Abstract
Background
Paternally expressed gene 10 (PEG10) is believed to be a key imprinted gene involved in placenta formation. However, its role in human folate-related spina bifida (SB) remains unclear.
Methods
The methylation status of the germline differentially methylated region (gDMR) in the PEG10/sarcoglycan epsilon (SGCE) imprinted cluster was compared between SB patients and control samples. Moreover, the influence of ectopic PEG10 expression on apoptosis was assessed to explore the underlying mechanisms related to folate deficiency-induced aberrant gDMR methylation in SB.
Results
The case group exhibited a significant increase in the methylation level of the gDMR and a marked reduction in the mRNA and protein expression of PEG10 compared with the control group. A prominent negative correlation was found between the folate level in brain tissue and gDMR methylation status (r = −0.62, P = 0.001). A cell model treated with a demethylating agent showed a significant elevation of PEG10 transcription level, as well as other imprinted genes in this cluster. In addition, the inhibition of PEG10 was found to be accompanied by aberrant activation of apoptosis in SB.
Conclusions
Our findings suggest that disturbed gDMR methylation of the PEG10/SGCE cluster due to folate deficiency is involved in SB through aberrant activation of apoptosis.
Impact
-
Disturbed genomic imprinting has been verified to be involved in neural tube defects (NTDs). However, little is known about the effect of ectopic expression of imprinted gene PEG10 on human NTDs.
-
Aberrant methylation status of the germline differentially methylated region (gDMR) of PEG10/SGCE cluster due to folate deficiency has been found to result in the inhibition of PEG10 and has a marked association with an increased occurrence of spina bifida.
-
Inhibited expression of PEG10 partly is found to be related to the abnormal activation of apoptosis in spina bifida.
Similar content being viewed by others
Introduction
Neural tube defects (NTDs), including spina bifida, anencephaly, and meningocele, are a class of severe structural birth defects with high disability and mortality. The incidence of NTDs is about 1 in every 1000 births and has dramatic regional variation. The etiology of NTDs is multifactorial and generally believed to arise from an interaction between genetics and environmental factors; among these, growing attention is being devoted to folate deficiency.1,2 Epidemiologic studies have verified that folic acid supplementation during peri-conception has the benefit of reducing NTDs in offspring by up to 70%, but the particular mechanism of action of folic acid in preventing NTDs is not fully understood.3 In addition to its participation in DNA synthesis, the involvement of folate in epigenetic modification through the methyl group provided by one-carbon metabolism has been an area of particular focus.
Genomic imprinting leads to the preferential expression of the maternal or paternal allele of certain genes. A series of fundamental studies have elucidated that imprinted genes are regulated by epigenetic mechanisms, especially DNA methylation occurs in the imprinting control region (ICR).4,5 Disruption of genomic imprinting during early embryonic development can have major consequences on embryogenesis. The specific DNA methylation marks at the ICR forms a differentially methylated region in the germline (gDMR) which influences the imprinting establishment of the whole imprinted cluster and the expression of multiple imprinted genes in the same imprinted cluster.6 The methylation status of gDMR is sensitive to the conditions of the surrounding environment, such as altered folate level.7 Our previous studies have revealed that aberrant methylation modification of gDMR owing to folate deficiency is involved in NTDs. By using folate-deficient cell and animal models, Chang et al. found that folate deficiency increased the expression of mesoderm-specific transcript/paternally expressed gene 1 (Mest/Peg1) via hypomethylation.8 In addition, Wang et al. further validated that germline imprinting at the GNAS gDMR would influence the establishment of imprinting in other regions of the whole imprinted gene cluster and be abolished upon treatment with a parental folate-deficient diet.9
Paternally expressed gene 10 (PEG10) is an imprinting gene that is derived from the Ty3/Gypsy retrotransposon family and located on human chromosome 7q21.3.10 PEG10 belongs to the PEG10/SGCE imprinted cluster, which includes multiple imprinted genes, such as PEG10, sarcoglycan epsilon (SGCE), calcitonin receptor (CALCR), tissue factor pathway inhibitor 2 (TFPI2), and protein phosphatase1, regulatory subunit 9A (PPP1R9A). Ono et al. indicated that deletion of PEG10 would cause early embryonic lethality, suggesting that PEG10 is essential for embryogenesis.11 However, how gDMR methylation modification of PEG10/SGCE cluster is affected by folate deficiency and how the PEG10 exerts this function during neural tube closure is unclear.
