Abstract
Background
Both spontaneous and treatment-induced clearance of hepatitis C virus (HCV) in adults have been associated with genetic polymorphisms in the interferon-λ genes. The aim of the present study was to confirm the association between the rs12979860 and evaluate the association between the rs368234815 and the rs4803217 single-nucleotide polymorphisms (SNPs) of the interferon-λ genes and the outcome of the infection in children.
Methods
Alleles and genotypes frequencies of 32 children, who presented spontaneous clearance of the virus and 135 children, with viral persistence were compared with ethnically matched controls obtained from the 1000 Genomes Project and the International HapMap Project databases.
Results
The frequencies of the C/C genotype of rs12979860, the TT/TT of the rs368234815 and the A/C of the rs4803217 were higher in the clearance group than in children with viral persistence (C/C versus T/T + C/T odds ratio (OR): 2.6; 90% confidence intervals (CI): 1.3–5; p = 0.01; TT/TT versus ΔG/TT + ΔG/ΔG OR: 2.8; 90% CI: 1.4–5.5; p = 0.01; and A/A versus A/C OR: 8.3; 90% CI: 1.5–45.9; p = 0.017, respectively) and with the ethnically matched controls.
Conclusions
The rs12979860, the rs368234815 and the rs4803217 SNPs are associated with spontaneous clearance of HCV in children.
Impact
-
Innate immune system response has a key role in the outcome of vertically acquired HCV infection in children.
-
The rs12979860, the rs368234815 and the rs4803217 SNPs are associated with spontaneous clearance of HCV in children.
-
Interferons-λ activate the Janus kinase–Stat pathway, which in turn induces several interferon-stimulated genes, leading to suppression of HCV replication both in vivo and in vitro.
Similar content being viewed by others
Introduction
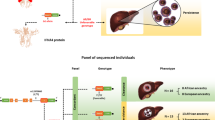
Interferons (IFN) are cytokines playing a major role in the innate immune response to viral infections (Fig. 1). IFN-λ belongs to the type III IFN family and maps on the human chromosome 19.1 IFN-λ induces antiviral activity and suppresses hepatitis C virus (HCV) replication in vivo and in vitro activating the Janus kinase–Stat pathway that inducts several IFN-stimulated genes, which are in turn responsible for the antiviral effect (Fig. 1).2 Both spontaneous and treatment-induced clearance of HCV in adults have been associated with certain genetic polymorphisms in the IFNL genes, such as the rs12979860, the rs368234815 and the rs4803217.3
A previous study by Ruiz-Extremera and collaborators has shown that the C allele and the C/C genotype of the rs12979860 single nucleotide polymorphism (SNP) were associated with spontaneous clearance of HCV infection in children.4 This association was confirmed by our group in subsequent multiple studies.5,6,7,8 No data are available on the impact of the SNPs rs368234815 and rs4803217 on the natural history of the infection in children.
The aim of the present study was to confirm the role of the SNP rs12979860, and to evaluate the role of the SNPs rs368234815 and rs4803217 in a large cohort of children with vertical acquired chronic HCV infection. Part of the children’s cohort was involved in previous studies by our group on the association between rs12979860 and spontaneous clearance of HCV.5,6,7,8 Allele and genotype frequencies in the cohort of patients described in the present study were compared with ethnically matched controls whose HCV status was unknown and obtained, using the data of the 1000 Genomes Project (1000GP)9 and the International HapMap Project (IHMP).10
Methods
Design of the study
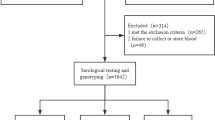
This is a retrospective study. All the children evaluated at the Meyer Children’s University Hospital of Florence between January 2011 to December 2019 during routine follow-up visits for vertical HCV infection were recruited if (1) born to a mother infected with HCV; (2) aged >30 months; (3) were hepatitis B surface antigen and human immunodeficiency virus antibodies negative; (4) were untreated against HCV; and (5) written informed consent for the study was provided by parents or guardians. At the time of the enrolment the following data were retrospectively collected: ethnic origin, source of infection, outcome of the infection (i.e., spontaneous clearance and chronic infection) and genotyping for the SNPs rs12979860, rs4803217 and rs368234815. The study was approved by the local ethical committee.
