Abstract
Diatoms are a diverse and ecologically important group of phytoplankton. Although most species are considered free living, several are known to interact with other organisms within the plankton. Detailed imaging and molecular characterization of any such partnership is, however, limited, and an appraisal of the large-scale distribution and ecology of such consortia was never attempted. Here, observation of Tara Oceans samples from the Benguela Current led to the detection of an epibiotic association between a pennate diatom and a tintinnid ciliate. We identified the diatom as Fragilariopsis doliolus that possesses a unique feature to form barrel-shaped chains, associated with seven different genera of tintinnids including five previously undescribed associations. The organisms were commonly found together in the Atlantic and Pacific Ocean basins, and live observations of the interaction have been recorded for the first time. By combining confocal and scanning electron microscopy of individual consortia with the sequencing of high-resolution molecular markers, we analyzed their distribution in the global ocean, revealing morpho-genetically distinct tintinnid haplotypes and biogeographically structured diatom haplotypes. The diatom was among the most abundant in the global ocean. We show that the consortia were particularly prevalent in nutrient-replete conditions, rich in potential predators. These observations support the hypothesis of a mutualistic symbiosis, wherein diatoms acquire increased motility and tintinnids benefit from silicification through increased protection, and highlight that such associations may be more prevalent than currently appreciated.
Similar content being viewed by others
Introduction
Marine phytoplankton are photosynthetic microbes responsible for around half of Earth’s net primary production [1]. Diatoms, a ubiquitous and predominant component of phytoplankton, are enveloped in a characteristic silica cell wall known as the frustule, and have been proposed to contribute around 40% of marine net primary productivity [2]. They serve as the basis of the marine food web and are significant players in global biogeochemical cycles [3, 4]. Diatoms are frequently reported to dominate phytoplankton communities in well-mixed coastal, as well as upwelling regions, where light and nutrients are available [5]. They are nonetheless frequent and diverse in open ocean oligotrophic systems [6] where their survival in such low-nutrient regions can depend on mutualistic associations with other plankton. For instance, some diatoms live in obligate or facultative symbioses with heterocystous N2-fixing cyanobacteria [7]. Diazotrophic bacteria such as Richelia intracellularis and Calothrix rhizosoleniae provide nitrogen in usable forms to the diatoms (e.g., Hemiaulus and Rhizosolenia spp.), which in return may provide structural protection to the cyanobacteria.
Planktonic diatoms have been described in numerous other biological interactions, involving a range of organisms across all domains of life, as well as viruses [8]. Beyond predation, competition, and parasitism, examples include endosymbiotic diatoms in nummulitid foraminifera (e.g., Thalassionema-related species in the foraminifera Heterostegina depressa [9]), in dinoflagellates (e.g., Galeidinium rugatum and Durinskia baltica [10, 11]), and pennate diatoms with attached bacteria [12], copepods [13, 14], other diatoms (e.g., Pseudo-nitzschia linea and Chaetoceros spp. [15]), Phaeocystis colonies [16], flagellated stramenopiles [17], and vorticellids [18].
On the other hand, tintinnids (Choreotrichida) are heterotrophic planktonic ciliates enveloped in a species-specific test composed of organic material, the lorica [19]. They represent one of the morphologically most diverse groups of planktonic protists [20], are abundant and ubiquitous throughout the water column, with concentrations ranging from 101 to 104 cells per liter in surface waters [21]. A few extracellular associations between tintinnids and live diatoms have been reported, such as ones involving the radial centric diatoms Chaetoceros spp. and Eutintinnus spp. either described as “phoretic commensalism”—wherein transport is believed to be the main benefit for diatoms [22]—or suggested as a form of obligate epiphytism enabling predation avoidance for tintinnids, and access to nutrients for diatoms [23]. The chain-forming pennate diatom Fragilariopsis doliolus was also recorded (as Pseudoeunotia doliolus) with Eutintinnus tenuis in material collected in 10 equatorial stations between the Galapagos archipelago and the Marquesas Islands [24]. Small chains of F. doliolus were previously found associated with Salpingella subconica near the Prince Edward Islands in the Southern Ocean with rates of association involving 3–30% of all F. doliolus and 35–83% of S. subconica cells encountered, as well as in the Benguela Current [25, 26]. These authors speculate that buoyancy and protection against mesozooplankton predation are the main advantages gained by the attachment of both partners. Of a different nature is the association of Laackmaniella and other tintinnids with apparently empty frustules of Fragilariopsis and other diatoms covering their lorica, also found in the Southern Ocean, for which it has been hypothesized that the ciliates retain diatom frustules following ingestion of the cellular contents, perhaps as a means of protection through camouflage [27, 28].
The many observations summarized above tend to indicate that diatom–tintinnid associations may be more prevalent than commonly thought. However, detailed molecular and morphological characterizations of these consortia are still lacking, as is a large-scale evaluation of their biogeography and ecology. The recent Tara Oceans expeditions have generated a worldwide, standardized amount of multidisciplinary information focusing on open ocean and size-fractionated plankton communities in the upper layer of the ocean [29, 30]. In the current study, morphological analysis of samples collected in the Benguela Current off South Africa (Station TARA_066) led to the initial observation and isolation of an epibiotic association between a pennate diatom and a tintinnid ciliate morphologically assigned to F. doliolus and Salpingella sp., respectively. The sequencing of a specific genetic marker within the ribosomal DNA (rDNA) gene loci, the short V9 region of the 18S rDNA gene, was performed to interrogate the Tara Oceans V9 metabarcoding data set. This revealed the biogeography of the organisms, thus highlighting nine stations in which sequence matches of both partners were high. We further used various microscopy methods and DNA sequencing of multiple genetic markers to analyze similar and new consortia within these nine other Tara Oceans stations. Additionally, the V4 region was sequenced because this marker resolves more accurately diatom diversity [31], and for similar reasons sequences of the ITS loci were generated for the tintinnids [32, 33]. Overall, a continuous sequencing from the beginning of 18S to D1–28S was completed to obtain highly resolved phylogenetic information. Live recordings of the interaction confirm its true existence off the Brazilian coast, and provide insights into its ecological significance. We further provide a detailed description of the pairing specificity, biogeography, and ecology of this prevalent interaction between an ecologically important diatom and heterotrophic ciliates.
Materials and methods
The sampling strategy used in the Tara Oceans expedition is described in Pesant et al. [34], and samples used for morphology and genomics are listed in Table S1.