Given the substantial evidence regarding the involvement of folate deficiency in NTDs and the important role of PEG10 in embryonic development, we designed this study to explore whether folate deficiency has an influence on PEG10 expression by affecting the gDMR methylation of the PEG10/SGCE cluster in spina bifida, a subtype of NTDs. In addition, we investigated the effect of the altered expression of PEG10 on the process of apoptosis in spina bifida.
Materials and methods
Samples collection
All samples were obtained from a long-term surveillance program investigating the high prevalence of NTDs in the Lüliang mountain area of northern China. Fetuses affected with NTDs were obtained from stillbirth deliveries and medical abortions based on a diagnosis using B-ultrasound. All case samples enrolled in this study had spina bifida and a definitive diagnosis was made by postmortem according to the International Classification of Disease, Tenth Revision, code Q05 spina bifida. Control samples, enrolled from the same region, were aborted for non-medical reasons and were confirmed to have no deformities by postmortem, furthermore, the risk factors that had been illustrated to be related to the occurrence of spina bifida, such as pregnant women age and gestational week, were taken into account so that the case group and the control group can be matched in these general characters. None of the pregnant women had received peri-conceptional folic acid supplementation. A total of 119 samples were collected from July 2005 to August 2010 for this study, including 53 controls and 66 cases. After the abortion procedure, anatomy was evaluated by certified pathologists from the Capital Institute of Pediatrics. The isolated samples were stored at −20 °C until they were transferred to the study laboratories. Information relating to the subjects in this study is shown in Table 1.
nCounter expression assay
The transcription level of PEG10 in human brain tissues was detected by the NanoString nCounter system (NanoString Technologies, Seattle, WA, USA), which measures the relative abundance of mRNA transcript via a hybridization assay and digital readouts of fluorescent probes. Total RNA was extracted following the manufacturer’s instructions (miRNeasy Mini Kit, Qiagen, Hilden, NRW, Germany) and gene-specific probes were designed by the manufacturer (NanoString Technologies). The probes were hybridized in solution to approximately 100 ng of total RNA of brain tissue for 18 h at 65 °C, then eluted and immobilized in the cartridge for data collection, which was performed on the nCounter Digital Analyzer (NanoString Technologies). Gene expression data were filtered using quality control (QC) criteria according to the manufacturer’s recommendations. Raw counts of QC-passed samples were normalized using three reference genes as internal controls (GAPDH, CLTC, and GUSB). All QC and normalization procedures were performed using nSolver Analysis Software v2.0. All data were log2-transformed before further analysis.
Western blotting
Blots were incubated with primary antibody [anti-PEG10 monoclonal antibody (ab181249, Abcam, Cambridge, UK; dilution, 1:1500), anti-CASPASE-3 monoclonal antibody (#9662, CST, USA; dilution, 1:1000), and anti-β-ACTIN monoclonal antibody (ab8226, Abcam, Cambridge, UK; dilution, 1:2000)] overnight at 4 °C, and then incubated with secondary anti-rabbit/mouse HRP-conjugated antibody (CST, USA; dilution, 1:5000) for 1 h at room temperature. The blots were developed with SuperSignal WestPico Chemiluminescence Substrate (Thermo, USA) and quantitated on a densitometer (Bio-Rad, Universal HoodII, USA) using Quantity One software.
Methylation analysis
Genomic DNA extraction and bisulfite treatment
Genomic DNA was extracted from 100-mg frozen brain tissue using a DNeasy Blood & Tissue Kit (Qiagen, Venlo, Netherlands). DNA with an OD260/OD280 absorbance ratio of 1.8–1.9 was used for subsequent work. A total of 1500 μg genomic DNA from each brain tissue sample underwent bisulfite treatment using the EZ DNA methylation kit (Zymo Research, Irvine, CA, USA) according to the manufacturer’s instructions.
Methylation measurement and analysis
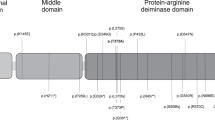
The MassARRAY platform with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (Agena, San Diego, CA, USA) was used to quantitatively analyze the methylation level of the gDMR in the PEG10/SGCE cluster. Polymerase chain reaction (PCR) primers were designed using the Agena epidesigner website (http://epidesigner.com). For each reverse primer, an additional T7 promoter tag for in vivo transcription was added, as well as a 10-mer tag on the forward primer to adjust for melting temperature differences. One pair of primers was used in this study: Left primer: aggaagagagGGGAAATTAAGTTTTAAAGGTTTGG; Right primer: cagtaatacgactcactatagggagaaggctCCAATTATAACTCCCATCAATTT. The amplification region is shown in Fig. 1. In total, six CpG units of the gDMR were examined in this region. The spectral methylation ratios were generated using Epityper software version 1.0.