Definitions
Vertical transmission of HCV: transmission from the HCV-infected mother to the foetus or to the child occurring during pregnancy or in the perinatal period.11 Vertical infection: HCV ribonucleic acid (HCV RNA) detected during the first year of life in at least two serum samples taken at least 3 months apart, and/or positive antibodies against HCV in children >18 months of age.11 Spontaneous clearance of HCV: polymerase chain reactions (PCR) for HCV RNA negative and HCV antibodies positive in at least three blood samples taken 6 months apart in children >30 months of age.12 Chronic HCV infection: PCR for HCV RNA and HCV antibodies positive in at least two serum samples taken at least 6 months apart in children >30 months of age.
Patients
One hundred and sixty-seven children were consecutively enroled of whom 32 (19.2%) spontaneously cleared the virus and 135 (80.8%) had chronic hepatitis C.
Comparison with subjects studied in the 1000GPand IHMP
The rs12979860/rs4803217 and or the rs368234815 SNPs allele and genotype frequencies were examined, and the concordance with the subjects studied in the 1000GP (http://www.1000genomes.org) and in the IHMP (https://www.genome.gov/10001688/international-hapmap-project), respectively, were evaluated.
Laboratory methods
PCR for HCV was performed as previously described.13 A third-generation enzyme-linked immunosorbent assay (Ortho Diagnostic System Inc., Raritan, NJ) was used to study HCV antibodies. Confirmation was obtained by western blot (Innogenetics, Zwijndrecht, Belgium). IFNL rs12979860, rs4803217 and rs368234815 SNPs genotyping was performed using Allelic Discrimination assays (Applied Biosystems), according to manufacturer’s instructions. IFNL rs12979860-T/C genotyping was performed, as previously described.6 IFNL rs4803217-T/G genotyping was performed using Taqman custom-designed primers and probes as follows: forward primer, CTGTGTGTCTGACCCTTCCG; reverse primer, TCCTGGAGGTGAGTTGGATTTAC; probe for allele T, CAATAAATTAAGACAAGTGGCTA (VIC); and probe for allele G, ATAAATTAAGCCAAGTGGCTA (FAM). rs368234815-TT/ΔG was genotyped with Taqman custom-designed primers and probes: forward primer, GCCTGCTGCAGAAGCAGAGAT; reverse primer, GCTCCAGCGAGCGGTAGTG; probe for allele TT, ATCGCAGAAGGCC (VIC); and probe for allele ΔG, ATCGCAGCGGCCC (FAM). Before the PCR reactions, DNA was added with Allelic Discrimination Assay Mix and TaqMan Universal PCR Master Mix to MicroAmp Optical 96-Well Reaction Plates (Applied Biosystems). For the real-time PCR reactions, the 7500 Fast Real-Time PCR System was used. After preheating for 10 min at 95 °C, 45 cycles of 15 s at 95 °C and 1 min at 60 °C followed. The ABI SDS software (Applied Biosystems) was used to analyse the data.
Statistical analysis
The SPSSX package (SPSS, Inc., Chicago, IL) was used to evaluate the data. Age was reported as median and interquartile range. Differences in frequencies were evaluated by chi2 or Fisher’s exact tests using two-tailed p values with a level of significance set at <0.05, and OR and 90% CI were calculated. Hardy–Weinberg equilibrium (HWE) was tested. A p value <0.05 defined a deviation from HWE.
Results
HCV-infected children
The characteristics of the 167 children enroled, according to the outcome of the infection, are described in Table 1. Thirty-two children (19.2%) presented spontaneous clearance of HCV and 135 (80.8%) developed a chronic infection.
Data extracted from the 1000GP and from the IHMP databases
rs12979860 and rs4803217 SNPs
The alleles and genotypes frequencies of 2504 subjects were extracted for the rs12979860 and the rs4803217 SNPs from the 1000GP database (Tables 2 and 3). In this cohort, 107 of 503 Europeans (21.3%) were Italians. The highest frequencies of the C allele and of the C/C genotype were found in East Asians (rs12979860: 92% and 85%, respectively, and rs4803217 92% and 84%, respectively). Italians had a lower frequency of the C allele and of the C/C genotype as compared with the remaining Europeans (rs12979860, C allele 60% and 71.5%, respectively, p = 0.002; C/C genotype: 35% and 51.8%, respectively, p = 0.002; rs4803217, C allele 61% and 71.5%, respectively, p = 0.003; and C/C genotype: 35% and 52%, respectively, p = 0.001).
rs368234815
The alleles and genotypes frequencies of 270 subjects were extracted for the rs368234815 SNP from the IHMP database (Table 4). The Asiatic population had the highest frequencies of the TT allele and of the TT/TT genotype (93% and 87%, respectively) and significantly higher than that found in Caucasians (TT allele: 68%, p = 0.001 and TT/TT genotype: 48%, p = 0.0001).