Morphological investigation of the consortium
Confocal laser-scanning microscopy (CLSM)
Consortia were analyzed from samples fixed on board Tara (in 2010) with a mix of formaldehyde (1% final concentration) and glutaraldehyde (0.25% final concentration) and imaged by CLSM (Leica TCS SP8), equipped with an HC PL APO 40×/1.10 W motCORR CS2 objective. Multiple fluorescent dyes were used to observe the cellular components of the ciliate and the microalgae, such as the nuclei (blue, Hoechst, Ex405/Em420-470) and the cellular membranes (green, DiOC6, Ex488/Em500-520), and cell surface (cyan, AlexaFluor 546 Ex552Em560-590). The autofluorescence of the chlorophyll was also visualized (red, Ex638/Em680-700). Sample preparation, staining, mounting, and CLSM protocols are detailed in Colin et al. [35]. Image processing and three-dimensional reconstructions were conducted with Fiji [36] and IMARIS (Bitplane) software. For consortia quantification, see Supplementary Information. Additional observations were made on samples from the Outpace Cruise in 2015; for details of sampling protocol and processing see Dolan et al. [37].
Scanning electron microscopy (SEM)
For SEM, we used 0.5 ml of 20–180 µm size fraction surface formaldehyde-fixed samples from Station TARA_102 placed on 3 µm pore size nucleopore filters, washed in distilled water, dehydrated in ethanol series (25, 50, 75, 95, and 100%) and critical point-dried. Dried filters were mounted on stubs, sputter-coated with gold-palladium, and observed using a JSM 6700F (JEOL Ltd, Tokyo, Japan).
Live recording of the consortia
Natural plankton samples were collected at a fixed station off Ubatuba, Brazil (23.52° S, 45.09° W) by means of horizontal tows of a 1.5 m long, 20 μm mesh sized net, between 21st June and 26th July 2017. Live samples were diluted with surface seawater, transferred to an insulated container, and immediately transported to the laboratory, where they were kept in a walk-in, temperature-controlled room set to match the ambient seawater temperature (21–23 °C). Upon no more than 1 h after sampling, aliquots of the diluted sample were pipetted onto a Fluorodish (WPI, Sarasota, USA) and observed under an Olympus IX73 inverted microscope equipped with a 40X objective and a Photron SA2 high speed camera. Image sequences were taken at 500 and 1000 frames per second (fps), and stored at 30 fps for visualization.
Station selection for isolation and molecular identification of the consortia
Universal PCR amplification of the V4-V9 18S rDNA subregion
Single consortia composed of the Fragilariopsis–Salpingella associations were isolated from formaldehyde (1%)–glutaraldehyde (0.25%)–fixed surface samples collected by a microplankton net (20–180 μm mesh size) in Station TARA_066 located in the Benguela Current in 2010 (Figures S1, S2). Using a glass micropipette, consortia were collected based on morphological traits of the barrel-shaped diatom, rinsed two to three times in a minimum volume of sterile artificial seawater, before proceeding to DNA extraction, universal-eukaryotic V4-V9 PCR amplification, cloning, plasmid purification, and sequencing (see Supplementary Information).
Detection in Tara Oceans stations
Sequencing results were assembled and trimmed to retain the V9 18S fragment and interrogated against the Tara Oceans ribotype database using BLAST [38]. The Tara Oceans nucleotide sequences are available at the European Nucleotide Archive (ENA) under the project PRJEB402 and PRJEB6610. Diatom isolated V9 sequences matched at 100% identity with the ribotype “f2f8b” assigned to “Raphid-pennate_X+sp”. Tintinnid isolated V9 sequences matched with three different tintinnid ribotypes and in majority with “a7cbc” and “b61a7” assigned to “Choreotrichia_X+sp.” (see Table S2 for complete barcode identifier). The search for these four sequences in the global metabarcoding data set revealed that they were widely distributed in various size fractions [39] and were positively correlated (Spearman rho = 0.37, p-value < 2.2e-16)(Figure S3a). Nine additional stations were chosen for further analysis, based on the simultaneous presence of over 30 copies of the diatom and tintinnid ribotypes (summed by assignation): Stations TARA_070, TARA_102, TARA_106, TARA_109, TARA_111, TARA_122, TARA_124, TARA_128, and TARA_139, in fraction 20–180 μm of surface samples where consortia were big enough to be identified and isolated, except TARA_128 in which we investigated the 5–20 μm fraction. Abundances of the four ribotypes are shown in Table S3 completed with an additional diatom barcode (5ecf4) later discovered in Station TARA_102.
Molecular and phylogenetic analysis of the diatom–tintinnid consortium
Advanced micromanipulation for cell isolation
A new method of isolation was applied to collect Fragilariopsis–Salpingella consortia from ethanol-preserved samples collected by a plankton net (20 μm mesh size) in the 10 selected Tara Oceans stations from the Indian, Atlantic, and Pacific Oceans. By using an OLYMPUS IX51 inverted microscope equipped with an Eppendorf manual microinjector CellTram® Air, single consortia were rinsed three times in 100% ethanol Labtech wells before being treated for DNA extraction according to steps in the MasterPure DNA and RNA purification kit (Epicenter). Samples used for molecular identification are available in Table S1.
Targeted PCR amplification of small subunit rDNA genes, internal transcribed spacers ITS1 and ITS2, 5.8S rDNA and 28S rDNA genes
To obtain different phylogenetic ribosomal markers for both partners, initial group-specific amplifications were conducted with the Phusion High-Fidelity DNA Polymerase (Finnzymes). Group-specific primers were designed and inspired from Bachy et al. [32] and McDonald et al. [40] and are shown in Table S4. For amplification protocol, see Supplementary Information. Amplicon sequences of both the diatom and the tintinnid were cleaned, trimmed, and assembled using Sequencher (version 5.4) and accession numbers are available in Table S1. Summaries of available imaging and molecular data for each station and consortia are available in Tables 1 and 2.
