Abstract
Thorarchaeota are a new archaeal phylum within the Asgard superphylum, whose ancestors have been proposed to play possible ecological roles in cellular evolution. However, little is known about the lifestyles of these uncultured archaea. To provide a better resolution of the ecological roles and metabolic capacity of Thorarchaeota, we obtained Thorarchaeota genomes reconstructed from metagenomes of different depth layers in mangrove and mudflat sediments. These genomes from deep anoxic layers suggest the presence of Thorarchaeota with the potential to degrade organic matter, fix inorganic carbon, reduce sulfur/sulfate and produce acetate. In particular, Thorarchaeota may be involved in ethanol production, nitrogen fixation, nitrite reduction, and arsenic detoxification. Interestingly, these Thorarchaeotal genomes are inferred to contain the tetrahydromethanopterin and tetrahydrofolate Wood–Ljungdahl (WL) pathways for CO2 reduction, and the latter WL pathway appears to have originated from bacteria. These archaea are predicted to be able to use various inorganic and organic carbon sources, possessing genes inferred to encode ribulose bisphosphate carboxylase-like proteins (normally without RuBisCO activity) and a near-complete Calvin–Benson–Bassham cycle. The existence of eukaryotic selenocysteine insertion sequences and many genes for proteins previously considered eukaryote-specific in Thorarchaeota genomes provide new insights into their evolutionary roles in the origin of eukaryotic cellular complexity. Resolving the metabolic capacities of these enigmatic archaea and their origins will enhance our understanding of the origins of eukaryotes and their roles in ecosystems.
Similar content being viewed by others
Introduction
The ‘Asgard’ archaea, a superphylum including Loki-, Thor-, Odin-, and Heimdallarchaeota, appear to be the closest archaeal relatives of eukaryotes [1]. Their genomes encode a variety of proteins previously considered eukaryote-specific, which have provided new insights into the archaeal origin hypothesis of eukaryotes [1]. Notably, among Asgard archaea, Thorarchaeota genomes uniquely encode proteins for eukaryotic membrane-trafficking machinery and vesicle biogenesis, indicating the potential position of Thorarchaeota in eukaryotic evolution [1]. Thorarchaeota were first identified from the sulfate-methane transition zone (SMTZ) in estuary sediments [2]. Genomic data show that they might have an important role in sedimentary biogeochemistry since their genomes include predicted genes for organic matter degradation, acetate production and sulfur reduction [2]. A very small number of available genomic bins (only 3 draft genomes with >70% completeness) have limited our understanding of their global ecological roles and metabolisms. For example, although a near-complete Wood–Ljungdahl pathway (WL pathway) was found in Thorarchaeota genomes, they lack the formate dehydrogenase responsible for the initial step in CO2 reduction, which has weakened the case for acetogenesis. This may be due to the incomplete genome-resolved metagenomics sequence coverage. The 16S rRNA gene surveys suggested that Thorarchaeota are broadly distributed in marine and freshwater sediments as well as mangrove sediments, microbial mats, sewage, and sinkhole sediments [1] (Supplementary Figure 1). Given that Thorarchaeota are thought to be descendants of the archaeal host that gave rise to eukaryotic cells, and little is known about their lifestyle, additional Thorarchaeota genomes are urgently needed to better resolve their evolutionary history and functional roles.
Materials and methods
Sample collection and processing
Samples were taken from Mai Po Nature Reserve, a coastal wetland located at Shenzhen River estuary and facing Deep Bay (Hau Hoi Wan) on September 12, 2014. The subsurface sediment samples (MP7, MP8 and MP9) were collected from site covering with mangrove forest (22°29.875’N, 114°01.767’E) at depth intervals of 0–2, 10–15 and 20–25 cm. The other subsurface sediment samples (MP10 and MP11) were taken at an intertidal mudflat (22°29.949’N, 114°01.656’E) with depths of 0–5 and 13–16 cm. The in situ bulk sediments were sealed into plastic bags immediately after collection, and stored in pre-cooled sampling box with ice cubes, then transported to laboratory within <4 h. For each sample, 5 g of wet sediments were taken for physicochemical parameters measurement (Supplementary Table 5). Redox potential, pH, organic matter and the concentrations of ammonium, nitrate and nitrite in pore water via centrifugation from the sediments were determined as described elsewhere [3]. The remaining sediment was stored in −20 °C for DNA isolation. DNA for metagenomics analysis was isolated from 5 g (wet weight) sediment per sample with the PowerSoil DNA Isolation Kit (MO BIO), following the manufacturer’s instructions.
Genomic assembly, binning and annotation
The raw shotgun sequencing metagenomic reads were dereplicated (100% identity over 100% lengths) and trimmed using Sickle (https://github.com/najoshi/sickle). Samples were assembled de novo to obtain five separate assemblies (MP7, MP8, MP9, MP10, and MP11). Whole genome de novo assemblies were performed using IDBA-UD [4] with the following parameters: -mink 65, -maxk 145, -steps 10. Initial binning was carried out using emergent self-organizing maps (ESOM) [5] on MP8 and MP11 assemblies with the reference Thorarchaeotal bins SMTZ1-83, SMTZ1-45 and SMTZ-45. The scaffolds in the area overlapping the reference were extracted for the mapping to all five assemblies. All the mapped raw reads of each sample then were forwarded for reassembly using IDBA-UD [4] with the following parameters: –mink 65, –maxk 145, –steps 3. MaxBin2 were used to automatically binning from the reassemblies [6]. Manual curation was adapted for reducing the genome contamination based on differential coverage, GC content, and the presence of duplicate genes. The completeness, contamination and strain heterogeneity of the genomes within bins were then estimated by using CheckM [7].
