Correction to: Signal Transduction and Targeted Therapy https://doi.org/10.1038/s41392-023-01335-5, published online 03 February 2023
In the process of collating the raw data, the authors noticed 7 inadvertent mistakes occurred in Figs. 3a, 3b, 3e, 5d, 6b, 6d and 6e that need to be corrected after online publication of the article.1 The correct data are provided as follows. The key findings of the article are not affected by these corrections. The original article has been corrected.
-
(1)
The plus signs in Fig. 3a should line up with the lane.

Fig. 3a E was ubiquitinated via K48-linked but not K6, K11, K27, K29, K33 or K63.
-
(2)
The plus signs and minus signs in Fig. 3b should line up with the lane.
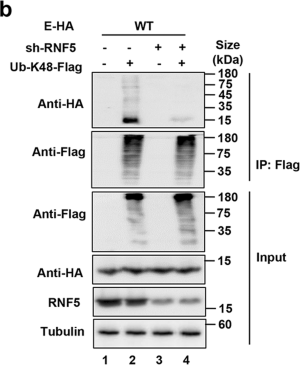
Fig. 3b Knockdown of RNF5 decreased K48-linked ubiquitination of E.
-
(3)
The plus signs and minus signs in Fig. 3e should line up with the lane.

Fig. 3e The EK63R mutant was recalcitrant to RNF5-mediated degradation.
-
(4)
Due to our negligence, the picture of another field of vision of Analog-1-1.5mg/kg-non-infected was mistakenly used as the picture of Analog-1-0mg/kg-non-infected in Fig. 5d. The correct picture of Analog-1-0mg/kg-non-infected is shown below.

Fig. 5d Representative images of H&E staining of lungs of differently treated mice.
-
(5)
The plus signs and minus signs in Fig. 6b should line up with the lane.

Fig. 6b Sensitivity of E mutants to RNF5-mediated degradation.
-
(6)
The first mutation site found in Alpha strain marked as “E8D B.1.1.7 (Alpha)” was overlapped with the arrows in Fig. 6d.

Fig. 6d A schematic of E showing the positions of variations found in SARS-CoV-2 variants.
-
(7)
The plus signs and minus signs in Fig. 6e should line up with the lane.

Fig. 6e All tested E alleles from different SARS-CoV-2 variants were sensitive to RNF5.
Reference
Li, Z. et al. The E3 ligase RNF5 restricts SARS-CoV-2 replication by targeting its envelope protein for degradation. Signal Transduct. Target. Ther. 8, 53 (2023).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, Z., Hao, P., Zhao, Z. et al. Correction To: The E3 ligase RNF5 restricts SARS-CoV-2 replication by targeting its envelope protein for degradation. Sig Transduct Target Ther 8, 85 (2023). https://doi.org/10.1038/s41392-023-01374-y
Published:
DOI: https://doi.org/10.1038/s41392-023-01374-y






