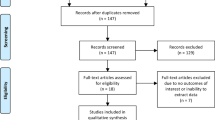
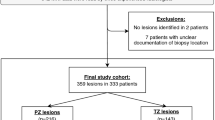
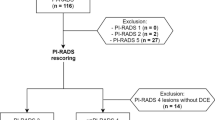
Abstract
Background
Protocol-based active surveillance (AS) biopsies have led to poor compliance. To move to risk-based protocols, more accurate imaging biomarkers are needed to predict upgrading on AS prostate biopsy. We compared restriction spectrum imaging (RSI-MRI) generated signal maps as a biomarker to other available non-invasive biomarkers to predict upgrading or reclassification on an AS biopsy.
Methods
We prospectively enrolled men on prostate cancer AS undergoing repeat biopsy from January 2016 to June 2019 to obtain an MRI and biomarkers to predict upgrading. Subjects underwent a prostate multiparametric MRI and a short duration, diffusion-weighted enhanced MRI called RSI to generate a restricted signal map along with evaluation of 30 biomarkers (14 clinico-epidemiologic features, 9 molecular biomarkers, and 7 radiologic-associated features). Our primary outcome was upgrading or reclassification on subsequent AS prostate biopsy. Statistical analysis included operating characteristic improvement using AUROC and AUPRC.
Results
The individual biomarker with the highest area under the receiver operator characteristic curve (AUC) was RSI-MRI (AUC = 0.84; 95% CI: 0.71–0.96). The best non-imaging biomarker was prostate volume-corrected Prostate Health Index density (PHI, AUC = 0.68; 95% CI: 0.53–0.82). Non-imaging biomarkers had a negligible effect on predicting upgrading at the next biopsy but did improve predictions of overall time to progression in AS.
Conclusions
RSI-MRI, PIRADS, and PHI could improve the predictive ability to detect upgrading in AS. The strongest predictor of clinically significant prostate cancer on AS biopsy was RSI-MRI signal output.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 4 print issues and online access
$259.00 per year
only $64.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The datasets generated during and/or analyzed during the current study are available as an R package at https://github.com/uclahs-cds/public-R-ASBiomarkerStratification. The datasets generated during and/or analyzed during the current study are also available from the corresponding author upon reasonable request.
References
Johansson J-E, Andrén O, Andersson S-O, Dickman PW, Holmberg L, Magnuson A, et al. Natural history of early, localized prostate cancer. JAMA. 2004;291:2713–9.
Mohler JL, Antonarakis ES, Armstrong AJ, D’Amico AV, Davis BJ, Dorff T, et al. Prostate Cancer, Version 2.2019, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2019;17:479–505.
Cooperberg MR, Zheng Y, Faino AV, Newcomb LF, Zhu K, Cowan JE, et al. Tailoring intensity of active surveillance for low-risk prostate cancer based on individualized prediction of risk stability. JAMA Oncol. 2020;6:e203187.
Detsky JS, Ghiam AF, Mamedov A, Commisso K, Commisso A, Zhang L, et al. Impact of biopsy compliance on outcomes for patients on active surveillance for prostate cancer. J Urol. 2020;204:934–40.
Klotz L, Loblaw A, Sugar L, Moussa M, Berman DM, Van der Kwast T, et al. Active Surveillance Magnetic Resonance Imaging Study (ASIST): results of a randomized multicenter prospective trial. Eur Urol. 2019;75:300–9.
Liss MA, Newcomb LF, Zheng Y, Garcia MP, Filson CP, Boyer H, et al. Magnetic resonance imaging for the detection of high grade cancer in the canary prostate active surveillance study. J Urol. 2020;204:701–6.
Chesnut GT, Vertosick EA, Benfante N, Sjoberg DD, Fainberg J, Lee T, et al. Role of changes in magnetic resonance imaging or clinical stage in evaluation of disease progression for men with prostate cancer on active surveillance. Eur Urol. 2020;77:501–7.