Genes were called by Prodigal with the ‘–p meta’ option [8]. Ribosomal RNA-coding regions (16S, 23S) were predicted with Barrnap (https://github.com/tseemann/barrnap). The KEGG server (BlastKOALA) [9], InterProScan database V60 [10], and BLASTP vs. non-redundant protein database retrieved on October 2016 (e-value cutoff of <1e−5) were used to annotate protein functions. In addition, all proteins were assigned to existing COGs and arCOGs by eggNOG-mapper [11]. PRED-SIGNAL [12] and PSORTb [13] were used to identify extracellular peptidases, and the dbCAN web server [14] was used for carbohydrate-active genes identification.
Thorarchaeotal genes were sent to Seblastian, a web server that focuses uniquely on eukaryotic selenoproteins [15]. SECIS and known selenoproteins were identified using the default parameters. Sec-specific tRNA were identified by Seblastian as well as by Secmarker [16].
Phylogenetic analyses
The concatenated 16S and 23S rRNA genes tree and the concatenated 55 ribosomal protein tree were generated using the methods and public data released [1]. The 16S and 23S rRNA gene sequences were aligned using Mafft-LINSi [17], trimmed with BMGE [18] (–m DNAPAM250:4 –g 0.5) and concatenated. Maximum-Likelihood phylogeny of the concatenated 16S and 23S rRNA genes tree was inferred with IQ-TREE with GTR+I+G4 model and ultrafast bootstrapping [19, 20] (–bb 1000). The list of 55 ribosomal proteins of selected archaea and eukaryotes refers to this study [1]. Mafft-LINSi was used to align each ribosomal protein, and BMGE (BLOSUM30 –b 3 –g 0.5) to trim the alignments. An SR4 recoding was performed on the alignment [21]. IQ-TREE with the mixture model (-m LG+C60+F) and with ultrafast bootstrapping [19] (–bb 1000) and Shimodaira-Hasegawa-like approximate likelihood-ratio test [22] (-alrt 1000) was run on the SR4 recoding alignment using a user-defined model referred as ‘C60SR4’ [1] to generate Maximum-Likelihood phylogeny of the 55 concatenated ribosomal protein tree. The Maximum-Likelihood phylogeny of RuBisCO and nifH protein trees were constructed by using RAxML under the LG plus gamma model of evolution (PROTGAMMALG in the RAxML model section), and with the number of bootstraps automatically determined (autoMRE).
Comparative genomic analyses
The Markov Cluster Algorithm (MCL) embedded in the anvi’o software (version 2.2.2, default parameters) [23] was used for protein clustering. ClusterVenn [24] was used for the visualization of orthologous protein clusters across the six Thorarchaeota genomes. The orthologous information was retrieved from eggNOG-mapper annotation results. The genome similarity values was generated and visualized by using OrthoANI [25].
Data availability
The Thorarchaeota genomic bins (MP8T_1, MP9T_1 and MP11T_1) supporting the results of this article are available in NCBI Genbank under the accession numbers: PJER00000000, PJES00000000, and PJET00000000, respectively.
Results and discussion
Genome reconstruction and phylogeny
Coastal mangrove forests are one of the most productive ecosystems. Although limited to tropical or subtropical coastlines and estuaries, they contribute to up to 15% of global carbon storage and provide nutrients and growth habitats for microorganisms, meio/macro-fauna, different flora and migratory birds [26,27,28]. Mai Po Nature Reserve (Hong Kong, China) is located at the Pearl River Estuary. It comprises subtropical mangroves, intertidal mudflats, fishponds and drainage channels. The area is between Shenzhen and Hong Kong, two of the largest cities in China, and a large amount of both domestic and industrial waste water are discharged by Shenzhen River and inland rivers of Hong Kong, consequently contaminating the sediments. Heavy metals (Cu, Pb, Hg, Cr, Ni, Cd, and As, etc), organic pollutants and anthropogenic nitrogen are the main pollutants detected [3, 29,30,31].
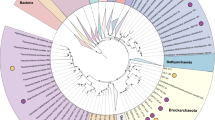
To date, no Asgard genomes have been found in mangrove ecosystems. We collected sediment cores from a mangrove field and an intertidal mudflat in Mai Po (MP) Nature Reserve, and sectioned at different depth layers: three from the mangrove field (MP7: 0–2 cm, MP8: 10–15 cm, and MP9: 20–25 cm), and two from intertidal mudflats (MP10: 0–5 cm, MP11: 13–16 cm). We recovered Thorarchaeota DNA reads from all five samples and reconstructed three high quality Thorarchaeota genomic bins (MP8T_1, MP9T_1 and MP11T_1), using de novo assembly and binning of metagenomics reads. These genomic bins range in size from ~3.5 to ~4.4 Mb, and are 85 to 92% complete, based on the presence of single-copy genes (Table 1). Phylogenetic analyses of both concatenated 55 archaeo-eukaryotic ribosomal proteins and concatenated 16S+23S rRNA genes confirmed the placement of these bins into the phylum Thorarchaeota and within the Asgard superphylum (Fig. 1a, b). Mapping of reads to all available Thorarchaeotal genomes (including MP8T_1, MP9T_1 and MP11T_1) and reassembly yielded a few scaffolds from Thorarchaeota in the top layer of the mangrove sediments (MP7T_1); however, a 50% complete Thorarchaeota genomic bin was reconstructed from the top layer of the intertidal mudflat sediment (MP10T_1). The abundance of reads assigned to Thorarchaeota increased with depth in the sediments (Fig. 1c), suggesting that Thorarchaeota prefer anoxic environments. Although oxygen was not measured at the sampling time, and macrofaunal burrowing activity in the mangroves may induce deeper oxygen penetration over depth [32, 33], we avoided any sampling from area with visible burrows to exclude this factor. For the further analysis, we focused on the three high quality Thorarchaeotal genomic bins (MP8T_1, MP9T_1 and MP11T_1) and compared to genomic bins from an estuary (SMTZ1-83, SMTZ1-45) [2] and a bay (AB25) [1].
