiol Mol Biol Rev. 2003;67:6–156.
Osterhout RE, Figueroa IA, Keasling JD, Arkin AP. Global analysis of host response to induction of a latent bacteriophage. BMC Microbiol. 2007;7:82.
Casjens SR, Hendrix RW. Bacteriophage lambda: early pioneer and still relevant. Virology. 2015;479–480C:310–30.
Geiduschek EP. Regulation of expression of the late genes of bacteriophage T4. Annu Rev Genet. 1991;25:437–60.
Geiduschek EP, Kassavetis GA. Transcription of the T4 late genes. Virol J. 2010;7:288.
Ling J, O’donoghue P, Soll D. Genetic code flexibility in microorganisms: novel mechanisms and impact on physiology. Nat Rev Micro. 2015;13:707–21.
Hurwitz BL, Hallam SJ, Sullivan MB. Metabolic reprogramming by viruses in the sunlit and dark ocean. Genome Biol. 2013;14:R123.
Hurwitz BL, U’ren JM. Viral metabolic reprogramming in marine ecosystems. Curr Opin Microbiol. 2016;31:161–8.
Samson JE, Magadan AH, Sabri M, Moineau S. Revenge of the phages: defeating bacterial defences. Nat Rev Microbiol. 2013;10:675–87.
Starosta AL, Lassak J, Jung K, Wilson DN. The bacterial translation stress response. FEMS Microbiol Rev. 2014;38:1172–201.
Spoerel N, Herrlich P, Bickle TA. A novel bacteriophage defence mechanism: the anti-restriction protein. Nature. 1979;278:30–4.
Hinton DM. Transcriptional control in the prereplicative phase of T4 development. Virol J. 2010;7:289.
Ackermann M. A functional perspective on phenotypic heterogeneity in microorganisms. Nat Rev Microbiol. 2015;13:497–508.
Young R. Phage lysis: three steps, three choices, one outcome. J Microbiol. 2014;52:243–58.
Oliveira H, Melo LDR, Santos SB, Nóbrega FL, Ferreira EC, Cerca N, et al. Molecular aspects and comparative genomics of bacteriophage endolysins. J Virol. 2013;87:4558–70.
Pene JJ, Murr PC, Barrow-Carraway J. Synthesis of bacteriophage phi29 proteins in Bacillus subtilis. J Virol. 1973;12:61–7.
Marraffini LA. CRISPR-Cas immunity in prokaryotes. Nature. 2015;526:55–61.
Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nat Rev Micro. 2010;8:317–27.
Seed KD. Battling phages: how bacteria defend against viral attack. PLoS Pathog. 2015;11:e1004847.
Vasu K, Nagaraja V. Diverse functions of restriction-modification systems in addition to cellular defense. Microbiol Mol Biol Rev. 2013;77:53–72.
Gottesman S. Proteases and their targets in Escherichia coli. Annu Rev Genet. 1996;30:465–506.
Mount DW. The genetics of protein degradation in bacteria. Annu Rev Genet. 1980;14:279–319.
Taylor K, Wegrzyn G. Replication of coliphage lambda DNA. FEMS Microbiol Rev. 1995;17:109–19.
Schulz F, Yutin N, Ivanova NN, Ortega DR, Lee TK, Vierheilig J, et al. Giant viruses with an expanded complement of translation system components. Science. 2017;356:82–85.
Porankiewicz J, Wang J, Clarke AK. New insights into the ATP-dependent Clp protease: Escherichia coli and beyond. Mol Microbiol. 1999;32:449–58.
Mahony J, Ainsworth S, van Sinderen D. Tale of the unseen phage. Bacteriophage. 2013;3:e25985.
Hare J, Ferrell JC, Witkowski TA, Grice AN. Prophage induction and differential RecA and UmuDAb transcriptome regulation in the DNA damage responses of Acinetobacter baumannii and Acinetobacter baylyi. PLoS ONE. 2014;9:e93861.
Fusco S, Liguori R, Limauro D, Bartolucci S, She Q, Contursi P. Transcriptome analysis of Sulfolobus solfataricus infected with two related fuselloviruses reveals novel insights into the regulation of CRISPR-Cas system. Biochimie. 2015;118:322–32.
Bekliz M, Colson P, La Scola B. The expanding family of virophages. Viruses. 2016;8:317.
Bolduc B, Youens-Clark K, Simon R, Hurwitz BL, Sullivan MB. iVirus: facilitating new insights in viral ecology with software and community data sets imbedded in a cyberinfrastructure. ISME J. 2016;11:7–14.
Bolduc B, Jang HB, Doulcier G, You Z, Roux S, Sullivan MB. vConTACT: an iVirus tool to classify double-stranded DNA viruses that infect Archaea and Bacteria. PeerJ. 2017;5:e3243.
Erez Z, Steinberger-Levy I, Shamir M, Doron S, Stokar-Avihail A, Peleg Y, et al. Communication between viruses guides lysis–lysogeny decisions. Nature. 2017;541:488–93.
Feiner R, Argov T, Rabinovich L, Sigal N, Borovok I, Herskovits AA. A new perspective on lysogeny: prophages as active regulatory switches of bacteria. Nat Rev Micro. 2015;13:641–50.
Barr JJ, Auro R, Furlan M, Whiteson KL, Erb ML, Pogliano J, et al. Bacteriophage adhering to mucus provide a non-host-derived immunity. Proc Natl Acad Sci. 2013;110:10771–6.