Folate measurement
The samples used for the nCounter expression assay were also subjected to brain tissue folate measurement using a competitive receptor binding immunoassay (Chemiluminescent Immunoenzyme Assay Access Immunoassay system II; Beckman Coulter, Krefeld, Germany). The detailed operation procedure has been described in previous research.12
Cell culture
The human colorectal cancer HCT-15 cell line was obtained from the China Infrastructure of Cell Line Resource (Beijing, China). Cells were cultured at 37 °C in a humidified 5% CO2 atmosphere in RPMI 1640 medium (Gibco, Gaithersburg, MD, USA) supplemented with 10% fetal bovine serum (Gibco, USA) and passaged every 2–3 days. Cells in the exponential growth phase were used for subsequent experiments. For demethylation studies, cultured cells were incubated for 72 h in 5μmol/L 5-Azacytidine (5-Aza; Sigma-Aldrich, St Louis, MO, USA), a methylation inhibitor, and the medium was changed daily.
RNA extraction and real-time PCR
RNA extraction and reverse transcription
Total RNA was extracted from cultured cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. RNA concentration and purity were determined by the OD260/OD280 absorbance ratio. Reverse transcription was performed with the RevertAid First Strand cDNA Synthesis Kit (ThermoFisher Scientific, Waltham, MA, USA) with oligo (dT), according to the instruction manual. cDNA was stored at −20 °C until required for use in real-time PCR.
Real-time PCR
To determine transcription of PEG10 (OMIM: 609810) and other genes in the PEG10/SGCE cluster, including SGCE (OMIM: 604149), CALCR (OMIM:114131), TFPI2 (OMIM:600033) and PPP1R9A (OMIM:602468), real-time PCR was performed to compare the mRNA expression levels of corresponding genes relative to the expression level of the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH; OMIM:138400). The primers were designed using PrimerExpress® software Version 3.0 (Applied Biosystems, Foster City, CA, USA) and are listed in Supplementary Table S1. Briefly, quantitative analysis of cDNA was performed using a 7500 Fast Real-Time PCR system (Applied Biosystems) with UltraSYBR Mixture (Cwbio, Beijing, China). Each PCR reaction consisted of 10-μL 2XSYBR green master mixture, 0.5 μL forward primer, 0.5 μL reverse primer, 1 μL cDNA template, and 8 μl H2O. Reactions were performed in triplicate. Thermal cycling conditions were: 50 °C for 20 s, 95 °C for 10 min, 40 cycles of 95 °C for 15 s, and 60 °C for 1 min. The expression levels of the target genes were calculated using the 2−ΔΔCt method, for which ΔCt = Cttarget gene − Ctgapdh.
Apoptosis analyses by flow cytometry
Adherent HCT-15 cells were rinsed twice with Dulbecco PBS (Gibco, USA) and digested with 0.25% trypsin. After centrifugation for 3 min at 1000 × g, the supernatant was removed, and the cells were resuspended with PBS and counted. Cell apoptosis was detected using the Annexin V-FITC Apoptosis Detection Kit (C1062; Biyotime, Shanghai, China) according to the manufacturer’s instructions. Cells were analyzed by a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA, USA). A total of 10,000 events were recorded for each sample.
Transcription factor prediction
Putative transcription factors that bound to the gDMR of PEG10/SGCE cluster were predicted by PROMO database (http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3).
Statistical analysis
Data were analyzed using SPSS 25.0 software (IBM Corporation, Armonk, NY, USA) and expressed as mean±standard deviation (SD). The means of two groups of samples were compared using the Student’s t-test, and the means of multiple groups of samples were compared using single-factor analysis of variance. Odds ratios (ORs), which were used to evaluate the incidence of spina bifida relative to the methylation level, were calculated with 95% confidence intervals (CIs). Adjusted ORs were calculated by logistic regression. Correlation analysis was performed using bivariate correlation to analyze the correlation between the methylation level and folate level of brain tissue. All P-values were two-sided, and P < 0.05 was considered to be significant and is presented as *P < 0.05, **P < 0.01, or ***P < 0.001.