Phylogenetic analyses of new Thorarchaeota lineages. a The concatenated 16S and 23S rRNA genes maximum-likelihood tree rooted with bacteria. b The concatenated 55 archaeo-eukaryotic ribosomal proteins from Archaea and Eukarya Maximum-Likelihood tree re-rooted with DPANN and Euryarchaeota. The bootstrap support values above 50, 70 and 85 are indicated with empty, gray and black filled circles, respectively. c The relative abundance of Thorarchaeota in the five samples in Mai Po. The numbers of mapped reads were normalized by total number of sequenced raw reads
Pan-genome analysis of Thorarchaeota
We used the Markov Cluster algorithm (MCL) to identify protein clusters present in all Thorarchaeota genomes [34]. Among the total 6969 protein clusters, ~ 56.0% (3902) were orthologous clusters present in at least two Thorarchaeota. Roughly 13.1% (913) core protein clusters were shared among all the six genomes, among which 296 protein clusters were single-copy genes (Supplementary Figure 2). Orthologous Average Nucleotide Identity (OrthoANI) values (Supplementary Figure 3) suggest that bins MP8T_1 and MP9T_1 belong to the same species, as this value exceeds 97%, while the nucleotide similarities of these two bins to other four Thorarchaeotal genomes ranged from 65.8 to 70.2%, indicating they are distantly related to the others. The OrthoANI value of bins MP11T_1 and SMTZ1-45 is 83.7%. Similar to other published Asgard archaea, the three Thorarchaeotal genomes of MP encode various proteins previously considered as eukaryote-specific including the proteins Sec23/24 and TRAPP domains only found in Thorarchaeota (Supplementary Table 1). In addition, bin MP8T_1 contains a vacuolar sorting-associated gene (vps62) [35, 36], which has not been previously reported in Asgard archaea. These genes encoded in Thorarchaeota are orthologues of cytoskeletal function, vesicular trafficking and endosomal sorting components, which are keys in the early evolutionary stages of eukaryogenesis, underlying the emergence of eukaryotic cellular complexity. Comparison of Thorarchaeota genomes (Fig. 2) revealed more bacterial type protein clusters for metabolic function (COG classification) than archaeal types, suggesting that Thorarchaeota acquired a large number of metabolic processes that originated from bacteria.
Carbon metabolism of Thorarchaeota
All the Thorarchaeota of Mai Po contain genes that encode proteins for a variety of extracellular protein degradation and assimilation processes, including extracellular peptidases, di/oligo peptide uptake, membrane transporters, intracellular aminopeptidases and proteases involved in the degradation of amino acids (Supplementary Table 2), consistent with the previous findings [2]. A limited number of enzymes with specific roles in carbohydrate degradation were also detected (Supplementary Table 3), which again is consistent with the previous suggestion that Thorarchaeota may prefer the heterotrophic life style in degradation of proteins and carbohydrates (Fig. 3). Among genes required for glycolysis, hexokinase is missing in the present six genomes. Genes identified in the SMTZ bins would only allow conversion of phosphoenolpyruvate to pyruvate by phosphoenolpyruvate synthase (PPS). However, the bins MP8T_1 and MP9T_1 not only contain pps genes but also pyruvate kinase (PK), which performs similar functions. The difference is that pyruvate kinase catalyzes an irreversible reaction with no ATP consumption, while phosphoenolpyruvate synthase performs a reversible reaction that requires one mole of ATP per mole of substrate [37]. It has been reported that transcriptional and translational regulation in response to trophic conditions differs between these two enzymes, which is shown by an increased expression level of pps in autotrophically and of pk in heterotrophically grown cells, respectively [38]. Thus, the co-existence of pk and pps may enable Thorarchaeota to adapt to various trophic/environmental conditions through regulating the reversible glycolysis pathway. The tidal wetland ecosystem is primarily considered as a sink of carbon sources, and can export a large amount of inorganic and organic carbon to estuary [39, 40], how the Thorarchaeota in this ecosystem control such potentially reversible glycolysis pathway in response to the organic carbon-rich environment and contribute to the marine carbon cycles are encouraged for further investigations.
Inferred physiological capabilities of Thorarchaeota phylum. Metabolic reconstruction of the Thorarchaeota genomic bin MP8T_1 based on the genes identified using the KEGG database, NCBI non-redundant protein database and eggNOG-mapper annotation. Dashed line indicates absence and solid line indicates presence in bin MP8T_1, and gray line indicates presence in other Thorarchaeota genomic bins. Details about the genes are provided in Supplementary Table 4
The Wood–Ljungdahl (WL) pathway is comprised of a set of enzymes for reducing CO2 and producing acetyl-CoA. The WL pathway can use either tetrahydrofolate (THF) and tetrahydromethanopterin (THMPT) as a C1 carrier, and they are involved with different enzymes, respectively. Generally, most bacterial acetogens use the THF-WL pathway, whereas archaeal methanogens use the THMPT-WL pathway [41]. The Thorarchaeota tend to contain most genes for both WL pathways. However, the absence of formate dehydrogenase (FDH) for initial CO2 reduction in all of our genomic bins suggests the Thorarchaeota identified here probably do not use THF-WL pathway for acetyl-CoA synthesis. Interestingly, all the six MP and SMTZ bins include predicted genes for a complete THMPT-WL pathway, including the formylmethanofuran dehydrogenase complex (fwdABCDEF), which catalyzes the first step of carbon fixation in methanogenesis. Thus, Thorarchaeota are probably able to reduce CO2 through the THMPT-WL pathway. The enzymes of the THMPT-WL pathway tend to oxidize acetyl-CoA that generated from butane oxidation and reversibly release of CO2 (Fig. 3) which has been shown in Candidatus Syntrophoarchaeum [42]. All the Thorarchaeota genomes encode a complete set of genes for butyryl-CoA oxidation to acetyl-CoA (Fig. 3) to allow utilization of the reverse THMPT-WL pathway to oxidize acetyl-CoA to CO2. Although all the present Thorarchaeota genomes except for MP11T_1 and SMTZ1-45 encode putative methylcobalamin:CoM methyltransferase, which is responsible for oxidation of butyl-CoM to butyryl-CoA [42], no known genes for alkyl-CoM formation were detected, leaving their butane oxidation capability unresolved.
