Abstract
Background
Genetic profiling of patients with prostate cancer could potentially identify mutations prone to castration-resistant prostate cancer (CRPC). Here, we aimed to identify the differences in genetic profiles of patients with hormone-sensitive prostate cancer (HSPC) and CRPC and stratify HSPC patients to identify mutations associated with CRPC progression.
Methods
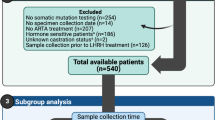
A total of 103 samples were collected, including 62 DNA samples from the tumor tissues of 59 HSPC patients and 41 cell-free DNA (cfDNA) samples from prostate cancer patients at different cancer stages. Targeted sequence was conducted on both the tissue DNA and cfDNA. The associations between mutations and clinical outcomes (CRPC-free time) were analyzed using χ2 test, logistic regression analysis, Kaplan–Meier analysis, and Cox regression analysis.
Results
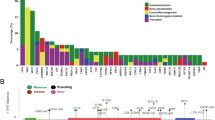
By comparing to that of cfDNA sequencing, the results from DNA sequencing of 1-needle (80%) and mixed 12-needle (77.8%) biopsies are highly comparable. FOXA1 (30.5%), CDK12 (23.7%), and TP53 (22.0%) were the top 3 most frequently mutated genes in HSPC patients; 50.8% (30/59) and 44.1% (26/59) HSPC patients had mutations in DDR and HRR pathway, respectively. Mutations in AR and APC as well as the members involved in the regulation of stem cell pluripotency and EMT pathway were often observed in CRPC samples. We established a panel of four genetic mutations (MSH2, CDK12, TP53, and RB1) to predict the risk of CRPC early progression with concordance index = 0.609 and the area under curve of the ROC curve as 0.838.
Conclusions
In this study, we demonstrated that the cfDNA can be used in genetic profiling in prostate cancer and our newly established panel is capable of predicting which mHSPC patient has a high risk of early CRPC progression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 4 print issues and online access
$259.00 per year
only $64.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The raw data in this study were freely available in Genome Sequence Archive (GSA) database with the project number PRJCA009634.
References
Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7–33.
Xia C, Dong X, Li H, Cao M, Sun D, He S, et al. Cancer statistics in China and United States, 2022: profiles, trends, and determinants. Chin Med J. 2022;135:584–90.
Harris WP, Mostaghel EA, Nelson PS, Montgomery B. Androgen deprivation therapy: progress in understanding mechanisms of resistance and optimizing androgen depletion. Nat Clin Pract Urol. 2009;6:76–85.
Spiess PE, Katz AE, Chin JL, Bahn D, Cohen JK, Shinohara K, et al. A pretreatment nomogram predicting biochemical failure after salvage cryotherapy for locally recurrent prostate cancer. BJU Int. 2010;106:194–8.
Koo KC, Park SU, Kim KH, Rha KH, Hong SJ, Yang SC, et al. Prognostic impacts of metastatic site and pain on progression to castrate resistance and mortality in patients with metastatic prostate cancer. Yonsei Med J. 2015;56:1206–12.
Tang DG. Understanding and targeting prostate cancer cell heterogeneity and plasticity. Semin Cancer Biol. 2022;82:68–93.
Li M, Nopparat J, Aguilar BJ, Chen YH, Zhang J, Du J, et al. Intratumor δ-catenin heterogeneity driven by genomic rearrangement dictates growth factor dependent prostate cancer progression. Oncogene. 2020;39:4358–74.
Teng PC, Huang SP, Liu CH, Lin TY, Cho YC, Lai YL, et al. Identification of DNA damage repair-associated prognostic biomarkers for prostate cancer using transcriptomic data analysis. Int J Mol Sci. 2021;22:11771.
Hamid AA, Gray KP, Shaw G, MacConaill LE, Evan C, Bernard B, et al. Compound genomic alterations of TP53, PTEN, and RB1 tumor suppressors in localized and metastatic prostate cancer. Eur Urol. 2019;76:89–97.
De Laere B, van Dam PJ, Whitington T, Mayrhofer M, Diaz EH, Van den Eynden G, et al. Comprehensive profiling of the androgen receptor in liquid biopsies from castration-resistant prostate cancer reveals novel intra-AR structural variation and splice variant expression patterns. Eur Urol. 2017;72:192–200.
Joseph JD, Lu N, Qian J, Sensintaffar J, Shao G, Brigham D, et al. A clinically relevant androgen receptor mutation confers resistance to second-generation antiandrogens enzalutamide and ARN-509. Cancer Discov. 2013;3:1020–9.
Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol. 2013;10:472–84.
Di Meo A, Bartlett J, Cheng Y, Pasic MD, Yousef GM. Liquid biopsy: a step forward towards precision medicine in urologic malignancies. Mol Cancer. 2017;16:80.