Results
Transcription and expression of PEG10 was significantly decreased in spina bifida brain tissues
The transcription level of PEG10 in brain tissue was measured in controls (n = 15) and spina bifida samples (n = 12) by NanoString nCounter Analysis System. The results showed that in the case group, the mRNA level of PEG10 (7.79 ± 0.38) was notably decreased compared with that of the control group (9.73 ± 0.39), P < 0.01 (Fig. 2a). Furthermore, western blot analysis for protein expression of PEG10 gene corroborated that at the mRNA level, PEG10 expression exhibited a marked down-regulation in the brain tissue of spina bifida samples (1.02 ± 0.06 in controls vs. 0.57 ± 0.07 in cases, P < 0.01) (Fig. 2b).
PEG10 hypermethylation was associated with a high occurrence of spina bifida
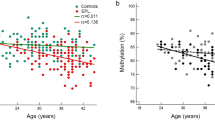
To investigate the possible epigenetic mechanism involved in the regulation of PEG10 expression, the gDMR methylation level in brain tissue was analyzed and compared between the control group and the case group. A total of 53 control samples and 66 spina bifida samples were enrolled in methylation analysis, and the methylation level was found to be significantly elevated in the case group compared with that of the control group (Fig. 3a). Further analysis of individual CpG sites in the gDMR was performed and revealed that methylation at almost every CpG site/unit in spina bifida samples was markedly higher than that of the control samples, with the exception of the first CpG site (Fig. 3b). Combined with the transcription analysis of PEG10, a negative relation was found between gDMR methylation and PEG10 transcription level, namely higher gDMR methylation but lower transcription level of PEG10 was observed in the case group (Fig. 3c).
a Mean methylation levels of PEG10/SGCE gDMR in controls and cases. b Methylation levels at all CpG sites in gDMR (shown in Fig. 1) were compared between samples from subjects. c Transcription level of PEG10 gene was significantly negatively correlated with the mean gDMR methylation level, r = −0.59. The left bar represents the gDMR methylation level, but the right bar represents the transcription level of PEG10 gene.
The relationship between the risk of spina bifida and the gDMR methylation level was assessed by adjusted logistical analysis (Table 2). Samples were divided according to the methylation level quartiles of the control group. Almost a quarter of the control samples were grouped into the lowest quartile (methylation level ≤ 42.79%), but only 3% of the cases were grouped into this classification. Conversely, the portion of case samples was higher than that of the control samples in the highest quartile (methylation level ≥ 54.00%), and high methylation level increased the risk of spina bifida by more than ten-fold compared with low methylation levels (OR: 9.93, 95 % CI: 1.86–53.07). These results suggest that hypermethylation in the gDMR of PEG10/SGCE cluster may be positively associated with the risk of spina bifida.
Folate insufficiency in spina bifida partly accounted for disturbed gDMR methylation modification
Given that folate is generally believed to be involved in the modification of methylation, the samples in which transcription analysis was performed were evaluated for folate levels and analyzed for the relationship with gDMR methylation. It was observed that the folate level in the case group was significantly lower than that in the control group (0.14 ± 0.01 in controls vs. 0.06 ± 0.01 in cases, P < 0.001) (Fig. 4a). Correlation analysis showed that gDMR methylation level had a significant negative correlation with the folate levels of brain tissue (r = −0.65, P < 0.001) (Fig. 4b).
a Folate levels of brain tissue in controls and cases were compared. controls: n = 15, cases: n = 12. b Correlation between folate levels of brain tissue and methylation level of PEG10. The abscissa represents the concentration of folate in brain tissue. The ordinate represents the ratio of methylation of each sample. The folate level had a notable negative correlation with the gDMR methylation level, r = −0.65, P < 0.001.
Transcription levels of imprinted genes in the PEG10/SGCE cluster were regulated by gDMR methylation
To investigate the regulatory potency of gDMR methylation on PEG10 transcription, a differential methylation model was constructed using HCT-15 cells treated with 5 μM methyltransferase inhibitor 5-Aza. Methylation analysis showed that the gDMR methylation level in the treated group was significantly reduced to only half of that in the control group (Fig. 5a). Accordingly, there were close to four-fold and three-fold increases in PEG10 mRNA and protein levels, respectively, in the treated group compared with the control group (Fig. 5b), suggesting that the expression of PEG10 was sensitive to the change in gDMR methylation.
a mRNA level of PEG10 gene was significantly negatively correlated with the mean gDMR methylation level, r = −0.98. The left bar represents the gDMR methylation level, but the right bar represents the transcription level of PEG10 gene. The treated group indicated that HCT-15 cells were treated with 5-Aza (5 μmol/L), control indicated that the cells which not treated with 5-Aza. b Western bolt analysis of protein level of PEG10 in treated group and control group. c–f Transcription levels of other imprinted genes in the PEG0/SGCE cluster, SGCE, CALCR, TFPI2, and PPP1R9A, were measured by real-time PCR in two groups. GAPDH was used as an internal control.