Phylogenetic analysis
For the tintinnids, contigs of the amplicons were obtained, and two matrices of 18S rDNA and ITS+5.8S+28S rDNA were built, including reference and outgroup sequences from Bachy et al. [32]. For diatoms, similar matrices were built including reference sequences from BLAST top hits in GenBank and reference sequences from Theriot et al. [41]. Sequences were aligned using MAFFT version 7 [42] and trimmed with Gblocks [43] before applying JmodelTest [44] to determine the best model of nucleotide substitution for each matrix. The general time-reversible model with gamma distribution of rate variation (GTR+G) was selected for the diatom ITS+5.8S+28S tree and the tintinnid 18S tree, which are, respectively, the best markers to anchor species at the genus level. Phylogenetic and Bayesian inferences were performed using PhyML 3.0 [45] and the program MrBayes [46], respectively (see supplementary information). Trees were visualized and edited using FigTree v.1.4.2. For ITS+5.8S+28S rDNA sequences, a statistical parsimony network was constructed and visualized with TCS software [47]. All alignments used for phylo-genetic inference are available in Table S5.
Environmental and community contextualization of the interaction
Partial least square analysis
We investigated correlations between environmental parameters collected during Tara Oceans (doi: 10.1594/PANGAEA.853810) and the read abundance of the two partners. As variables were shown to be multicolinear, partial least square regression analysis was conducted on range-transformed oceanographic data as predictors and Hellinger-transformed read abundance data of corresponding barcodes as responses. Samples from all size fractions in surface depth were selected to increase statistical signal. Analysis was performed using the « plsdepot » package in R version 3.3.0. Predictors and responses used in the analysis as well as resulting regression coefficients are available in Table S6.
Results and Discussion
Discovery of a widespread diatom–tintinnid interaction
Confocal microscopy analysis of microplankton from Tara Oceans Station TARA_066 in the Benguela Current (off South Africa) revealed an interaction between a diatom and a tintinnid (Figs. 1a, b). Isolation of individual consortia by micromanipulation led to the identification of the 18S rDNA sequences of both partners, including the V9 hypervariable region (Materials and methods section). A search for the same V9–18S rDNA sequences in the Tara Oceans global metabarcoding data set from 126 sampling stations [39] revealed nine additional stations with high sequence abundance of both partners, which were therefore chosen for further investigation. Visual evidence of Fragilariopsis–Salpingella consortia was established in all the selected samples along with the discovery of new ciliate genera involved in the association (Table 1).
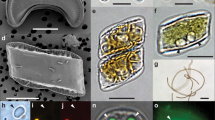
Light, electron, and confocal laser-scanning microscopy observations of Fragilariopsis–Salpingella associations and interface. a, b, d, e Salpingella faurei at Station TARA_066. c Typical smooth lorica and trumpet shaped oral opening of Salpingella decurtata at Station TARA_139. d Amphorides laackmanni at Station TARA_106. f Salpingella curta at Station TARA_066. g-i SEM pictures of a diatom–tintinnid consortium from Station TARA_102 g-h. Panel h is a close up view of the contact zone in g where the arrow indicates membranelles. The diatom frustule valves display the specific features from Fragilariopsis doliolus: transverse striae (s) with two alternating rows of poroids (p) at the base of the interstriae (i), the eccentric raphe system (r), lying at the junction of valve face and proximal mantle (m). Panel i shows a full barrel of diatom chain taken at Station TARA_102. j Three-dimensional (3D) reconstruction of the epibiosis involving Salpingella faurei imaged with confocal laser-scanning microscopy at Station TARA_066 labeling DNA (Hoechst, blue), chloroplasts (chlorophyll autofluorescence, red), membranes (DiOC6, green), cell surface (AlexaFluor 546, cyan). Scale bar = 10 μm except in c where scale bar = 20 μm
Morphological diversity of diatom–tintinnid consortia
Diatom-associated ciliates displayed diverse morphologies, with at least seven different ciliate genera including five that had never been described with F. doliolus before (see Supplementary Information taxonomic details). Light microscopy analysis of specimens from the Benguela Current (Figs. 1a, b, d, e) were identified as Salpingella faurei whereas those from the tropical North Pacific Ocean (Fig. 1c) were identified as Salpingella decurtata. The diatom was also associated with Salpingella curta in the tropical southeastern Pacific Ocean (Station TARA_102, data not shown) and Eutintinnus spp. in the Marquesas Islands area (Fig. 2). New associations involving five ciliate genera (Figs. 2a–e) are reported here for the first time and in some cases consortia have been seen with pennate diatoms other than F. doliolus (Figs. 2h–l). Regarding morphology of the diatom, SEM revealed barrel-shaped chains and diatom frustule valves displaying transverse striae with two alternating rows of poroids at the base of the interstriae, the latter rising externally above the level of the valve, as well as a strongly eccentric raphe system, lying at the junction of the valve face and proximal mantle (Fig. 1g). These ultrastructural features, along with the asymmetric valve shape, clearly distinguish the pennate diatom F. doliolus from congeneric species, which are all symmetrical bilaterally and have smooth external valve faces [48, 49].
High diversity of pennate–tintinnid interactions in the open ocean observed by light microscopy and CLSM. a Protorhabdonella tintinnid at Station TARA_124. b Ascampbelliella tintinnid at Station TARA_110. c Dictyocysta tintinnid at Station TARA_066. d Amphorellopsis tintinnid at Station TARA_066. e Unknown tintinnid at Station TARA_066. f, g Eutintinnus tintinnid at Station TARA_124 with f one or g two diatom chains. h-l Tintinnid interactions with other diatom species (h, j and l probably Thalassionemataceae) extracted from the Outpace cruise (date: 28/03/2015; Lat: 18°25,15’S; Long: 165°56,38’W between New Caledonia and Tahiti). Scale bar = 10 μm
Several features of the consortia are noteworthy. Contrary to other cases of diatom–tintinnid associations in which lorica are generally coated with empty diatom frustules, the diatom cell content was clearly intact (e.g., Fig. 1j). Tintinnid cells were also observed within the lorica, with rare exceptions including Eutintinnus and Protorhabdonella specimens, in which it was most likely the fixative (lugol, formol, or glutaraldehyde-paraformaldehyde) that induced cell loss of the ciliate, as intact cells were repeatedly observed in ethanol-preserved samples. Colonies of F. doliolus differed in cell number, from a few adjacent cells surrounding less than half of the tintinnid (Fig. 1d) to a nearly complete barrel (Fig. 1i) totally hiding the ciliate. Moreover, we differentiated cases in which the Salpingella lorica opening was located just at the end of the diatom cell (Figs. 1b, d, e, g, h, i, j), or not (Figs. 1a, c, f), independently of the diatom–tintinnid size ratio. At times, two diatom colonies were observed on the same Eutintinnus cell (Fig. 2g). No tintinnids were ever found associated with an entirely empty frustule chain of F. doliolus. Three-dimensional reconstructions from CLSM images also showed a tight adherence of both lorica and frustule with no evidence of any specific attachment structure (Figure S1).