Annala M, Vandekerkhove G, Khalaf D, Taavitsainen S, Beja K, Warner EW, et al. Circulating tumor DNA genomics correlate with resistance to abiraterone and enzalutamide in prostate cancer. Cancer Discov. 2018;8:444–57.
Wyatt AW, Azad AA, Volik SV, Annala M, Beja K, McConeghy B, et al. Genomic alterations in cell-free DNA and enzalutamide resistance in castration-resistant prostate cancer. JAMA Oncol. 2016;2:1598–606.
Cornford P, Bellmunt J, Bolla M, Briers E, De Santis M, Gross T, et al. EAU-ESTRO-SIOG guidelines on prostate cancer. Part II: treatment of relapsing, metastatic, and castration-resistant prostate cancer. Eur Urol. 2017;71:630–42.
Sabari JK, Offin M, Stephens D, Ni A, Lee A, Pavlakis N, et al. A prospective study of circulating tumor DNA to guide matched targeted therapy in lung cancers. J Natl Cancer Inst. 2019;111:575–83.
Takami H, Fukuoka K, Fukushima S, Nakamura T, Mukasa A, Saito N, et al. Integrated clinical, histopathological, and molecular data analysis of 190 central nervous system germ cell tumors from the iGCT Consortium. Neuro-Oncol. 2019;21:1565–77.
Onken MD, Worley LA, Dávila RM, Char DH, Harbour JW. Prognostic testing in uveal melanoma by transcriptomic profiling of fine needle biopsy specimens. J Mol Diagn. 2006;8:567–73.
McCoy P, Mangiola S, Macintyre G, Hutchinson R, Tran B, Pope B, et al. MSH2-deficient prostate tumours have a distinct immune response and clinical outcome compared to MSH2-deficient colorectal or endometrial cancer. Prostate Cancer Prostatic Dis. 2021;24:1167–80.
Ballhausen A, Przybilla MJ, Jendrusch M, Haupt S, Pfaffendorf E, Seidler F, et al. The shared frameshift mutation landscape of microsatellite-unstable cancers suggests immunoediting during tumor evolution. Nat Commun. 2020;11:4740.
Mateo J, Seed G, Bertan C, Rescigno P, Dolling D, Figueiredo I, et al. Genomics of lethal prostate cancer at diagnosis and castration resistance. J Clin Investig. 2020;130:1743–51.
Stopsack KH, Nandakumar S, Wibmer AG, Haywood S, Weg ES, Barnett ES, et al. Oncogenic genomic alterations, clinical phenotypes, and outcomes in metastatic castration-sensitive prostate cancer. Clin Cancer Res. 2020;26:3230–8.
Reimers MA, Yip SM, Zhang L, Cieslik M, Dhawan M, Montgomery B, et al. Clinical outcomes in cyclin-dependent kinase 12 mutant advanced prostate cancer. Eur Urol. 2020;77:333–41.
Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23:703–13.
Stuopelyte K, Sabaliauskaite R, Bakavicius A, Haflidadóttir BS, Visakorpi T, Väänänen RM, et al. Analysis of AR-FL and AR-V1 in whole blood of patients with castration resistant prostate cancer as a tool for predicting response to abiraterone acetate. J Urol. 2020;204:71–78.
Choudhury AD, Werner L, Francini E, Wei XX, Ha G, Freeman SS, et al. Tumor fraction in cell-free DNA as a biomarker in prostate cancer. JCI Insight. 2018;3:e122109.
Kohli M, Li J, Du M, Hillman DW, Dehm SM, Tan W, et al. Prognostic association of plasma cell-free DNA-based androgen receptor amplification and circulating tumor cells in pre-chemotherapy metastatic castration-resistant prostate cancer patients. Prostate Cancer Prostatic Dis. 2018;21:411–8.
Fu Y, Wang A, Zhou J, Feng W, Shi M, Xu X, et al. Advanced NSCLC patients with EGFR T790M harboring TP53 R273C or KRAS G12V cannot benefit from osimertinib based on a clinical multicentre study by tissue and liquid biopsy. Front Oncol. 2021;11:621992.
Vandekerkhove G, Struss WJ, Annala M, Kallio HML, Khalaf D, Warner EW, et al. Circulating tumor DNA abundance and potential utility in de novo metastatic prostate cancer. Eur Urol. 2019;75:667–75.
Jacob F, Salinas RD, Zhang DY, Nguyen PTT, Schnoll JG, Wong SZH, et al. A patient-derived glioblastoma organoid model and biobank recapitulates inter- and intra-tumoral heterogeneity. Cell. 2020;180:188–204.e122.
Abida W, Armenia J, Gopalan A, Brennan R, Walsh M, Barron D, et al. Prospective genomic profiling of prostate cancer across disease states reveals germline and somatic alterations that may affect clinical decision making. JCO Precis Oncol. 2017;2017:PO.17.00029.