Furthermore, the transcription levels of other imprinted genes in the PEG10/SGCE cluster, including SGCE, CALCR, TFPI2, and PPP1R9A, were analyzed and the results were found to resemble those of PEG10 in that mRNA levels of these genes were notably increased in the treated group compared with the control group (Fig. 5c–f).
Altered PEG10 expression resulted in aberrant activation of apoptosis
PEG10 has been identified as an inhibitor of apoptotic processes.13 Therefore, further analysis was performed to evaluate the effect of altered expression of the PEG10 gene on apoptosis both in human brain tissue and cell samples.
Western blotting analysis in brain tissue of spina bifida samples indicated that the expression of apoptosis executioner CASPASE-3 had a nearly eight-fold increase compared with that of the control samples (0.20 ± 0.07 in controls vs. 1.56 ± 0.14 in cases, P < 0.001), suggesting abnormal activation of apoptosis occurred in spina bifida (Fig. 6a). In HCT-15 cells, a marked decrease of CASPASE-3 expression was found in the treated group (1.69 ± 0.07 in controls vs. 0.95 ± 0.12 in the treated group, P < 0.001) (Fig. 6b), in addition, through flow cytometry, we also found that the proportion of apoptotic cells was significantly decreased in cells treated with 5-Aza, in which activated expression of PEG10 was also observed (Fig. 6c).
a Expression levels of apoptosis marker, CASPASE-3, in fetal brain tissues isolated from patients with spina bifida or controls were measured by western blot. b Western blot analysis of CASPASE-3 expression in 5-Aza-treated cells and controls. c Flow cytometry analysis of cell apoptosis between 5-Aza-treated cells and controls.
Discussion
Genome imprinting has been confirmed to play a crucial role in embryogenesis, especially in brain development.5 Findings from our study showed that folate deficiency is linked to the observed aberrant low expression level of PEG10, a maternal allele imprinted gene, in the brain tissue of spina bifida cases through regulation of the methylation level in the gDMR of PEG10/SGCE cluster. Results in a cell model verified the effects of altered gDMR methylation on PEG10 gene expression regulation, as well as other imprinted genes in the same cluster. Moreover, results in this study indicated that suppression of PEG10 may result in ectopic activation of apoptosis in spina bifida. Our data provide experimental evidence supporting that aberrant gDMR methylation modification of PEG10/SGCE cluster mediated by folate deficiency is an attribute of spina bifida.
Retroviral elements have been commonly considered as “junk DNA” and even potentially harmful for host organisms.14 However, a series of knockout mouse models indicated that at least some imprinted retrotransposons play essential roles in mammalian developmental systems as endogenous genes.15 For example, PEG10, derived from the Ty3/Gypsy family of retrotransposons,10 has been shown to be involved in placental formation.11 Murphy et al found that the methylation profile of the PEG10/SGCE gDMR was largely consistent across human conceptual tissues, such as the placenta and brain, suggesting that PEG10 may have a role in brain function during mammalian embryogenesis, although few studies have focused on the activity of PEG10 in brain development.16 Our study investigated the role of PEG10 in the occurrence of spina bifida and found that expression of PEG10 was significantly down-regulated in the brain tissue of spina bifida samples. As a paternally expressed genes that are generally known to promote growth,17 the suppression of PEG10 was speculated to partly account for the neural tube closure disorders, namely NTDs.