Live recording of diatom–tintinnid consortia in natural coastal waters
Live recordings of the consortium from fresh samples illustrate the consortium’s mobility and occurrence off the Brazilian coast. The tintinnid motility profits both partners and enables forward (Video S1) and backward (Video S2) swimming, as well as changing directions when the ciliate membranelles encounter other organisms such as the dinoflagellate Ceratium (Video S3). The diatom serves as an external physical envelope that enables the tintinnid to sense co-occurring species (such as a centric diatom in Video S4) before direct contact with the lorica. A further hypothesis is that attached ciliates profit from changes in fluid dynamics of the feeding current, the so-called anchor effect, which leads to steeper velocity gradients and higher flow rates close to the lorica [50]. Moreover, even though F. doliolus cells possess a raphe, it may not be useful without a solid surface, so the diatom may benefit from the interaction by increased motility. As no attachment structure has been observed, this reinforces the idea that a strong biofilm composed of extracellular polymeric substances may seal both organisms. These are, to our knowledge, the first live recordings of such an association.
Molecular identification of the interacting partners
A total of 24 individual diatom–tintinnid consortia were isolated by micromanipulation from the 10 Tara Oceans stations, representing a range of oceanic regions (Table 1 and Materials and methods section). We focused on Salpingella sp. to test for genetic specificity because it occurred in all the chosen stations. Following DNA extraction, group-specific PCR amplification was performed to sequence a series of rDNA molecular markers (18S, ITS1+2, 5.8S, 28S-D1) from both the diatom and the tintinnid in each consortium. Sequences were obtained for at least one of the partners, yielding sequences for 21 diatoms and 18 tintinnids, and we could genetically identify both partners for 15 different consortia originating from six stations (Table 2).
ITS, 5.8S, and 28S molecular markers were chosen to perform the diatom phylogeny because the 18S rDNA has been considered a relatively poor marker for studies of Fragilariopsis and Pseudo-nitzschia phylogeny (but see Lim et al. [51]). The diatom sequences of the isolated specimens formed a monophyletic group (bootstrap value 96%) branching within the Fragilariopsis genus (bootstrap value 91%) (Fig. 3a). Fragilariopsis appears as monophyletic and branched within the paraphyletic Pseudo-nitzschia genus (Fig. 3a). These first publicly available sequences of F. doliolus (identified by our morphological analysis) obtained from field material revealed that the species is distinct from the previously known clades in the genus Fragilariopsis, which reflects the unique above-mentioned morphological characteristics of the species, as well as its much wider and more temperate distribution compared with its congeners [6].
Phylogenetic identification of the diatom and tintinnid partners. a Diatom tree. Maximum likelihood rooted phylogenetic tree of diatom ITS + 5.8 S + 28 SrDNA sequences (854 aligned positions). Reference sequences were extracted from top assigned BLAST hits in NCBI. Numbers at nodes are percentage bootstrap values (1000X). The branch rooting of the tree has been shortened to increase clarity, and isolates are colored according to their respective stations of origin. b Tintinnid tree. Maximum likelihood rooted phylogenetic tree of choreotrich SSU-rDNA sequence, based on 1455 aligned positions. Reference sequences were extracted from [20, 32]. Numbers at nodes are percentage bootstrap values (1000X) (values < 70% are omitted). Bayesian posterior probabilities higher than 0.90 are indicated by filled circles. Symbiotic tintinnid isolates are colored according to their respective stations of origin, indicated in the map in panel a
Molecular phylogeny of the isolated tintinnid 18S rDNA confirmed the identification of Salpingella genus, as all Salpingella sequences (ours and the one previously published) grouped in a monophyletic clade with strong support (bootstrap value 97.6%; posterior probability 1). In spite of the morphological differences of the isolated specimens, we could not distinguish Salpingella species at the genetic level (bootstrap value 67%), indicating the limited resolution of the available 18S sequence information or illustrating a morphological plasticity of the Salpingella species (Fig. 3b).
To gain further insights into the genetic diversity of the species involved in these consortia, statistical parsimony networks of the ITS1+2 regions, together with 5.8S and the D1 region of 28S were undertaken. F. doliolus sequences diverged up to nine nucleotides over a 727-bp alignment using 20 sequences (Fig. 4a). Three major haplotypes were distinguished, with haplotypes 1 and 2 being composed of sequences originating from stations belonging to distant provinces. Sequences originating from the Benguela Current (Station TARA_066) all grouped together compared with the rest of the sequences and were closest to haplotype 3, showing the existence of region-specific genetic variants of F. doliolus.
Phylogeographic patterns of both partners based on haplotype similarity networks. Sequences are from ITS+5.8S+28S rDNA. a Haplotypes from the diatom Fragilariopsis doliolus displayed more genetic diversity and biogeographical structuring particularly for sequences originating from the Benguela Current, such as from Station TARA_066 (stations and color codes are indicated in panel a). b The tintinnid Salpingella sp. haplotype network displays two main clusters (haplotypes 1 and 2) of sequences coming from diverse ocean regions, but with low genetic diversity overall. In each network, a white node represents one mutation, and the size of the colored circles are proportional to the abundances of sequences within a haplotype. Haplotypes are colored by the stations of origin of sequences composing it, and labeled by the “TI” identifiers. Ocean Province: SAO (South Atlantic Ocean), SPO (South Pacific Ocean)
The tintinnid network based on 17 sequences of 721 bp revealed two major haplotypes, composed of sequences from geographically distant stations (Fig. 4b). Both haplotypes can be found simultaneously in two distant stations (Station TARA_066 and TARA_106). Moreover, haplotype 1 corresponded to isolated tintinnids with a long lorica under light microscopy (similar to Figs. 1c, f), whereas haplotype 2 generally corresponded to a shorter morphotype (similar to Figs. 1a, b). This reveals that the morphological similarity of haplotypes within the isolated tintinnids is not a function of geographical separation. However, lorica length is known to be a variable character [52]. Besides TI_823 and TI_884, the other tintinnid sequences diverged from the major haplotypes by fewer than five nucleotides. The combinations of tintinnid haplotypes with F. doliolus haplotypes were not specific.