Feng ZY, Wang L, Min XD, Wang SG, Wang GP, Cai J. Prostate cancer detection with multiparametric magnetic resonance imaging: Prostate Imaging Reporting and Data System Version 1 versus Version 2. Chin Med J (Engl). 2016;129:2451–9.
Besasie BD, Sunnapwar AG, Gao F, Troyer D, Clarke GD, White H, et al. Restriction spectrum imaging-MRI to improve prostate cancer imaging in men on active surveillance. J Urol. 2021;206:44–51.
Quon JS, Moosavi B, Khanna M, Flood TA, Lim CS, Schieda N. False positive and false negative diagnoses of prostate cancer at multi-parametric prostate MRI in active surveillance. Insights Imaging. 2015;6:449–63.
White NS, McDonald C, Farid N, Kuperman J, Karow D, Schenker-Ahmed NM, et al. Diffusion-weighted imaging in cancer: physical foundations and applications of restriction spectrum imaging. Cancer Res. 2014;74:4638–52.
Newcomb LF, Zheng Y, Faino AV, Bianchi-Frias D, Cooperberg MR, Brown MD, et al. Performance of PCA3 and TMPRSS2:ERG urinary biomarkers in prediction of biopsy outcome in the Canary Prostate Active Surveillance Study (PASS). Prostate Cancer Prostatic Dis. 2019;22:438–45.
Markowski MC, Boorjian SA, Burton JP, Hahn NM, Ingersoll MA, Maleki Vareki S, et al. The microbiome and genitourinary cancer: a collaborative review. Eur Urol. 2019;75:637–46.
Tomlins SA, Day JR, Lonigro RJ, Hovelson DH, Siddiqui J, Kunju LP, et al. Urine TMPRSS2:ERG plus PCA3 for individualized prostate cancer risk assessment. Eur Urol. 2016;70:45–53.
Vasarainen H, Salman J, Salminen H, Valdagni R, Pickles T, Bangma C, et al. Predictive role of free prostate-specific antigen in a prospective active surveillance program (PRIAS). World J Urol. 2015;33:1735–40.
Seibert TM, Fan CC, Wang Y, Zuber V, Karunamuni R, Parsons JK, et al. Polygenic hazard score to guide screening for aggressive prostate cancer: development and validation in large scale cohorts. BMJ. 2018;360:j5757.
Halstuch D, Ber Y, Margel D. Screening, active surveillance, and treatment of localized prostate cancer among carriers of germline BRCA mutations. Eur Urol Focus. 2020;6:212–4.
Ahmad AE, Mohammed A, Bhindi B, Richard PO, Fadaak K, Leao R, et al. Serum adipokines as predictors for the outcome of prostate biopsies at early stage prostate cancer diagnosis. Cancer Manag Res. 2019;11:10043–50.
Schwen ZR, Mamawala M, Tosoian JJ, Druskin SC, Ross AE, Sokoll LJ, et al. Prostate Health Index and multiparametric magnetic resonance imaging to predict prostate cancer grade reclassification in active surveillance. BJU Int. 2020;126:373–8.
Friedman JH. Greedy function approximation: a gradient boosting machine. Ann Stat. 2001;29:1189–232.
Kuhn M. Building predictive models in R using the caret package. J Stat Softw Artic. 2008;28:1–26.
Greenwell B, Boehmke B, Cunningham J, Developers G gbm: generalized boosted regression models. 2020.
P’ng C, Green J, Chong LC, Waggott D, Prokopec SD, Shamsi M, et al. BPG: seamless, automated and interactive visualization of scientific data. BMC Bioinformatics. 2019;20:42.
Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez J-C, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77.
Therneau T, Atkinson B. rpart: recursive partitioning and regression trees. 2019.
Kinsella N, Stattin P, Cahill D, Brown C, Bill-Axelson A, Bratt O, et al. Factors influencing men’s choice of and adherence to active surveillance for low-risk prostate cancer: a mixed-method systematic review. Eur Urol. 2018;74:261–80.