DMRs characterize imprinted genes by carrying parental allele-specific methylation profiles. In the present study, we focused on the unique gDMR of PEG10/SGCE cluster18 and found that the methylation level of gDMR was notably elevated in spina bifida specimens, suggesting that there was an impaired epigenetic plasticity of the PEG10/SGCE imprinted cluster. Given the methylation pattern of gDMR is established during gametogenesis and escapes epigenome re-programming which occurred in the early stage of implantation, any epigenetic defects in gDMRs induced by malnutrition during gametogenesis will lead to the epigenetic inheritance of diseases,6 environmental interferences in the etiology of impaired imprinting have been documented.7 Folate is an essential micronutrient throughout life, particularly in the early stages of human development. A link has been established between inadequate folate intake during peri-conception and increased risk of NTDs incidence, and folic acid supplementation has been verified to result in a substantial reduction of NTDs by nearly 5% in Shanxi province, China, the research field of this study.19 Because folate acts as a one-carbon carrier, it plays a crucial role in providing an adequate source of methyl groups for the methylation of DNA, therefore gDMR methylation status was altered accordingly in response to folate insufficiency. Our previous studies have confirmed that aberrant methylation modification in other gDMRs of imprinted genes had a remarkable association with the increased occurrence of NTDs.8,9,20 More meaningfully, folic acid supplementation has been verified that it had an effect on rescuing the damaged steady state of gDMR methylation.21
The association analysis of the current study showed that brain tissue folate levels exhibited a significantly negatively associated with PEG10/SGCE gDMR methylation. Combined with the risk assessment analysis, these results illustrated that the aberrant gDMR hypermethylation of the PEG10/SGCE imprinted cluster was significantly positively associated with an increased risk of spina bifida. Although the methylation difference of gDMR between the case group and control group is only 4%, this is basically consistent with the difference of specific gene methylation between NTDs and the control group shown in several of our previous studies.8,9,12,22 Folate deficiency has been verified to participate in the formation of NTDs by dysregulating the methylation modification of these genes in the early stage of embryonic development. We speculate that the inhibition of these genes by extremely methylation abnormalities will be almost equivalent to the effect of gene knockout, which may lead to more serious early embryonic death. Moreover, the hierarchical analysis of methylation level showed that the proportion of the two groups was also significant. Therefore, we propose that folate deprivation-related hypermethylation of the PEG10/SGCE gDMR is a potential risk factor of spina bifida.
To elucidate the influence of altered methylation of the PEG10/SGCE gDMR in spina bifida, we constructed a demethylation model using HCT-15 cells treated with 5-Aza to investigate whether PEG10 expression was affected by gDMR methylation. The results showed an about 30% reduction of the methylation level in the cell model and the transcription level of PEG10 was notably negatively associated with gDMR methylation level, which verified the result in the human specimen. Furthermore, because imprinted genes reside mostly, but not exclusively, in clusters throughout the mammalian genome, the parent-of-origin-specific acquisition of DNA methylation marks at gDMR would influence the expression of imprinted genes in the whole cluster in embryos.23,24,25 In the present study, mRNA expression of other imprinted genes in the PEG10/SGCE cluster, including SGCE, CALCR, TFPI2, and PPP1R9A, was also found to be significantly increased in the group treated with methylation inhibitor, that implied the vital role of gDMR of PEG10/SGCE cluster as hallmarks indicative in imprinting regulation.
The classical view indicates that DNA methylation has a negative effect on transcription factor–DNA interactions and has generally been believed to play a repressive role in gene transcription.26 The gDMR of the PEG10/SGCE cluster exists in the promoter region between the PEG10 and SGCE genes.27 On the basis of the analysis by the PROMO database, multiple transcription factors (TFs) were predicted to have binding sites in the gDMR of PEG10/SGCE, particularly, the TFs which contain the CG sites/units in the DNA binding sites, such as E2F-1, P53, PAX5, and XBP-1. Aberrant gDMR hypermethylation probably influenced the binding of TFs, and thereby further influenced the expression regulation of PEG10. E2F-1 has been verified to have a trans-activating capacity to promote the up-regulation of PEG10 expression, and in turn cause proliferation, migration, and invasion in cancer cells.28,29 Further research is necessary to investigate the effect of TF regulation on PEG10 expression in spina bifida.
Although methylation dysregulation associated with folate deficiency has been generally confirmed to be involved in the occurrence of NTDs, how folate deficiency affects methylation modification is still an open question and needs further mechanism research. Several of our previous studies have revealed different regulations of folate on DNA methylation, including hypomethylation of genomic and LINE-1 gene,22 and dysregulated methylation modification of other specific genes including hypomethylation of MEST/PEG18 and GNAS,9 while hypermethylation of IFG220 and another oncogene GLI2.12 This phenomenon is very similar to the findings in tumors that genome instability and hypomethylation was found in oncogenes, while hypermethylation was found in tumor suppressor genes,30 which means that the effect of folate deficiency on methylation modification varies with the function of a specific gene.