We further explored the genetic diversity of both partners of the consortia by focusing on the V9 subregion (Table S5), a marker widely used in metabarcoding surveys [53]. The complete V9 diatom sequences displayed three variable positions out of 130 bp with respect to other Fragilariopsis sequences available at NCBI, in positions 58, 70, and 86. Fragilariopsis species therefore display a 98% similiarity for this molecular marker. A typical 97% identity clustering of OTUs from a metabarcoding survey would have grouped together V9 sequences from F. doliolus and F. curta or F. cylindrus, yet none of the latter two species could possibly interact with tintinnids the way F. doliolus does due to the unique barrel-shape morphology of its chain. The V9 tintinnid sequences displayed one significantly variable site at position 55 and the sequences displayed a 3-bp difference with the V9 sequence available at NCBI of a S. acuminata isolate FG304. Additionally, tintinnids belonging to haplotype 1 (except TI_887) displayed a T at position 55, whereas tintinnids belonging to haplotype 2 displayed a C, reinforcing the existence of two haplotypes revealed with the parsimony network.
Geographic distribution and ecological context of the consortia
Among the 15 complete F. doliolus V9 sequences obtained, 11 matched the Tara Oceans metabarcode f2f8b (Table 2). Significantly, barcode f2f8b, which had been assigned to “Raphid_pennate_X+sp.”, was among the most abundant unassigned diatom barcodes in the primary Tara Oceans data set, with 301,093 reads over the 293 samples from 46 sampling sites within the micro- and meso-plankton size fractions (containing organisms between 20–180 μm and 180–2000 μm, respectively) [6], representing 24.5% of the abundance of all the “Raphid-pennate_X+sp.” sequences in the top 100 unassigned barcodes. This single barcode was nearly as abundant as all barcodes assigned to the Pseudo-nitzschia genus (down to 80% of sequence identity), which represents the 7th most abundant diatom genus with 305,115 reads, based on Malviya et al. [6]. In light of the extended Tara Oceans data set [39], encompassing 126 Tara Oceans stations, f2f8b is the second most prevalent Fragilariopsis sequence with a total abundance of 414,113 reads. It is found throughout all Tara Oceans stations, with higher abundances reported in the southern hemisphere (0°S–40°S) and occurrences both in coastal and open ocean stations (Fig. 5a). By comparison, one diatom V9 matched at 100% identity the barcode “53cf4” that totaled 853 reads across the whole Tara Oceans metabarcoding data set and displayed a distribution similar to that of f2f8b (Figure S3b). The other three complete V9 sequences from isolated diatom cells were not found in the Tara Oceans metabarcode data set. It is possible that the V9 microdiversity was not captured in the Tara Oceans data due to sequencing biases or filtering of the metabarcode database, which was performed by removal of metabarcodes present in less than three reads and two distinct samples [38].
Abundance of the symbiotic diatom and tintinnid V9 rDNA sequences across the 126 Tara Oceans stations and environmental predictors of each symbiotic partner. a Absolute abundance of the most prevalent V9 rDNA sequences from the symbiotic tintinnid (a7cbc and b61a7) and diatom (f2f8b) in the Tara Oceans global metabarcoding data set [39]. Abundances were extracted from samples of microplankton (20–180 μm size fraction) collected in sub-surface waters [34]. Regions framed with a gray dashed line in the Mediterranean Sea, Red Sea, and South Pacific Ocean have been magnified on the right for more clarification. Absolute abundances were transformed according to the log(Abundance+1) formula. b Circle of correlations. The abundance of the diatom (red) and tintinnid (blue) V9 rDNA barcodes were predicted using the Tara Oceans metabarcoding and environmental parameters (green) data sets from surface water samples. The most prevalent tintinnid barcode a7cbc is more abundant in high density samples, whereas the main abiotic driver for the second tintinnid barcode b61a7 is chlorophyll. Regression coefficients are available in Table S6
In total, the tintinnid V9 sequences matched at 100% identity to three unique barcodes in the Tara Oceans data set: a7cbc, b61a7, and deb2a, all of which had been assigned to “Choreotrichia_XX+sp”. Barcode a7cbc was particularily abundant and widely distributed, with an abundance of 106,292 reads over the complete Tara Oceans metabarcoding data set (Fig. 5a). B61a7 had a total abundance of 24,839 reads and deb2a of 3660 reads, the latter being restricted to the Cape Agulhas region (Fig. 5a and Figure S3c). All the tintinnids belonging to haplotype 1 displayed a V9 sequence corresponding to b61a7, whereas all the tintinnids belonging to haplotype 2 displayed a V9 corresponding to a7cbc. The pairing of the V9 sequences belonging to the two partners within the consortium was not specific, coherent with the conclusions of the haplotype combinations (Table S4).
Sequences with 100% identity to the specific diatom and tintinnid ribotypes were extracted from all the size fractions from surface samples of the 126 Tara Oceans stations and analyzed with respect to oceanographic variables such as temperature, nitrate, phosphate, and silicate through partial least square analysis (Fig. 5b). F2f8b and 53cf4 diatom barcode abundances were explained by temperature (regression coefficients of 0.21 and 0.02, respectively), nitrate (0.23 and 0.01), and density (−0.25 and −0.02), a measure derived from temperature and salinity. Abundance of the tintinnid barcode a7cbc was predicted by temperature with a negative (−0.20) regression coefficient, followed by oxygen (0.23) and density (0.18). B61a7 (tintinnid) was explained by chlorophyll (−0.18), nitrate (−0.18), and phosphate (−0.16) (Table S6). F2f8b (diatom) and a7cbc (tintinnid), the most abundant barcodes of each partner, showed opposite signs with respect to the majority of abiotic predictors, yet the organisms were paired together in similar samples. Consequently, these results suggest that conditions favoring one partner are opposite than for the other, and that other factors impact the distribution of the consortia, such as biotic interactions. Moreover, the consortia reported in this study occurred in nutrient-replete regions (Figure S4), illustrated by the higher nitrate concentrations (Welch two sample t-test p-value < 0.05), contrary to other symbiotic partnerships in the ocean that often occur in oligotrophic conditions [7]. For example, Station TARA_066 is located in the Benguela Current, station TARA_070 is situated at the limit of the nitrate plume originating from the Benguela Current flowing northwest, Stations TARA_102, 106, and 109 are in the Peru Current, Station TARA_124 benefits from the island mass effect of the Marquesas archipelago, and Station TARA_139 is located in the North Pacific Equatorial Countercurrent. Taken together, these data suggest that access to nutrients is not the primary benefit for the diatoms in these consortia. However, diatoms use nutrients from the surface layer and assimilate them through photosynthesis, which can lead to the production of exudates of particulate matter that tintinnids can potentially benefit from directly or indirectly [54].