Alleles and genotypes frequencies in the cohort of HCV-infected children
The frequencies of the rs12979860, rs368234815 and rs4803217 alleles and genotypes in the HCV cohort according to the outcome of the infection are described in Table 5.
Comparison of the allele frequencies
rs12979860
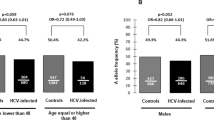
The frequency of the rs12979860 C allele was greater among children with spontaneous HCV clearance than in those chronically infected (75% versus 56% p = 0.005; OR: 2.3; 90% CI: 1.4–3.9) and to the Italians enroled in the 1000GP database (75% versus 60%, p = 0.03; OR: 1.9; 90% CI: 1.1–3.2), but was similar to that found in subjects of European ancestry (75% versus 69%, p = 0.3). Conversely, the T-allele frequency was greater in children with chronic hepatitis C than in the European controls (44% versus 31%, p = 0.001; OR: 1.7; 90% CI: 1.3–2.2).
rs368234815
The frequency of the rs368234815 TT allele was greater among children with spontaneous HCV clearance than in those chronically infected (75% versus 54%; p = 0.002; OR: 2.5; 90% CI: 1.5–4.3), but was similar to that found in subjects of Caucasian origin (75% versus 68%, p = 0.3). Conversely, the ΔG-allele frequency was greater in children with chronic hepatitis C as compared with Caucasians from the IHMP (46% versus 32%, p = 0.003; OR: 1.8; 90% CI: 1.2–2.5).
rs4803217
The frequency of the C allele of rs4803217 SNP was 48.4% among children with spontaneous clearance of HCV and 39.4% among those with chronic infection (p = 0.19). The frequency of the C allele was lower in children who spontaneously cleared the infection than that observed in the Europeans (48.4% versus 69%, p = 0.001; OR: 0.4; 90% CI: 0.2–0.6) and in Italians from the 1000GP database (48.4% versus 61%, p = 0.080; OR: 0.6; 90% CI: 0.3–0.9%). Conversely, the A-allele frequency was significantly greater in children with chronic hepatitis C as compared with Europeans (60.6% versus 31%, p = 0.001 OR: 3.4; 90% CI: 2.7–4.3%) and with Italians (60.6% versus 39%, p = 0.001 OR: 2.3; 90% CI: 1.7–3.2%).
Comparison of genotype frequencies
Genotypes were in HWE in the children with HCV infection (p = 0.63), for Rs12979860 (p = 0.6), rs368234815 (p = 0.59) and Rs4803217 (p = 0.58) genotypes, in Europeans and Italians from the 1000GP database and in Caucasians from the IHMP database (Tables 2–4).
Rs12979860
Children with the C/C genotype had higher clearance rate than children with the C/T and T/T genotype combined (p = 0.01; OR: 2.6; 90% CI: 1.3–5), and with the T/T genotype (p = 0.007; OR: 10; 90% CI: 1.81–60.8). The clearance rate of HCV was similar between the C/T and T/T, and between the C/C and the C/T genotypes. The frequency of the C/C genotype of the children with spontaneous clearance of HCV was similar to that of Europeans (53.1% and 48%, respectively, p = 0.5), but greater to that of Italians from the 1000GP database (53.1% versus 35%, p = 0.05; OR: 2.1; 90% CI: 1.1–4.1).
Rs368234815
The clearance rate of HCV was similar between the TT/TT and ΔG/TT genotypes. Children with the TT/TT genotype had higher clearance than those with ΔG/ΔG (p = 0.004; OR: 12.6; 90% CI: 2.2–72.3), and with the ΔG/ΔG and ΔG/TT genotypes taken together (p = 0.01; OR: 2.8; 90% CI: 1.4–5.5). The frequency of the TT/TT genotype was similar in children with spontaneous clearance of HCV and Caucasians from the IHMP (53.1% and 48%, respectively, p = 0.6).