As an oncogene, PEG10 has been implicated in the inhibition of apoptosis in a variety of tumors.13 Okabe et al. found that PEG10 was a target protein of SIAH, a key mediator of apoptosis, and could decrease cell death in hepatocellular carcinomas.31 Moreover, a viability decrease was observed in Chinese hamster ovary (CHO-K1) cells with PEG10 interacted with activin receptor-like kinase 1 (ALK1), a transforming growth factor (TGF)- β signaling receptor.32 In the present study, we further analyzed the influence of ectopic expression of PEG10 on apoptosis, and the results showed that aberrant activation of apoptosis was found in both brain tissue of spina bifida samples and non-treated HCT-15 cells with PEG10 suppression. Apoptosis plays an important role in the morphogenesis and homeostasis of the developing central nervous system in vertebrates, and accumulated evidence from gene knockout models has elucidated that the inactivation of specific apoptosis regulators is involved in NTDs.33 Our results suggest that inhibited expression of PEG10 induced by gDMR hypermethylation owe to folate insufficiency may attribute to spina bifida partly via the aberrant activation of apoptosis.
Conclusions
Our data indicate that gDMR methylation dysregulation of the PEG10/SGCE cluster mediated by folate insufficiency is involved in spina bifida. Our findings represent a novel strategy for interfering with the occurrence of NTDs by folate supplementation.
References
Avagliano, L. et al. Overview on neural tube defects: from development to physical characteristics. Birth Defects Res. 111, 1455–1467 (2019).
Greene, N. D. & Copp, A. J. Neural tube defects. Annu. Rev. Neurosci. 37, 221–242 (2014).
Czeizel, A. E. & Dudas, I. Prevention of the first occurrence of neural-tube defects by periconceptional vitamin supplementation. N. Engl. J. Med. 327, 1832–1835 (1992).
Bartolomei, M. S., Oakey, R. J. & Wutz, A. Genomic imprinting: an epigenetic regulatory system. PLoS Genet. 16, e1008970 (2020).
Perez, J. D., Rubinstein, N. D. & Dulac, C. New perspectives on genomic imprinting, an essential and multifaceted mode of epigenetic control in the developing and adult brain. Annu. Rev. Neurosci. 39, 347–384 (2016).
Abramowitz, L. K. & Bartolomei, M. S. Genomic imprinting: recognition and marking of imprinted loci. Curr. Opin. Genet. Dev. 22, 72–78 (2012).
Monk, D., Mackay, D. J. G., Eggermann, T., Maher, E. R. & Riccio, A. Genomic imprinting disorders: lessons on how genome, epigenome and environment interact. Nat. Rev. Genet. 20, 235–248 (2019).
Chang, S. et al. The effect of folic acid deficiency on Mest/Peg1 in neural tube defects. Int. J. Neurosci. 131, 468–477 (2021).
Wang, L. et al. Altered gnas imprinting due to folic acid deficiency contributes to poor embryo development and may lead to neural tube defects. Oncotarget 8, 110797–110810 (2017).
Ono, R. et al. A retrotransposon-derived gene, Peg10, is a novel imprinted gene located on human chromosome 7q21. Genomics 73, 232–237 (2001).
Ono, R. et al. Deletion of Peg10, an imprinted gene acquired from a retrotransposon, causes early embryonic lethality. Nat. Genet. 38, 101–106 (2006).
Lu, X. L. et al. Sonic hedgehog signaling affected by promoter hypermethylation induces aberrant Gli2 Expression in spina bifida. Mol. Neurobiol. 53, 5413–5424 (2016).
Xie, T. et al. Peg10 as an oncogene: expression regulatory mechanisms and role in tumor progression. Cancer Cell Int. 18, 112 (2018).
Garcia-Montojo, M., Doucet-O’Hare, T., Henderson, L. & Nath, A. Human endogenous retrovirus-K (Hml-2): a comprehensive review. Crit. Rev. Microbiol. 44, 715–738 (2018).
Peters, J. The role of genomic imprinting in biology and disease: an expanding view. Nat. Rev. Genet 15, 517–530 (2014).
Murphy, S. K., Huang, Z. & Hoyo, C. Differentially methylated regions of imprinted genes in prenatal, perinatal and postnatal human tissues. PLoS ONE 7, e40924 (2012).