To explore biotic factors that may influence the interaction, we used the metabarcode data to examine the occurrence of the consortia with tintinnid competitors (oligotrichs) and potential predators (copepods), both known to impact tintinnid distribution [55], as well as to prey on diatoms. The total barcode abundance of oligotrichs and copepods was assessed in 126 stations sampled during the Tara Oceans expedition (Figure S5a,b). Barcode abundance of oligotrichs did not display an overlapping pattern with the diatom–tintinnid association. However, significantly higher copepod abundance was found in samples in which the association was observed (Welch two sample t-test p-value < 0.05), suggesting that high predation pressure may favor occurrence of the consortia. Flow cytometry counts of bacteria and pico-eukaryotes, potential prey for the tintinnids, were not significantly different in stations where the consortia were found (Figure S5c). Although tintinnids can feed on diatoms [27, 56], most of the tintinnids involved in the consortia are characterized by small lorica oral diameters suggesting that small prey items of pico- and nano-sizes are likely to be their preferred prey [57] and could potentially benefit from increased abundance of bacteria and nanoflagellate bacteriovores in the diatom phycosphere. As a complementary non-targeted approach, we searched for the tintinnid and diatom V9 sequences within a global ocean co-occurrence network [58]. Although this revealed the presence of the tintinnid a7cbc sequence, it did not co-occur with the relevant diatom species (see Supplementary Table S7 and Figure S6).
Finally, the ecological importance of the diatom–tintinnid association was quantified through actual cell counts in different stations using the 20–180 µm size fractions by CLSM and light microscopy (Fig. 6) and lugol-fixed samples (Supplementary Information), allowing us to evaluate the prevalence of free-living and associated partners, as well as quantification at species level. Up to 700 diatom–tintinnid consortia per ml of net sample were enumerated, equivalent to ~40 to 50 consortia/L of seawater in samples from Station TARA_066 in the Benguela Current and TARA_102 in the upwelling Peruvian west coastal Pacific current (Figs. 6a, b). Diatom-associated Salpingella represented up to 95% of all Salpingella lorica (Figure S7) and constituted over 50% of the tintinnid community (Table S8). Conversely, over 74% of F. doliolus chains present in a sample could be associated to tintinnids, indicating that when both partners co-occur the association is dominant as compared with the abundances of free partners. Notwithstanding, we also found some stations in which both partners were free living. For example, in Station TARA_137 we found free Salpingella sp. in the absence of F. doliolus (data not shown). In Stations TARA_106 and TARA_109, we found low abundances of F. doliolus (<10 chains/L) and Salpingella sp., yet very few associations were detected (Fig. 6).
Quantification of diatom–tintinnid consortia across the ocean based on observations of net samples and converted in consortia/L of seawater (20–180 μm fraction). a CLSM counts of both partners and their physiological state in samples from eight Tara Oceans stations. b Counts from light microscopy observations. Four different species of tintinnids displaying associations with F. doliolus were counted across nine different formol-fixed samples in which the association was the most abundant (stations in the South and the North Pacific as well as the Benguela Current). The abundance of the four species per liter of seawater are shown, depending on whether they are bound to F. doliolus or not
Conclusions
Our study investigates the epibiotic association between the diatom F. doliolus and tintinnid ciliates, revealing the widespread occurrence, ecological importance, and morphogenetic characteristics of the Fragilariopsis–Salpingella consortia, as well as reporting five new tintinnid genera involved in the interaction.
This study is significant because it reveals that F. doliolus possesses a unique morphological feature, being able to form barrel-shaped chains enabling dynamic interactions with tintinnids. The species is highly abundant and distributed worldwide, and also displays biogeographic structuring evidenced by the first molecular data available for this species. Moreover, analysis of the Salpingella sp. brings to light two major haplotypes that are not related to geographic separation yet correspond to phenotypic features. The combination of morphological and molecular studies reveals the presence of viable diatom and tintinnid cells in a motile consortium, a widespread distribution in nutrient-replete and predator-rich conditions, as well as a long-term nonspecific stability. These observations support the hypothesis of a mutualistic symbiosis, wherein diatoms would acquire increased motility and tintinnids benefit from silicification through increased protection. Our study not only highlights an abundant and ubiquitous marine microbial interaction by integrating data from the single cell to the global ocean, but also explores potential benefits underlying such an association that further studies will help elucidate.
References
Field CB, Behrenfeld MJ, Randerson JT, Falkowski P. Primary production of the biosphere: integrating terrestrial and oceanic components. Science. 1998;281:237–40.
Nelson DM, Tréguer P, Brzezinski MA, Leynaert A, Quéguiner B. Production and dissolution of biogenic silica in the ocean: revised global estimates, comparison with regional data and relationship to biogenic sedimentation. Glob Biogeochem Cycles. 1995;9: 359–72.
Falkowski PG. The ocean’s invisible forest. Sci Am. 2002;287: 54–61.
Smetacek V. Biological oceanography: diatoms and the silicate factor. Nature. 1998;391:224–25.
Morel FMM, Price NM. The biogeochemical cycles of trace metals in the oceans. Science. 2003;300:944–47.
Malviya S, Scalco E, Audic S, Vincent F, Veluchamy A, Poulain J, et al. Insights into global diatom distribution and diversity in the world’s ocean. Proc Natl Acad Sci USA. 2016;113:E1516–25.
Foster RA, Kuypers MMM, Vagner T, Paerl RW, Musat N, Zehr JP. Nitrogen fixation and transfer in open ocean diatom-cyanobacterial symbioses. ISME J. 2010;5:1484–93.
Tomaru Y, Takao Y, Suzuki H, Nagumo T, Nagasaki K. Isolation and characterization of a single-stranded RNA virus infecting the bloom-forming diatom C haetoceros socialis. Appl Environ Microbiol. 2009;75:2375–81.