Rs4803217
No C/C genotype was found in the HCV cohort. Children with the A/C genotype had higher clearance than those with A/A (p = 0.01; OR: 8.3; 90% CI: 1.5–45.9). The frequency of the A/C genotype was greater in children with spontaneous clearance of HCV (96.9%) as compared to Europeans (42%, p = 0.02; OR: 7.3; 90% CI: 1.3–40) and Italians from the 1000GP database (52%, p = 0.02; OR: 7.7; 90% CI: 1.3–44.2).
Discussion
This study demonstrates that the SNPs of IFNL3 and IFNL4 influence the spontaneous viral clearance of vertical acquired HCV infection. In particular, it confirms that the genotypes rs12979860 C/C and, for the first time, demonstrates that the rs368234815 TT/TT of the IFNL4 and the genotype rs4803217 A/C of the IFNL3 are associated with a significantly higher probability of spontaneous clearance of the infection compared to the subjects with alternative alleles and genotypes.
The SNP rs12979860 (C/T) is located in the first intron of the novel human IFNL4 gene.14 Previous genome-wide association studies in adults have shown that the rs12979860 CC genotype was associated with higher spontaneous and treatment induced clearance of the virus.15,16 The rs12979860 is located 367 base pairs from rs368234815 (TT/ΔG; previously known as ss469415590) a functional dinucleotide variant located in the first exon of IFNL4.17,18 The IFNL4 rs368234815-ΔG allele creates an open reading frame for the IFN-λ4 protein, whereas the alternative rs368234815-TT allele eliminates the IFN-λ4 protein.17 The rs368234815-ΔG and the -TT alleles are negatively and positively associated with spontaneous clearance of HCV, respectively.3 Finally, the rs4803217 SNP is located in the 3′ untranslated region of IFNL3. Even though data on rs4803217 SNP and HCV clearance are limited, it is reported that rs4803217-C allele is associated with a decreased degradation of IFN-λ3 mRNA and with a favourable outcome of the infection.19 The present study, confirming previous results in adults, supports the hypothesis that both IFNL3 and IFNL4 promote spontaneous clearance of HCV, regardless of patient’s age.
The value of the results of the present study was confirmed by the comparisons of alleles and genotypes frequencies among children with chronic infection, spontaneous clearance and, where available, with ethnically matched controls. In the context of genome-wide association studies, it is important to select the proper reference population that is usually the one geographically closer.20,21 If the genetic structure is not is not properly evaluated, the associations found can be spurious as demonstrated in the present article. In the present study, for example, according to the 1000GP database, for the rs12979860 SNP the C allele was more common in East-Asians when compared with the Europeans and, among these, with Italians. While for the rs4803217 A/C genotype, the correlation with spontaneous clearance was confirmed by the comparison of the frequency observed in the cohort of children with spontaneous clearance with the genotype frequencies of both Europeans and Italians extracted from the 1000GP database, for rs12979860 C/C genotype, the correlation was confirmed only when the frequency observed in the cohort of children with spontaneous clearance was compared with that of Italians selected as reference. For rs368234815 SNP, the data extracted from the IHMP database were limited to the Caucasian population with no specific data from southern Europe or Italy. This is the possible explanation why the highest prevalence of the TT/TT genotype in children who presented spontaneous clearance of the infection could not be confirmed by the comparison, with the cohort of matched controls extracted by the IHMP database. It is well known that within Europe there is a major axis of genetic variation from North to South21 that is confirmed by the results obtained for the rs12979860 SNP according to the 1000GP database.