Argyraki, M. et al. In-utero stress and mode of conception: impact on regulation of imprinted genes, fetal development and future health. Hum. Reprod. Update 25, 777–801 (2019).
Monk, D. et al. Comparative analysis of human chromosome 7q21 and mouse proximal chromosome 6 reveals a placental-specific imprinted gene, Tfpi2/Tfpi2, which requires Ehmt2 and Eed for allelic-silencing. Genome Res. 18, 1270–1281 (2008).
Liu, J. et al. Prevalence and trend of neural tube defects in five counties in Shanxi Province of Northern China, 2000 to 2014. Birth Defects Res. A Clin. Mol. Teratol. 106, 267–274 (2016).
Wu, L. et al. Altered methylation of Igf2 Dmr0 is associated with neural tube defects. Mol. Cell Biochem. 380, 33–42 (2013).
Toschi, P. et al. Maternal peri-conceptional undernourishment perturbs offspring sperm methylome. Reproduction 159, 513–523 (2020).
Wang, L. et al. Relation between hypomethylation of long interspersed nucleotide elements and risk of neural tube defects. Am. J. Clin. Nutr. 91, 1359–1367 (2010).
Edwards, C. A. et al. The evolution of imprinting: chromosomal mapping of orthologues of mammalian imprinted domains in monotreme and marsupial mammals. BMC Evol. Biol. 7, 157 (2007).
Tucci, V., Isles, A. R., Kelsey, G. & Ferguson-Smith, A. C. Genomic imprinting and physiological processes in mammals. Cell 176, 952–965 (2019).
Kelsey, G. & Feil, R. New insights into establishment and maintenance of DNA methylation imprints in mammals. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368, 20110336 (2013).
Yin, Y. et al. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science 356, eaaj2239 (2017).
Suzuki, S. et al. Retrotransposon silencing by DNA methylation can drive mammalian genomic imprinting. PLoS Genet. 3, e55 (2007).
Peng, Y. P. et al. Peg10 overexpression induced by E2f-1 promotes cell proliferation, migration, and invasion in pancreatic cancer. J. Exp. Clin. Cancer Res. 36, 30 (2017).
Wang, C. et al. Peg10 directly regulated by E2fs might have a role in the development of hepatocellular carcinoma. FEBS Lett. 582, 2793–2798 (2008).
Meng, H. et al. DNA methylation, its mediators and genome integrity. Int. J. Biol. Sci. 11, 604–617 (2015).
Okabe, H. et al. Involvement of Peg10 in human hepatocellular carcinogenesis through interaction with Siah1. Cancer Res. 63, 3043–3048 (2003).
Lux, A. et al. Human retroviral Gag- and Gag-Pol-like proteins interact with the transforming growth factor-beta receptor activin receptor-like kinase 1. J. Biol. Chem. 280, 8482–8493 (2005).
Cecconi, F., Piacentini, M. & Fimia, G. M. The involvement of cell death and survival in neural tube defects: a distinct role for apoptosis and autophagy? Cell Death Differ. 15, 1170–1177 (2008).
Acknowledgements
We gratefully acknowledge all the participants in the study. We thank Emma Longworth-Mills, Ph.D., from LiwenBianji (Edanz) (www.liwenbianji.cn/) for editing the English text of a draft of this manuscript.
Funding
This project was supported by the National Natural Science Fund of China (81701452, 81670802), the National Natural Science Fund of Beijing (7202018), the National Key Research and Development Program (2016YFC1000502), and the CAMS Initiative for Innovative Medicine (2016-I2M-1-008).
Author information
Authors and Affiliations
Contributions
X.L., T.Z., and L.Wang designed the research; X.L., S.Y., J.M., and P.P. conducted the research; S.W., C.S., L.Wu, and S.C. provided essential reagents or materials; X.L. and S.W. analyzed data or performed statistical analysis; X.L. wrote the manuscript; L.Wang revised the manuscript; X.L. and L.Wang had primary responsibility for the final content, and all authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Consent statement
The study protocol was reviewed and approved by the local ethics committee and institute review board of the Capital Institute of Pediatrics, and all participants submitted informed consent.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Lu, X., Yang, S., Jie, M. et al. Folate deficiency disturbs PEG10 methylation modifications in human spina bifida. Pediatr Res 92, 987–994 (2022). https://doi.org/10.1038/s41390-021-01908-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41390-021-01908-6