The last universal common ancestor of archaea is thought to be a methanogen that contained the WL pathway and used THMPT as C1 carrier [43]. Nevertheless, we did not detect any genes encoding methyl-CoM reductase (mcr) in any Thorarchaeota genomes of this study. The loss of mcr and methanogenesis and the presence of an archaeal WL pathway has been reported in various newly found archaeal lineages [2, 44,45,46,47,48]. Despite this loss, some archaea have retained some or all enzymes of the archaeal WL pathway, possibly a remnant of their ancestral methane-cycling lifestyle [49, 50]. Moreover, some others have adapted to different environmental conditions, e.g., aerobic environments, with the help of lateral gene transfer from bacteria throughout their evolution process [51, 52]. Interestingly, orthologous groups of the genes associated with the THF-WL pathway in Thorarchaeota genomes are all bacterial orthologs. Typically, bacterial acetogens use THF as the cofactor for methyl synthesis, whereas methanogens use THMPT instead [41]. In bacterial metabolism, folate is not only central to the acetyl-CoA pathway, but also more generally the universal C1 carrier for amino acid, cofactor and nucleotide biosynthesis as well as providing the methyl groups for modified bases and ribosome methylation [41]. Archaea generally possess THMPT as a C1 carrier, except that halophiles possess THF, and Methanosarcina barkeri has both THMPT and THF pathways [41]. Interestingly, among the archaea in Asgard superphylum (Supplementary Figure 4), Lokiarchaeotal bin (CR4) also contains both of the pathways, and the Odinarchaeotal bin (LCB_4) contains only THMPT, whereas the three Heimdallarchaeotal bins (AB_125, LC2, and LC3) contain only THF. The diversity of predicted CO2 assimilation pathways in the Asgard archaea suggests diverse metabolic capacities within this group. Understanding how they converge to the transition role between prokaryotes and eukaryotes on the phylogenetic tree will require more evolutionary and physiological evidence.
The ADP-forming subunit of acetyl-CoA synthetase (ACD), which is thought to catalyze acetate production in archaea, was found in all Mai Po and SMTZ Thorarchaeota. The gene for enzyme ACD is commonly found in archaeal genomes [45] as well as in a few bacterial genomes recently [53, 54]. The alternative acetogenesis pathway involves two genes, phosphate acetyltransferase (PTA) and acetate kinase (ACK) [44], both of which were missing in all the present Thorarchaeota bins. In archaea, the ack/pta pathway is currently only found in the methanogenic genus Methanosarcina and phylum Bathyarchaeota [44, 55]. He et al. [44] experimentally verified the ability of Bathyarchaeota to produce acetate using ack/pta pathway by heterologous expression. The ethanol fermentation pathway is believed to be non-existent [2] because of the absence of aldehyde dehydrogenase (ALD), the key enzyme that responsible for the reversible conversion of acetate to aldehyde. However, alternatively, aldehyde ferredoxin oxidoreductase (AOR) as well as NAD(P)-dependent alcohol dehydrogenase (AdhA) are found in the bins MP8T_1 and MP9T_1. The former enzyme could perform similar function as AOR. Such AOR/ADH pathway is considered to be an efficient way for alcohol production, and the hyperthermophilic archaeon Pyrococcus furiosus with single gene insertion proved to be the first archaea that is capable of significant alcohol formation [56].
Three out of six Thorarchaeotal genomic bins appear to contain a near-complete Calvin–Benson–Bassham (CBB) pathway, which is the pathway for carbon fixation using ribulose bisphosphate carboxylase (RubisCO). The bins SMTZ1-83, SMTZ1-45, MP8T_1 and AB25 contain a gene encoding a RubisCO homolog. Phylogenetic analysis of these predicted proteins revealed that the Thorarchaeal RubisCOs form a distinct clade within the Type IV RubisCO (RubisCO-like protein/RLP). While Type I-III RuBisCOs enable light-independent CO2 incorporation into sugars derived from nucleotides like adenosine monophosphate (AMP) [57,58,59], Type IV are probably involved in methionine salvage pathway [57, 60]. Similar to the recently designated archaeal class Hadesarchaea [46], the six Thorarchaeotal bins do not contain the genes for phosphoribulokinase (PRK) in the CBB pathway [61]. The gene prk is commonly absent in archaea, with a few exceptions in methanogens [61,62,63]. However, unlike Thorarchaeota, Hadesarchaea have a Type III RubisCO [46], which is only found in archaea [64]. In addition, a set of intermediate Type II/III RuBisCO were identified in the recently described archaeal phylum Verstraetearchaeota, predicted to be methylotrophic methanogens [65]; however, no CBB pathway genes were found. Interestingly, except for Thorarchaeota, other archaeal members in Asgard superphylum all encode Type III/IV RuBisCO genes, but are still predicted to be missing the CBB pathway (Supplementary Figure 5). If the presence of a Type III RuBisCO and CBB pathway in Hadesarchaea indeed broaden our understanding of archaeal inorganic carbon fixation [46], the co-existence and functionality of RLP and CBB pathway in Thorarchaeota is still mysterious. It is believed that all the other RuBisCOs and RLP are originated from the Type III RuBisCO in archaeal methanogens [58]. Thus, the Thorarchaeota may have abandoned their ancient autotrophic lifestyle as their RLP evolved from its Type III origin, while retaining other CBB genes. However, we cannot rule out other possible functions in relation to sulfur metabolism for RLP, e.g., functions as 5-methylthio-D-ribulose-1-phosphate (MTR-1P) isomerase that bridges S-adenosylmethionine (SAM)-dependent polyamine biosynthesis to isoprenoid biosynthesis [66, 67].