Holzmann M, Berney C, Hohenegger J. Molecular identification of diatom endosymbionts in nummulitid Foraminifera. Symbiosis. 2006;42:93–101.
Schnepf E, Elbrächter M. Dinophyte chloroplasts and phylogeny—a review. Grana. 1999;38:81–97.
Takano Y, Hansen G, Fujita D, Horiguchi T. Serial replacement of diatom endosymbionts in two freshwater dinoflagellates, P eridiniopsis spp. (Peridiniales, Dinophyceae). Phycologia. 2008;47:41–53.
Kaczmarska I, Ehrman JM, Bates SS, Green DH, Léger C, Harris J. Diversity and distribution of epibiotic bacteria on Pseudo-nitzschia multiseries (Bacillariophyceae) in culture, and comparison with those on diatoms in native seawater. Harmful Algae. 2005;4:725–41.
Fernandes LF, Calixto-Fere M. Morfologia e distribuição de duas diatomáceas (Bacillariophyta) epizóicas no Brasil. Acta Bot Bras. 2012;26:836–41.
Gárate-Lizárraga I, Muñetón-Gómez MS. Primer registro de la diatomea epibionte Pseudohimantidium pacificum y de otras asociaciones simbióticas en el Golfo de California. Acta Bot Mex. 2009;88:31–45.
Ruggiero MV, Sarno D, Barra L, Kooistra WHCF, Montresor M, Zingone A. Diversity and temporal pattern of Pseudo-nitzschia species (Bacillariophyceae) through the molecular lens. Harmful Algae. 2015;42:15–24.
Sazhin AF, Felipe Artigas L, Nejstgaard JC, Frischer MF. The colonization of two Phaeocystis species (Prymnesiophyceae) by pennate diatoms and other protists: a significant contribution to colony biomass. Phaeocystis, major link in the biogeochemical cycling of climate-relevant elements. Biogeochemistry. 2007; 83: 137.
Gómez F, Moreira D, Benzerara K, López-García P. S olenicola setigera is the first characterized member of the abundant and cosmopolitan uncultured marine stramenopile group MAST-3. Environ Microbiol. 2010;13:193–202.
Nagasawa S, Warren A. Redescription of V orticella o ceanica Zacharias, 1906 (Ciliophora: Peritrichia) with notes on its host, the marine planktonic diatom C haetoceros coarctatum Lauder,1864. Hydrobiologia. 1996;337:27–36.
Agatha S, Laval-Peuto M, Simon P. The Tintinnid Lorica. In: Dolan JR, Montagnes DJS, Agatha S, Coats WD, Stoecker DK, editors. The Biology and Ecology of Tintinnid Ciliates: Models for Marine Plankton. West Sussex: Wiley-Blackwell; 2013. p. 17–41.
Bachy C, Dolan JR, López-García P, Deschamps P, Moreira D, Accuracy of protist diversity assessments: morphology compared with cloning and direct pyrosequencing of 18S rRNA genes and ITS regions using the conspicuous tintinnid ciliates as a case study. ISME J. 2012a;7:244–55.
McManus GB, Santoferrara LF. Tintinnids in microzoolankton communities. In: Dolan JR, Montagnes DJS, Agatha S, Stoecker DK, editors. The Biology and Ecology of Tintinnid Ciliates: Models for Marine Plankton. Oxford: Wiley-Blackwell; 2012. p. 198–213.
Pavillard J. Observations sur les Diatomées (2e série). Bull De la Société Bot De Fr. 1916;60:126–33.
Gómez F. On the consortium of the tintinnid Eutintinnus and the diatom Chaetoceros in the Pacific ocean. Mar Biol. 2007;151:1899–906.
Pavillard J. Péridiniens et diatomées pélagiques recueillis par Alain Gerbault entre les Iles Marquises et les Iles Galapagos. Bull. Inst. Océanogr. Monaco. 1935;669:1–8.
Froneman PW, Pakhomov EA, Meaton V. Observations on the association between the diatom, Fragilariopsis doliolus Wallich, and the tintinnid, Salpingella subconica Kofoid. S Afr J Sci. 1998;94:202–202.
Decelle J, Colin S and Foster RA. Photosymbioses in marine planktonic protists. In Ohtsuka S, Suzaki T, Horiguchi T, Suzuki N, and Not F, editors. Marine Protists. Springer, Tokyo; 2015.
Gowing MM, Garrison DL. Abundance and feeding ecology of larger protozooplankton in the ice edge zone of the Weddell and Scotia seas during the austral winter. Deep Sea Res Part A Oceanogr Res Pap. 1992;39:893–919.
Wasik A, Mikolajczyk E, Ligowski R. Agglutinated loricae of some Baltic and Antarctic Tintinnina species (Ciliophora). J Plankton Res. 1996;18:1931–940.
Karsenti E, Acinas SG, Bork P, Bowler C, De Vargas C, Raes J, et al. A holistic approach to marine eco-systems biology. PLoS Biol. 2011;9:e1001177.
Bork P, Bowler C, de Vargas C, Gorsky G, Karsenti E, Wincker P. Tara Oceans studies plankton at planetary scale. Science. 2015;348:873–873.
Zimmermann J, Jahn R, Gemeinholzer B. Barcoding diatoms: evaluation of the V4 subregion on the 18S rRNA gene, including new primers and protocols. Org Divers Evol. 2011;11:173–92.
Bachy C, Gómez F, López-García P, Dolan JR, Moreira D. Molecular phylogeny of tintinnid ciliates (Tintinnida, Ciliophora). Protist. 2012b;163:873–87.
Santoferrara L, McManus GB, Alder VA. Utility of genetic marker sand morphology for species discrimination within the order Tintinnida (Ciliophora, Spirotrichea). Protist. 2013;164: 24–36.
Pesant S, Not F, Picheral M, Kandels-Lewis S, Le Bescot N, Gorsky G, et al. Open science resources for the discovery and analysis of Tara Oceans data. Sci data. 2015;2:150023.
Colin S, Coelho LP, Sunagawa S, Bowler C, Karsenti, Bork EP, et al. Quantitative 3D-imaging for cell biology and ecology of environmental microbial eukaryotes. Elife. 2017;6:pii: e26066. (accepted in eLife with minor revisions).
Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–82.