Mirzaei MK, Maurice CF. Menage a trois in the human gut: interactions between host, bacteria and phages. Nat Rev Microbiol. 2017;15:397–408.
Santiago-Rodriguez TM, Naidu M, Abeles SR, Boehm TK, Ly M, Pride DT. Transcriptome analysis of bacteriophage communities in periodontal health and disease. BMC Genom. 2015;16:549.
Weitz JS, Stock CA, Wilhelm SW, Bourouiba L, Coleman ML, Buchan A, et al. A multitrophic model to quantify the effects of marine viruses on microbial food webs and ecosystem processes. ISME J. 2015;9:1352–64.
O’sullivan L, Buttimer C, Mcauliffe O, Bolton D, Coffey A. Bacteriophage-based tools: recent advances and novel applications. F1000Res. 2016;5:2782.
Dalmasso M, Hill C, Ross RP. Exploiting gut bacteriophages for human health. Trends Microbiol. 2014;22:399–405.
Ando H, Lemire S, Pires DP, Lu TK. Engineering modular viral scaffolds for targeted bacterial population editing. Cell Syst. 2015;1:187–96.
Daly RA, Borton MA, Wilkins MJ, Hoyt DW, Kountz DJ, Wolfe RA, et al. (2016). Microbial metabolisms in a 2.5-km-deep ecosystem created by hydraulic fracturing in shales. Nat Microbiol, 1:16146.
Dedrick RM, Marinelli LJ, Newton GL, Pogliano K, Pogliano J, Hatfull GF. (2013). Functional requirements for bacteriophage growth: gene essentiality and expression in mycobacteriophage Giles. Mol Microbiol 88:577–89.
Heilmann S, Sneppen K, Krishna S. (2012). Coexistence of phage and bacteria on the boundary of self-organized refuges. Proc Natl Acad Sci 109:12828–833.
Thompson LR, Zeng Q, Chisholm SW. (2016). Gene Expression Patterns during Light and Dark Infection of Prochlorococcus by Cyanophage. PLoS One 11: e0165375.
Kaplan AH, Manchester M, Swanstrom R. (1994). The activity of the protease of human immunodeficiency virus type 1 is initiated at the membrane of infected cells before the release of viral proteins and is required for release to occur with maximum efficiency. J Virol 68:6782–786.
Hope TJ. (2007). Bridging efficient viral infection. Nat Cell Biol 9:243–244.
Zhu S, Watanabe M, Kirkpatrick E, Murray AB, Sok R, Karst SM. (2015). Regulation of Norovirus Virulence by the VP1 Protruding Domain Correlates with B Cell Infection Efficiency. J Virol 90:2858–867.
Devhare PB, Desai S, Lole KS. (2016). Innate immune responses in human hepatocyte-derived cell lines alter genotype 1 hepatitis E virus replication efficiencies. Sci Rep 6:26827.
Schelker M, Mair CM, Jolmes F, Welke R-W, Klipp E, Herrmann A, et al. (2016). Viral RNA Degradation and Diffusion Act as a Bottleneck for the Influenza A Virus Infection Efficiency. PLoS Comput Biol 12:e1005075.
Iwamoto M, Cai D, Sugiyama M, Suzuki R, Aizaki H, Ryo A, et al. (2017). Functional association of cellular microtubules with viral capsid assembly supports efficient hepatitis B virus replication. Sci Rep 7:10620.
Jin J, Cheng J, He Z, Ren Y, Yu X, Song Y, et al. (2017). Different Origins of Newcastle Disease Virus Hemagglutinin-Neuraminidase Protein Modulate the Replication Efficiency and Pathogenicity of the Virus. Front Microbiol 8:1607.
Mayer BT, Krantz EM, Swan D, Ferrenberg J, Simmons K, Selke S, et al. (2017). Transient Oral Human Cytomegalovirus Infections Indicate Inefficient Viral Spread from Very Few Initially Infected Cells. J Virol 91. e-pub ahead of print, https://doi.org/10.1128/JVI.00380-17.
Acknowledgements
We give special thanks to the lab for constructive feedback, Dr. Simon Roux (OSU) for bioinformatics support and the University of Arizona (UA) and OSU for computational storage. We thank Lacey Orsini (UA) and OSU’s undergraduates, especially Aubrey Jensen, for experimental support. As well, we thank Ryan Sontag (BSD at PNNL), and Dr. Karin Holmfeldt for isolating the phage–host system. A portion of the research was performed using EMSL, a DOE Office of Science User Facility sponsored by the Office of Biological and Environmental Research (User Award No. 47930 to MBS). This work was also funded by the Gordon and Betty Moore Foundation grants (GBMF No. 2631, 3305, 3790) to MBS.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Howard-Varona, C., Hargreaves, K.R., Solonenko, N.E. et al. Multiple mechanisms drive phage infection efficiency in nearly identical hosts. ISME J 12, 1605–1618 (2018). https://doi.org/10.1038/s41396-018-0099-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-018-0099-8
This article is cited by
-
Benzo[a]pyrene stress impacts adaptive strategies and ecological functions of earthworm intestinal viromes
The ISME Journal (2023)
-
Protist impacts on marine cyanovirocell metabolism
ISME Communications (2022)
-
Towards an integrative view of virus phenotypes
Nature Reviews Microbiology (2022)
-
Life and death in the soil microbiome: how ecological processes influence biogeochemistry
Nature Reviews Microbiology (2022)
-
Spatial patterns in phage-Rhizobium coevolutionary interactions across regions of common bean domestication
The ISME Journal (2021)