Nitrogen metabolism in Thorarchaeota
Nitrogen fixation is an important microbial process that converts atmospheric N2 to ammonia in mangrove ecosystems. All six Thorarchaeotal genomes contain predicted genes for nitrogen fixation protein NifH and nitrogenase cofactors, suggesting Thorarchaeota can use nitrogen gas as a nitrogen source for amino acid biosynthesis and/or release ammonium for mangroves. Phylogenetic analysis revealed that the Thorarchaeotal nifH genes are of archaeal origin, and form a distinct group along with other nifH genes from newly designated archaeal groups, e.g., Theionarchaea, Bathyarchaeota and Altiarchaeales (Supplementary Figure 6). Other nitrogen fixation genes, e.g., nifU, nifS, nifX and nifB involved in the assembly and incorporation of iron and molybdenum into the nitrogenase subunits were identified in all Thorarchaeota genomes except for MP11T_1, which is missing nifX (Supplementary Table 4). Nitrite can also be a nitrogen sources for Thorarchaeota, as suggested by the detection of nitrite reductase (NADH) large subunit (nirB) that can convert nitrite to ammonia. These genes were detected in all Thorarchaeotal genomes except for MP11T_1. The nirB gene was detected in Thermococcus sp., Halogeometricum sp. and recently in Bathyarchaeota subgroups 1 and 7/17 [45]. However, no genes encoding enzymes catalyzing nitrate reduction were detected in Thorarchaeota, implying that the nitrite does not derived from nitrate reduction.
Arsenic transformation in Thorarchaeota
Arsenic and selenium are both known as “essential toxins” [68]. In prokaryotes, these two elements are readily metabolized and participate in a full range of metabolic functions including assimilation, methylation, detoxification and anaerobic respiration [68]. MP8T_1, MP9T_1 and SMTZ1-45 encode a full arsenic efflux detoxification pathway, consisting of phosphate transporters, arsenate reductase and putative arsenical pump-driving ATPase. These three genes cooperate to accumulate and reduce As (V) to As (III), then pump As (III) out of the cells. In addition, putative arsenite S-adenosylmethyltransferase (ArsM) were found in all Mai Po Thorarchaeotal genomic bins as well as in SMTZ1-45, indicating that Thorarchaeota possess an arsenic methylation pathway. The first identification and characterization of an archaeal arsM was reported in a methanogen (Methanosarcina acetivorans) recently [69]. It is arguable whether the methylation is a detoxification mechanism since it may produce more toxic methylated intermediates, e.g., monomethylarsenite and dimethylarsenite. However, it is proved that the two intermediates do not accumulate in the cells expressing arsM, and the cells produce trimethylarsine gas as the final product [70, 71]. The identification of both arsenic efflux detoxification and methylation pathways in Thorarchaeota provides novel insights into the role of Thorarchaeota in the arsenic biogeochemical cycle. Specifically, in mangrove sediments, high levels of As were found around the world ranging from 0.52 to 70 mg kg–1 [72]. A recent survey in the same area as the present study reported up to 93 mg kg–1 arsenic in mangrove sediment [29]. It provides an environmental implication that Thorarchaeota not only adapt to sediment condition with high As, but also could be possibly applied to bioremediation of As-contaminated sediment or water.
Selenocysteine-encoding system in Thorarchaeota
Selenocysteine (Sec) is a cysteine analog with selenium replacing sulfur. Selenoproteins are a rare class of proteins that possess a Sec residue, and require a specific set of genes dedicated to Sec synthesis and insertion: SelA, selenocysteine synthase; SelB, a special translation factor that binds guanine nucleotides and recognizes selenocystyl-tRNASec; SelC, a Sec-specific tRNA (tRNASec); and SelD, the selenophosphate synthase [68]. Selenoproteins are not present in all organisms, but scatteredly distributed among the three domains of life [73]. The known functions of selenoproteins are, e.g., redox homeostasis, electron transport/energy metabolism, compound detoxification and oxidative protein folding in bacteria [74]; hydrogenotrophic methanogenesis, Sec biosynthesis in archaea [74, 75]; and very diverse for eukaryotic selenoproteins [76]. Thorarchaeota bin SMTZ1-83 contains all the four components, and the other five Thorarchaeotal bins only lack SelA. A search for known selenoproteins in Thorarchaeotal genomes on the Seblastian server did not return any significant hits [15]. Sec insertion sequences (SECIS) are RNA structures found in the selenoprotein transcripts as the main signals for Sec insertion. Generally, the sequence or structure of SECIS are distinct between the three domains of life [77]. However, recently, conserved RNA structures of SECIS in Lokiarchaeota were found resemble the eukaryotic SECIS [74]. Similar to Lokiarchaeota, multiple eukaryotic-like SECIS were identified in each of the Thorarchaeotal genomes (Supplementary Figure 7). It indicates that Thorarchaeota may encode currently unknown families of selenoproteins. Subsequently, the possessions of eukaryotic-like SECIS in other Asgard archaea (Odinarchaeota and Heimdallarchaeaota) were expected as their close affiliation with eukaryotes [1], however, neither eukaryotic-like SECIS nor tRNASec were detected. The presences of tRNASec in other archaea are only available in Methanococcales and Methanopyrus kandleri, which are relatively rare compare to that in bacteria and eukaryotes [16]. It is unclear whether the selenocysteine-encoding system emerged prior to the divergence of Loki-/Thorarchaeota and Odin-/Heimdallarchaeota, and the possibility of horizontal gene transfer after the division of these Asgard archaea are open.