Wu YM, Cieślik M, Lonigro RJ, Vats P, Reimers MA, Cao X, et al. Inactivation of CDK12 delineates a distinct immunogenic class of advanced prostate cancer. Cell. 2018;173:1770.e1714.
Antonarakis ES, Isaacsson Velho P, Fu W, Wang H, Agarwal N, Sacristan Santos V, et al. CDK12-altered prostate cancer: clinical features and therapeutic outcomes to standard systemic therapies, poly (ADP-ribose) polymerase inhibitors, and PD-1 inhibitors. JCO Precis Oncol. 2020;4:370–81.
Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–34.
Pomerantz MM, Spisák S, Jia L, Cronin AM, Csabai I, Ledet E, et al. The association between germline BRCA2 variants and sensitivity to platinum-based chemotherapy among men with metastatic prostate cancer. Cancer. 2017;123:3532–9.
Tang T, Wang LA, Wang P, Tong D, Liu G, Zhang J, et al. Case report: co-existence of BRCA2 and PALB2 germline mutations in familial prostate cancer with solitary lung metastasis. Front Oncol. 2020;10:564694.
Liu Q, Tong D, Liu G, Yi Y, Xu J, Yang X, et al. A novel BRCA2 mutation in prostate cancer sensitive to combined radiotherapy and androgen deprivation therapy. Cancer Biol Ther. 2018;19:669–75.
Piazza A, Heyer WD. Homologous recombination and the formation of complex genomic rearrangements. Trends Cell Biol. 2019;29:135–49.
Mateo J, Boysen G, Barbieri CE, Bryant HE, Castro E, Nelson PS, et al. DNA repair in prostate cancer: biology and clinical implications. Eur Urol. 2017;71:417–25.
Shafi AA, Yen AE, Weigel NL. Androgen receptors in hormone-dependent and castration-resistant prostate cancer. Pharmacol Ther. 2013;140:223–38.
Murillo-Garzón V, Kypta R. WNT signalling in prostate cancer. Nat Rev Urol. 2017;14:683–96.
Tang F, Xu D, Wang S, Wong CK, Martinez-Fundichely A, Lee CJ, et al. Chromatin profiles classify castration-resistant prostate cancers suggesting therapeutic targets. Science. 2022;376:eabe1505.
Li P, Yang R, Gao WQ. Contributions of epithelial-mesenchymal transition and cancer stem cells to the development of castration resistance of prostate cancer. Mol Cancer. 2014;13:55.
Cheng Q, Butler W, Zhou Y, Zhang H, Tang L, Perkinson K, et al. Pre-existing castration-resistant prostate cancer-like cells in primary prostate cancer promote resistance to hormonal therapy. Eur Urol. 2022;81:446–55.
Liu Q, Tong D, Liu G, Xu J, Do K, Geary K, et al. Metformin reverses prostate cancer resistance to enzalutamide by targeting TGF-β1/STAT3 axis-regulated EMT. Cell Death Dis. 2017;8:e3007.
Acknowledgements
The authors thank all patients in this study for their collaboration and the Geneplus (Beijing, China) for conducting the sequencing.
Funding
This work was supported by the National Natural Science FoundatiPRon of China (Grant Nos: 82172807 and 82172721) and University Research Project of Army Medical University (2017XYY07, 2018XLC1014, 2019CXLCB006 and 17BJZ13).
Author information
Authors and Affiliations
Contributions
QL and JJ were responsible for designing the study, data interpretation, writing the manuscript, and approving the final version. ZW was responsible for the acquisition of data, data analysis, data interpretation, writing of the manuscript, and statistical analysis. XY contributed to the acquisition of data, data analysis, acquisition of data, and statistical analysis. PT conducted the acquisition of data and data analysis. JQ contributed to data interpretation. DZZ contributed to the manuscript writing. TT, YW, SP, SW, WL, LW, YZ, JZ, KL, ZS, and JX contributed to the acquisition of data.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
This study was approved by the Committee for Ethics of Daping hospital (No. 2018_28). Informed consent was obtained from every patient enrolled in this study.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, Z., Yan, X., Tang, P. et al. Genetic profiling of hormone-sensitive and castration-resistant prostate cancers and identification of genetic mutations prone to castration-resistant prostate cancer. Prostate Cancer Prostatic Dis 26, 180–187 (2023). https://doi.org/10.1038/s41391-022-00618-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41391-022-00618-2
This article is cited by
-
Demonstrating Bioequivalence for Two Dose Strengths of Niraparib and Abiraterone Acetate Dual-Action Tablets Versus Single Agents: Utility of Clinical Study Data Supplemented with Modeling and Simulation
Clinical Pharmacokinetics (2024)
-
Therapeutic, diagnostic and prognostic values of TRIM proteins in prostate cancer
Pharmacological Reports (2023)
-
Performance of clinical risk scores and prediction models to identify pathogenic germline variants in patients with advanced prostate cancer
World Journal of Urology (2023)