Dolan JR, Gimenez A, Cornet-Barthaux V. Community structure of tintinnid ciliates of the microzooplankton in the South East Pacific Ocean: comparison of a high primary productitivity with a typical oligotrophic site. J Eukaryot Microbiol. 2016;63: 813–22.
de Vargas C, Audic S, Henry N, Decelle J, Mahe F, Logares R, et al. Eukaryotic plankton diversity in the sunlit ocean. Science. 2015;348:1261605.
Richter DJ, Watteaux R, Vannier T, Leconte J, Reygondeau G, Maillet N, et al. Global plankton biogeography is shaped via ocean circulation dynamics. (Manuscript in preparation)
McDonald SM, Sarno D, Zingone A. Identifying P seudo-nitzschia species in natural samples using genus-specific PCR primers and clone libraries. Harmful Algae. 2007;6:849–60.
Theriot EC, Ashworth M, Ruck E, Nakov T, Jansen R. A preliminary multigene phylogeny of the diatoms (Bacillariophyta): challenges for future research. Plant Ecol Evol. 2010;143:278–96.
Katoh K, Standley DM. MAFFT: iterative refinement and mdditional methods. Methods Mol Biol Mult Seq Alignment Methods. 2013;1079:131–46.
Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol USYB. 2007;56:564–77.
Darriba D, Taboada GL, Doallo R, Posada D. JModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9:772.
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–21.
Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP. Bayesian inference of phylogeny and its impact on evolutionary biology. Science. 2001;294:2310–314.
Clement M, Posada D, Crandall KA. TCS: a computer program to estimate gene genealogies. Mol Ecol. 2000;9:1657–60.
Medlin LK, Sims PA. The transfer of Pseudoeunotia doliolus to Fragilariopsis. Beih Zur Nova Hedwig. 1993;106:323–34.
Round FE, Crawford RM, Mann DG. The diatoms, Biology and Morphology of the Genera. Cambridge: Cambridge University Press; 1990.
Jonsson PR, Johansson M, Pierce RW. Attachment to suspended particles may improve foraging and reduce predation risk for tintinnid ciliates. Limnol Oceanogr. 2004;49:1907–914.
Lim HC, Teng ST, Lim PT, Wolf M, Leaw CP. 18S rDNA phylogeny of P seudo-nitzschia (Bacillariophyceae) inferred from sequence-structure information. Phycologia. 2016;55:134–46.
Laval-Peuto M, Brownlee DC. Identification and systematics of the Tintinnina (Ciliophora): evaluation and suggestion for improvement. Ann. Inst. Océanogr., Paris. 1986;62:69–84.
Amaral-Zettler LA, Mccliment EA, Ducklow HW, Huse SM. A method for studying protistan diversity using massively parallel sequencing of V9 hypervariable regions of small-subunit ribosomal RNA genes. PLoS ONE. 2009;4:e6372.
Gügi B, Costaouec T, Burel C, Lerouge P, Helbert W, Bardor M. Diatom-specific oligosaccharide and polysaccharide structures help to unravel biosynthetic capabilities in diatoms. Mar Drugs. 2015;13:5993–6018.
Dolan JR, Claustre H, Carlotti F, Plounevez S, Moutin T. Microzooplankton diversity: relationships of tintinnid ciliates with resources, competitors and predators from the Atlantic coast of Morocco to the eastern Mediterranean. Deep Sea Res Part I: Oceanogr Res Pap. 2002;49:1217–232.
Verity PG, Villareal TA. The relative food value of diatoms, dinoflagellates, flagellates, and cyanobacteria for tintinnid ciliates. Arch Für Protistenkd. 1986;131:71–84.
Dolan JR. Morphology and ecology in tintinnid ciliates of the marine plankton: correlates of lorica dimensions. Acta Protozool. 2010;49:235–344.
Lima-Mendez G, Faust K, Henry N, Decelle J, Colin S, Carcillo F, et al. Determinants of community structure in the global plankton interactome. Science. 2015;348:1262073.
Acknowledgements
We thank the commitment of the following people and sponsors who made the Tara Oceans project possible: Centre National de la Recherche Scientifique and the Groupement de Recherche GDR3280, European Molecular Biology Laboratory, Génoscope/Commissariat à l’Energie Atomique, the French Government “Investissements d’Avenir” programs OCEANOMICS (ANR-11-BTBR-0008), FRANCE GENOMIQUE (ANR-10-INBS-09-08), MEMO LIFE (ANR-10-LABX-54), Paris Sciences et Lettres Research University (ANR-11-IDEX-0001-02), the European Union Framework Programme 7 (MicroB3/No.287589), Agnès b., the Veolia Environment Foundation, Region Bretagne, World Courier, Illumina, Cap l’Orient, the Electricité de France Foundation EDF Diversiterre, Fondation pour la Recherche sur la Biodiversité, the Prince Albert II de Monaco Foundation, Etienne Bourgois, and the Tara schooner, its captains and crew. Tara Oceans would not exist without continuous support from 23 institutes (oceans.taraexpeditions.org). CB acknowledges funding from a European Research Council Advanced Grant Award (Diatomite: 294823), the LouisD Foundation of the Institut de France, and a Fellowship from the Radcliffe Institute for Advanced Study at Harvard University. We thank Rainer Pepperkok from EMBL for providing access to the Advanced Light Microscopy Facility (ALMF). FJV acknowledges the Fondation de la Mer, Charles Bachy, Thibaut Pollina, and Yann Thomas. This article is contribution 67 of Tara Oceans.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Vincent, F.J., Colin, S., Romac, S. et al. The epibiotic life of the cosmopolitan diatom Fragilariopsis doliolus on heterotrophic ciliates in the open ocean. ISME J 12, 1094–1108 (2018). https://doi.org/10.1038/s41396-017-0029-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-017-0029-1
This article is cited by
-
Algal blooms in the ocean: hot spots for chemically mediated microbial interactions
Nature Reviews Microbiology (2024)
-
N-linked glycosylation enzymes in the diatom Thalassiosira oceanica exhibit a diel cycle in transcript abundance and favor for NXT-type sites
Scientific Reports (2021)
-
Prevalence of epibiosis in plankton community of the Indian EEZ: a review
Symbiosis (2021)
-
Genome-enabled phylogenetic and functional reconstruction of an araphid pennate diatom Plagiostriata sp. CCMP470, previously assigned as a radial centric diatom, and its bacterial commensal
Scientific Reports (2020)
-
Tara Oceans: towards global ocean ecosystems biology
Nature Reviews Microbiology (2020)