In this study, we reconstructed three Thorarchaeota genomes from mangrove and mudflat sediments to resolve the role of this new archaeal phylum in biogeochemical cycling. In addition to the previously described metabolic capabilities, including organic matter degradation, inorganic carbon fixation, sulfur/sulfate reduction and acetate production, they also appear to be involved in ethanol production, nitrogen fixation and nitrite reduction, as well as arsenic detoxification. The RuBisCO protein and near-complete CBB cycle genes reveal potential carbon metabolic versatility in the Thorarchaeota. Thorarchaeota are predicted to contain THMPT-WL and THF-WL pathways, and the latter appears to have originated from bacteria. The presence of eukaryotic-like selenocysteine insertion sequences, as well as a collection of proteins previously considered eukaryote-specific, in Thorarchaeotal genomes, provides new insights into the origin of eukaryotic cellular complexity. In conclusion, our results enrich current knowledge of the lifestyle and the metabolic capacity of Thorarchaeota, begin to resolve the modern ecological functions of this new archaeal phylum, but also pave the way for advancing our understandings on the metabolism of ancestral archaea host of eukaryogenesis [78].
References
Zaremba-Niedzwiedzka K, Caceres EF, Saw JH, B ckstr m D, Juzokaite L, Vancaester E, et al. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature. 2017;541:353–8.
Seitz KW, Lazar CS, Hinrichs K-U, Teske AP, Baker BJ. Genomic reconstruction of a novel, deeply branched sediment archaeal phylum with pathways for acetogenesis and sulfur reduction. ISME J. 2016;10:1696–705.
Zhou Z, Meng H, Liu Y, Gu J-D, Li M. Stratified bacterial and archaeal community in mangrove and intertidal wetland mudflats revealed by high throughput 16S rRNA gene sequencing. Front Microbiol. 2017;8:2148. https://doi.org/10.3389/fmicb.2017.02148.
Peng Y, Leung HCM, Yiu SM, Chin FYL. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 2012;28:1420–8.
Dick GJ, Andersson AF, Baker BJ, Simmons SL, Thomas BC, Yelton AP, et al. Community-wide analysis of microbial genome sequence signatures. Genome Biol. 2009;10:R85.
Wu Y-W, Simmons BA, Singer SW. MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics. 2016;32:605–7.
Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–55.
Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119.
Kanehisa M, Sato Y, Morishima K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol. 2016;428:726–31.
Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–40.
Huerta-Cepas J, Forslund K, Coelho LP, Szklarczyk D, Jensen LJ, Mering von C, et al. Fast genome-wide functional annotation through orthology assignment by eggNOG-Mapper. Mol Biol Evol. 2017; 34:2115-22.
Bagos PG, Tsirigos KD, Plessas SK, Liakopoulos TD, Hamodrakas SJ. Prediction of signal peptides in archaea. Protein Eng Des Sel. 2008;22:27–35.
Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, et al. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics. 2010;26:1608–15.
Yin Y, Mao X, Yang J, Chen X, Mao F, Xu Y. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012;40:W445–W451.
Mariotti M, Lobanov AV, Guigó R, Gladyshev VN. SECISearch3 and Seblastian: new tools for prediction of SECIS elements and selenoproteins. Nucleic Acids Res. 2013;41:e149.
Santesmasses D, Mariotti M, Guigó R. Computational identification of the selenocysteine tRNA (tRNASec) in genomes Gough J (ed). PLoS Comput Biol. 2017;13:e1005383.
Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80.
Criscuolo A, Gribaldo S. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol. 2010;10:210.
Minh BQ, Nguyen MAT, Haeseler von A. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 2013;30:1188–95.
Nguyen L-T, Schmidt HA, Haeseler von A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2014;32:268–74.
Susko E, Roger AJ. On reduced amino acid alphabets for phylogenetic inference. Mol Biol Evol. 2007;24:2139–50.
Guindon SP, Dufayard J-FO, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–21.
Eren AM, Esen ÖC, Quince C, Vineis JH, Morrison HG, Sogin ML, et al. Anvi’o: an advanced analysis and visualization platform for ‘omics data. PeerJ. 2015;3:e1319.
Wang Y, Coleman-Derr D, Chen G, Gu YQ. OrthoVenn: a web server for genome wide comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 2015;43:W78–W84.
Ouk Kim Y, Chun J, Lee I, Park S-C. OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol. 2016;66:1100–3.
Alongi DM. Carbon cycling and storage in Mangrove Forests. Annu Rev Mar Sci. 2014;6:195–219.
Bhattacharyya A, Majumder NS, Basak P, Mukherji S, Roy D, Nag S, et al. Diversity and distribution of archaea in the Mangrove sediment of Sundarbans. Archaea. 2015;2015:1–14.
Yan B, Hong K, Yu ZN. Archaeal communities in mangrove soil characterized by 16S rRNA gene clones. J Microbiol. 2006;44:566–71.
Li R, Chai M, Qiu GY. Distribution, fraction, and ecological assessment of heavy metals in sediment-plant system in Mangrove Forest, South China Sea. PLoS ONE. 2016a;11:e0147308.
Li R, Xu H, Chai M, Qiu GY. Distribution and accumulation of mercury and copper in mangrove sediments in Shenzhen, the world’s most rapid urbanized city. Environ Monit Assess. 2016b;188:87.
Liang Y, Wong MH. Spatial and temporal organic and heavy metal pollution at Mai Po Marshes Nature Reserve, Hong Kong. Chemosphere. 2003;52:1647–58.
Cai L, Lin J, Li H. Macroinfauna communities in an organic-rich mudflat at Shenzhen and Hong Kong, China. Bull Mar Sci. 2001;69:1129–38.
Luo L, Gu J-D. Influence of macrofaunal burrows on extracellular enzyme activity and microbial abundance in subtropical mangrove sediment. Microb Ecol. 2016;52:1807–10.
van Dongen S, Abreu-Goodger C. Using MCL to extract clusters from networks. Methods Mol Biol. 2012;804:281–95.