Spontaneous clearance of HCV has been demonstrated to be dependent on both the viral genotype, as it is more common for genotype 322 and on the host’s immune system (Table A, provided as Supplementary file).23 Children presenting spontaneous clearance of HCV following vertical transmission of the infection have higher blood aminotransferase levels in their first year of life than those who develop chronic infection.24 Since the values of transaminases are an expression of immune-mediated hepatic damage, it is possible to hypothesize that the outcome of the infection transmitted vertically is strictly dependent on the action of the immune system. Previous studies have shown that the number of natural killer (NK) cells and their differentiation phenotype influence the outcome of HCV infection in children after vertical transmission. The cytolytic function of NK cells characterized phenotypically by the presence of CD56 (dim), in particular, contributes to the elimination of HCV in children with spontaneous clearance.23 The role of IFNL4 and of IFNL3 has been shown to be fundamental in the response to pharmacologic therapy with pegylated IFN and in spontaneous viral clearance in adults.25
In the present study, the IFNL4 rs12979860 C allele and the C/C genotype were more common in children with spontaneous HCV clearance than in those with chronic infection. The results confirm previous studies performed in adults25 and children.4,5 Subjects with the protective rs12979860 C/C genotype have higher serum type III IFN levels, which has potent antiviral properties.26 Furthermore, patients with CC genotype have higher levels of inhibitory receptors on the surface of their NK cells and fewer lymphocytes that expose the ligand for tumour-induced necrosis factor-α apoptosis.27 The association between SNPs of rs12979860 of the IFNL4 gene and spontaneous and treatment-induced clearance of HCV were already supported by paediatric data4,5, but the present study adds new insights evaluating the impact of the IFNL4 rs368234815 and of the IFNL3 rs4803217 polymorphisms on the natural history of the infection.
The IFNL4 rs368234815 TT/TT genotype was found to be associated with HCV clearance, while the ∆G allele was more frequent in children with chronic infection. Although IFN-λ4 can be generated only by individuals who carry the IFNL4 rs368234815 ΔG allele, the present study supports that this variant is associated with the failure to clear HCV infection, as previously demonstrated in adults.28,29 The paradoxical functions of IFN-λ4, which appears to induce antiviral activity yet impair effective clearance of HCV,30 has been correlated recently with the possible preactivation of the Janus kinase–Stat signalling that leads to impaired HCV clearance by both IFN-α and IFN-λ.31
The present study shows that the the IFNL3 rs4803217 A/C genotype is associated with spontaneous clearance of HCV infection in Italian children. It should be noted that surprisingly, and without any clear explanation, the C/C genotype was not identified in any of the patients enroled although it was described in 34% of the Italians enroled in the 1000GP database. It has been demonstrated that the Rs4803217 C allele promotes spontaneous clearance dictating an increasing transcript stability, when compared to the A allele.19 Furthermore, in vitro studies demonstrated the expression of IFNL3 associated with the C allele is higher.32 Although the C allele was not associated with spontaneous clearance in our data, a clear correlation between the presence of the A allele and of the AA genotype in children with chronic infection was demonstrated.
In conclusion, this study provides insights in the immunological mechanism of the natural history of HCV infection in children.
References
Kotenko, S. V. et al. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat. Immunol. 4, 69–77 (2003).
Urban, T., Charlton, M. R. & Goldstein, D. B. Introduction to the genetics and biology of interleukin-28B. Hepatology 56, 361–366 (2012).
O’Brien, T. R. et al. Comparison of functional variants in IFNL4 and IFNL3 for association with HCV clearance. J. Hepatol. 63, 1103–1110 (2015).
Ruiz-Extremera, A. et al. Genetic variation in interleukin 28B with respect to vertical transmission of hepatitis C virus and spontaneous clearance in HCV-infected children. Hepatology 53, 1830–1838 (2011).
Indolfi, G. et al. Interleukin 28B rs12979860 single-nucleotide polymorphism predicts spontaneous clearance of hepatitis C virus in children. J. Pediatr. Gastroenterol. Nutr. 58, 666–668 (2014).
Indolfi, G. et al. Comparative analysis of rs12979860 SNP of the IFNL3 gene in children with hepatitis C and ethnic matched controls using 1000 Genomes Project data. PLoS ONE 9, e85899 (2014).
Indolfi, G., Sambrotta, M., Moriondo, M., Azzari, C. & Resti, M. Genetic variation in interleukin-28B locus is associated with spontaneous clearance of HCV in children with non-1 viral genotype infection. Hepatology 54, 1490–1491 (2011).
Indolfi, G., Azzari, C. & Resti, M. Polymorphisms in the IFNL3/IL28B gene and hepatitis C, from adults to children. World J. Gastroenterol. 20, 9245–9252 (2014).
Abecasis, G. R. et al. A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073 (2010).
The International HapMap Consortium. A haplotype map of the human genome. Nature 437, 1299–1320 (2005).
Indolfi, G., Azzari, C. & Resti, M. Perinatal transmission of hepatitis C virus. J. Pediatr. 163, 1549–1552. e1541 (2013).
Indolfi, G. et al. Hepatitis C virus infection in children and adolescents. Lancet Gastroenterol. Hepatol. 4, 477–487 (2019).