Bokhorst LP, Alberts AR, Rannikko A, Valdagni R, Pickles T, Kakehi Y, et al. Compliance rates with the Prostate Cancer Research International Active Surveillance (PRIAS) protocol and disease reclassification in noncompliers. Eur Urol. 2015;68:814–21.
Van Hemelrijck M, Ji X, Helleman J, Roobol MJ, van der Linden W, Nieboer D, et al. Reasons for discontinuing active surveillance: assessment of 21 centres in 12 countries in the Movember GAP3 Consortium. Eur Urol. 2019;75:523–31.
Willemse PM, Davis NF, Grivas N, Zattoni F, Lardas M, Briers E, et al. Systematic review of active surveillance for clinically localised prostate cancer to develop recommendations regarding inclusion of intermediate-risk disease, biopsy characteristics at inclusion and monitoring, and surveillance repeat biopsy strategy. Eur Urol. 2022;81:337–46.
Lange JM, Gulati R, Leonardson AS, Lin DW, Newcomb LF, Trock BJ, et al. Estimating and comparing cancer progression risks under varying surveillance protocols. Ann Appl Stat. 2018;12:1773–95.
Acknowledgements
First, thank you to all the patients who altruistically participated in this research and to all the coordinators and lab personnel who are vital to move research but are not named on the manuscripts.
Funding
MAL is supported through the DOD Prostate Cancer Research Program (PCRP) Physician Research Training Award. This work was supported by the Office of the Assistant Secretary of Defense for Health Affairs through the Prostate Cancer Research Program under Award No. W81XWH-15-1-0441. Opinions, interpretations, conclusions, and recommendations are those of the authors and are not necessarily endorsed by the Department of Defense. The work was also funded by an NCI Early Detection Research Network (EDRN) Associate Member Grant (U24CA086368) and by EDRN Biomarker Development Laboratories (U01CA214194, U24CA115102, and U01CA214170), by the ITCR (U24CA249265) and by NCI CCSG grants to UCLA (P30CA016042) and UTHSC (P30CA054174). Beckman-Coulter Life Sciences supplied reagents for the PHI test. JJT’s research is funded in part by the Prostate Cancer Foundation Young Investigator Award and a SPORE Career Enhancement Program (CA186786). The funder played no role in the experimentation or authorship of the manuscript.
Author information
Authors and Affiliations
Contributions
Conceptualization: MAL, IMT. Data curation: MAL, BB, RJL, TLJ-P, DAT, AGS, JJT, JS. Formal analysis: SEE, AL. Funding acquisition: MAL, RJL, OJS, AMC, DWC, PCB. Investigation: MAL, OJS, DAT. Methodology: MAL, PCB. Project administration: MAL, BB. Resources: RJL, JJT, LJS, MAL, PCB. Software: MAL, GDC, PCB. Supervision: MAL, IMT, RJL, PCB. Validation: MAL, PCB. Visualization: MAL, SEE, AL, PCB. Writing—original draft: MAL, SEE, BB. Writing—review and editing: SEE, BB, AL, OJS, DAT, GDC, AGS, RGL, TLJ-P, LJS, DWC, JJT, JS, AMC, IMT, MAL, PCB.
Corresponding authors
Ethics declarations
Competing interests
JJT and AMC are co-founders and have equity in Lynx Dx, which has licensed the urine biomarkers mentioned in this study from Hologic and the University of Michigan. JJT has leadership roles in Lynx Dx. The University of Michigan has been issued a patent on ETS gene fusions in prostate cancer on which AMC is a co-inventor.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Eng, S.E., Basasie, B., Lam, A. et al. Prospective comparison of restriction spectrum imaging and non-invasive biomarkers to predict upgrading on active surveillance prostate biopsy. Prostate Cancer Prostatic Dis 27, 65–72 (2024). https://doi.org/10.1038/s41391-022-00591-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41391-022-00591-w
This article is cited by
-
Recent advances and future perspectives in the therapeutics of prostate cancer
Experimental Hematology & Oncology (2023)