Bonangelino CJ. Genomic screen for vacuolar protein sorting genes in Saccharomyces cerevisiae. Mol Biol Cell. 2002;13:2486–501.
Wiederhold E, Gandhi T, Permentier HP, Breitling R, Poolman B, Slotboom DJ. The yeast vacuolar membrane proteome. Mol Cell Proteom. 2008;8:380–92.
Herzberg O, Chen CCH, Liu S, Tempczyk A, Howard A, Wei M, et al. Pyruvate site of pyruvate phosphate dikinase: crystal structure of the enzyme−phosphonopyruvate complex, and mutant analysis. Biochemistry. 2002;41:780–7.
Tjaden B, Plagens A, Dorr C, Siebers B, Hensel R. Phosphoenolpyruvate synthetase and pyruvate, phosphate dikinase of Thermoproteus tenax: key pieces in the puzzle of archaeal carbohydrate metabolism. Mol Microbiol. 2006;60:287–98.
Maher DT, Santos IR, Golsby-Smith L, Gleeson J, Eyre BD. Groundwater-derived dissolved inorganic and organic carbon exports from a mangrove tidal creek: the missing mangrove carbon sink? Limnol Ocean. 2013;58:475–88.
Bauer JE, Cai W-J, Raymond PA, Bianchi TS, Hopkinson CS, Regnier PAG. The changing carbon cycle of the coastal ocean. Nature. 2013;504:61–70.
Sousa FL, Martin WF. Biochemical fossils of the ancient transition from geoenergetics to bioenergetics in prokaryotic one carbon compound metabolism. Biochim Biophys Acta. 2014;1837:964–81.
Laso-Pérez R, Wegener G, Knittel K, Widdel F, Harding KJ, Krukenberg V, et al. Thermophilic archaea activate butane via alkyl-coenzyme M formation. Nature. 2016;539:396–401.
Borrel G, Adam PS, Gribaldo S. Methanogenesis and the Wood–Ljungdahl pathway: an ancient, versatile, and fragile association. Genome Biol Evol. 2016;8:1706–11.
He Y, Li M, Perumal V, Feng X, Fang J, Xie J, et al. Genomic and enzymatic evidence for acetogenesis among multiple lineages of the archaeal phylum Bathyarchaeota widespread in marine sediments. Nat Microbiol. 2016;1:16035.
Lazar CS, Baker BJ, Seitz K, Hyde AS, Dick GJ, Hinrichs K-U, et al. Genomic evidence for distinct carbon substrate preferences and ecological niches of Bathyarchaeota in estuarine sediments. Environ Microbiol. 2016;18:1200–11.
Baker BJ, Saw JH, Lind AE, Lazar CS, Hinrichs K-U, Teske AP, et al. Genomic inference of the metabolism of cosmopolitan subsurface Archaea, Hadesarchaea. Nat Microbiol. 2016;1:16002.
Probst AJ, Weinmaier T, Raymann K, Perras A, Emerson JB, Rattei T, et al. Biology of a widespread uncultivated archaeon that contributes to carbon fixation in the subsurface. Nat Commun. 2014;5:5497.
Sousa FL, Neukirchen S, Allen JF, Lane N, Martin WF. Lokiarchaeon is hydrogen dependent. Nat Microbiol. 2016;1:16034.
Bapteste É, Brochier C, Boucher Y. Higher-level classification of the Archaea: evolution of methanogenesis and methanogens. Archaea. 2005;1:353–63.
Vornolt J, Kunow J, Stetter KO, Thauer RK. Enzymes and coenzymes of the carbon monoxide dehydrogenase pathway for autotrophic CO2 fixation in Archaeoglobus lithotrophicus and the lack of carbon monoxide dehydrogenase in the heterotrophic A. profundus. Arch Microbiol. 1995;163:112–8.
Nelson-Sathi S, Dagan T, Landan G, Janssen A, Steel M, McInerney JO, et al. Acquisition of 1,000 eubacterial genes physiologically transformed a methanogen at the origin of Haloarchaea. Proc Natl Acad Sci USA. 2012;109:20537–42.
Becker EA, Seitzer PM, Tritt A, Larsen D, Krusor M, Yao AI, et al. Phylogenetically driven sequencing of extremely halophilic archaea reveals strategies for static and dynamic osmo-response. PLoS Genet. 2014;10:e1004784.
Parizzi LP, Grassi MCB, Llerena LA, Carazzolle MF, Queiroz VL, Lunardi I, et al. The genome sequence of Propionibacterium acidipropionici provides insights into its biotechnological and industrial potential. BMC Genom. 2012;13:562.
Schmidt M, Schönheit P. Acetate formation in the photoheterotrophic bacterium Chloroflexus aurantiacus involves an archaeal type ADP-forming acetyl-CoA synthetase isoenzyme I. FEMS Microbiol Lett. 2013;349:171–9.
Rother M, Metcalf WW. Anaerobic growth of Methanosarcina acetivorans C2A on carbon monoxide: an unusual way of life for a methanogenic archaeon. Proc Natl Acad Sci USA. 2004;101:16929–34.
Basen M, Schut GJ, Nguyen DM, Lipscomb GL, Benn RA, Prybol CJ, et al. Single gene insertion drives bioalcohol production by a thermophilic archaeon. Proc Natl Acad Sci USA. 2014;111:17618–23.
Tabita FR, Hanson TE, LI H, Satagopan S, Singh J, Chan S. Function, structure, and evolution of the RubisCO-like proteins and their RubisCO homologs. Microbiol Mol Biol Rev. 2007;71:576–99.
Tabita FR, Hanson TE, Satagopan S, Witte BH, Kreel NE. Phylogenetic and evolutionary relationships of RubisCO and the RubisCO-like proteins and the functional lessons provided by diverse molecular forms. Philos Trans R Soc B. 2008;363:2629–40.