Indolfi, G. et al. Alanine transaminase levels in the year before pregnancy predict the risk of hepatitis C virus vertical transmission. J. Med. Virol. 78, 911–914 (2006).
Ge, D. et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature 461, 399–401 (2009).
Shi, X. et al. IL28B genetic variation is associated with spontaneous clearance of hepatitis C virus, treatment response, serum IL-28B levels in Chinese population. PLoS ONE 7, e37054 (2012).
Indolfi, G., Thorne, C., El Sayed, M. H., Giaquinto, C. & Gonzalez-Peralta, R. P. The challenge of treating children with hepatitis C virus infection. J. Pediatr. Gastroenterol. Nutr. 64, 851–854 (2017).
Prokunina-Olsson, L. et al. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat. Genet. 45, 164–171 (2013).
Biswas, A. et al. Interferon λ3 gene (IL28B) is associated with spontaneous or treatment-induced viral clearance in hepatitis C virus-infected multitransfused patients with thalassemia. Transfusion 57, 1376–1384 (2017).
McFarland, A. P. et al. The favorable IFNL3 genotype escapes mRNA decay mediated by AU-rich elements and hepatitis C virus-induced microRNAs. Nat. Immunol. 15, 72–79 (2014).
Novembre, J. et al. Genes mirror geography within Europe. Nature 456, 98–101 (2008).
Stathias, V. et al. Exploring genomic structure differences and similarities between the Greek and European HapMap populations, implications for association studies. Ann. Hum. Genet 76, 472–483 (2012).
Bortolotti, F. et al. Long-term course of chronic hepatitis C in children, from viral clearance to end-stage liver disease. Gastroenterology 134, 1900–1907 (2008).
Indolfi, G. et al. Altered natural killer cells subsets distribution in children with hepatitis C following vertical transmission. Aliment. Pharmacol. Ther. 43, 125–133 (2016).
Resti, M. et al. Clinical features and progression of perinatally acquired hepatitis C virus infection. J. Med. Virol. 70, 373–377 (2003).
Lange, C. M. & Zeuzem, S. IL28B single nucleotide polymorphisms in the treatment of hepatitis C. J. Hepatol. 55, 692–701 (2011).
Langhans, B. et al. Interferon-lambda serum levels in hepatitis C. J. Hepatol. 54, 859–865 (2011).
Golden-Mason, L. et al. Natural killer inhibitory receptor expression associated with treatment failure and interleukin-28B genotype in patients with chronic hepatitis C. Hepatology 54, 1559–1569 (2011).
Alves, C. F. et al. Interferon lambda 4 (IFNL4) gene polymorphism is associated with spontaneous clearance of HCV in HIV-1 positive patients. Genet. Mol. Biol. 39, 374–379 (2016).
Xie, X., Zhang, L. & Chen, Y. Z. Association between IFNL4 rs368234815 polymorphism and sustained virological response in chronic hepatitis C patients undergoing PEGylated interferon/ribavirin therapy, a meta-analysis. Hum. Immunol. 77, 609–615 (2016).
O’Brien, T. R., Prokunina-Olsson, L. & Donnelly, R. P. IFN-lambda4, the paradoxical new member of the interferon lambda family. J. Interferon Cytokine Res. 34, 829–838 (2014).
Ferraris, P. et al. Cellular mechanism for impaired hepatitis C virus clearance by interferon associated with IFNL3 gene polymorphisms relates to intrahepatic interferon-lambda expression. Am. J. Pathol. 186, 938–951 (2016).
Knapp, S., Meghjee, N., Cassidy, S., Jamil, K. & Thursz, M. Detection of allele specific differences in IFNL3 (IL28B) mRNA expression. BMC Med. Genet. 15, 104 (2014).
Author information
Authors and Affiliations
Contributions
All the authors provided substantial contributions to conception and design of the study, acquisition, analysis and interpretation of data. G.I. and G.M. drafted the article. All the authors revised the article critically for important intellectual content and provided the final approval of the version to be published.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Consent statement
The written informed consent was obtained from the parent or guardian before participating in the study.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Mangone, G., Serranti, D., Bartolini, E. et al. SNPs of the IFNL favour spontaneous clearance of HCV infection in children. Pediatr Res 91, 1516–1521 (2022). https://doi.org/10.1038/s41390-021-01557-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41390-021-01557-9