Wrighton KC, Castelle CJ, Varaljay VA, Satagopan S, Brown CT, Wilkins MJ, et al. RubisCO of a nucleoside pathway known from Archaea is found in diverse uncultivated phyla in bacteria. ISME J. 2016;10:2702–14.
Ashida H, Saito Y, Nakano T, Tandeau de Marsac N, Sekowska A, Danchin A, et al. RuBisCO-like proteins as the enolase enzyme in the methionine salvage pathway: functional and evolutionary relationships between RuBisCO-like proteins and photosynthetic RuBisCO. J Exp Bot. 2007;59:1543–54.
Berg IA, Kockelkorn D, Ramos-Vera WH, Say RF, Zarzycki J, Hügler M, et al. Autotrophic carbon fixation in archaea. Nat Rev Micro. 2010;8:447–60.
Mueller Cajar O, Badger MR. New roads lead to RubisCO in archaebacteria. BioEssays. 2007;29:722–4.
Reysenbach AL, Flores GE. Electron microscopy encounters with unusual thermophiles helps direct genomic analysis of Aciduliprofundum boonei. Geobiology. 2008;6:331–6.
Sato T, Atomi H, Imanaka T. Archaeal type III RuBisCOs function in a pathway for AMP metabolism. Science. 2007;315:1003–6.
Vanwonterghem I, Evans PN, Parks DH, Jensen PD, Woodcroft BJ, Hugenholtz P, et al. Methylotrophic methanogenesis discovered in the archaeal phylum Verstraetearchaeota. Nat Microbiol. 2016;1:16170.
Erb TJ, Evans BS, Cho K, Warlick BP, Sriram J, Wood BM, et al. A RubisCO-like protein links SAM metabolism with isoprenoid biosynthesis. Nat Chem Biol. 2012;8:926–32.
North JA, Sriram J, Chourey K, Ecker CD, Sharma R, Wildenthal JA, et al. Metabolic regulation as a consequence of anaerobic 5-methylthioadenosine recycling in Rhodospirillum rubrum. Mbio. 2016;7:e00855–16.
Stolz JF, Basu P, Santini JM, Oremland RS. Arsenic and selenium in microbial metabolism. Annu Rev Microbiol. 2006;60:107–30.
Wang P-P, Sun G-X, Zhu Y-G. Identification and characterization of arsenite methyltransferase from an archaeon, Methanosarcina acetivorans C2A. Environ Sci Technol. 2014;48:12706–13.
Qin J, Rosen BP, Zhang Y, Wang G, Franke S, Rensing C. Arsenic detoxification and evolution of trimethylarsine gas by a microbial arsenite S-adenosylmethionine methyltransferase. Proc Natl Acad Sci USA. 2006;103:2075–80.
Yin XX, Chen J, Qin J, Sun GX, Rosen BP, Zhu YG. Biotransformation and volatilization of arsenic by three photosynthetic cyanobacteria. Plant Physiol. 2011;156:1631–8.
Wu G-R, Hong H-L, Yan C-L. Arsenic accumulation and translocation in Mangrove (Aegiceras corniculatum L.) grown in arsenic contaminated soils. Int J Environ Res Public Health. 2015;12:7244–53.
Mariotti M, Santesmasses D, Capella-Gutierrez S, Mateo A, Arnan C, Johnson R, et al. Evolution of selenophosphate synthetases: emergence and relocation of function through independent duplications and recurrent subfunctionalization. Genome Res. 2015;25:1256–67.
Mariotti M, Lobanov AV, Manta B, Santesmasses D, Bofill A, Guigó R, et al. Lokiarchaeota marks the transition between the archaeal and eukaryotic selenocysteine encoding systems. Mol Biol Evol. 2016;33:2441–53.
Stock T, Rother M. Selenoproteins in archaea and gram-positive bacteria. Biochim Biophys Acta. 2009;1790:1520–32.
Lobanov AV, Hatfield DL, Gladyshev VN. Eukaryotic selenoproteins and selenoproteomes. Biochim Biophys Acta. 2009;1790:1424–8.
Krol A. Evolutionarily different RNA motifs and RNA-protein complexes to achieve selenoprotein synthesis. Biochimie. 2002;84:765–74.
Spang A, Saw JH, Jørgensen SL, Zaremba-Niedzwiedzka K, Martijn J, Lind AE, et al. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature. 2015;521:173–9.
Acknowledgements
This work was supported by National Natural Science Foundation of China (No. 31622002, 41506163) to M.L., a China Postdoctoral Science Foundation (No. 2017M612718), a Natural Science Foundation of Guangdong Province, China (No. 2017A030310296) and a National Natural Science Foundation of China (No. 31700430) to Y.L., and a Sloan Fellowship in Ocean Science to B.J.B.
Author contributions:
Y.L. and M.L. conceived the study. Z.C.Z. and J.D.G. sampled in the field, revised and reviewed the paper. Y.L. and J.P. analyzed the data. B.J.B. interrupted the metabolic capabilities. Y.L., M.L., Z.C.Z. and B.J.B. wrote the paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest:
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Liu, Y., Zhou, Z., Pan, J. et al. Comparative genomic inference suggests mixotrophic lifestyle for Thorarchaeota. ISME J 12, 1021–1031 (2018). https://doi.org/10.1038/s41396-018-0060-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-018-0060-x
This article is cited by
-
Long-read assembled metagenomic approaches improve our understanding on metabolic potentials of microbial community in mangrove sediments
Microbiome (2023)
-
Potential for mercury methylation by Asgard archaea in mangrove sediments
The ISME Journal (2023)
-
Inference and reconstruction of the heimdallarchaeial ancestry of eukaryotes
Nature (2023)
-
Three families of Asgard archaeal viruses identified in metagenome-assembled genomes
Nature Microbiology (2022)
-
Genomic and transcriptomic dissection of Theionarchaea in marine ecosystem
Science China Life Sciences (2022)