Abstract
Background
Infants with moderate and severe neonatal encephalopathy (NE) frequently suffer from long-term adverse outcomes. We hypothesize that the urinary metabolome of newborns with NE reflects the evolution of injury patterns observed with magnetic resonance imaging (MRI).
Methods
Eligible patients were newborn infants with perinatal asphyxia evolving to NE and qualifying for therapeutic hypothermia (TH) included in the HYPOTOP trial. MRI was employed for characterizing brain injury. Urine samples of 55 infants were collected before, during, and after TH. Metabolic profiles of samples were recorded employing three complementary mass spectrometry-based assays, and the alteration of detected metabolic features between groups was assessed.
Results
The longitudinal assessment revealed significant perturbations of the urinary metabolome. After 24 h of TH, a stable disease pattern evolved characterized by the alterations of 4–8% of metabolic features related to lipid metabolism, metabolism of cofactors and vitamins, glycan biosynthesis and metabolism, amino acid metabolism, and nucleotide metabolism. Characteristic metabolomic fingerprints were observed for different MRI injury patterns.
Conclusions
This study shows the potential of urinary metabolic profiles for the noninvasive monitoring of brain injury of infants with NE during TH.
Impact
-
A comprehensive approach for the study of the urinary metabolome was employed involving a semi-targeted capillary electrophoresis–time-of-flight mass spectrometry (TOFMS) assay, an untargeted ultra-performance liquid chromatography (UPLC)–quadrupole TOFMS assay, and a targeted UPLC-tandem MS-based method for the quantification of amino acids.
-
The longitudinal study of the urinary metabolome identified dynamic metabolic changes between birth and until 96 h after the initiation of TH.
-
The identification of altered metabolic pathways in newborns with pathologic MRI outcomes might offer the possibility of developing noninvasive monitoring approaches for personalized adjustment of the treatment and for supporting early outcome prediction.
Similar content being viewed by others
Introduction
Perinatal asphyxia is defined as the interruption of blood flow and gas exchange to the fetus in the perinatal period. Prolonged hypoxia–ischemia can trigger a cascade of events leading to injury to vital organs, especially the brain (i.e., neonatal encephalopathy, NE). NE following birth asphyxia is a major cause of early neonatal death, cerebral palsy, or neurodevelopmental disability in term neonates, affecting ~0.5–1.5 per 1000 live births in high-income countries and ~1 million infants every year worldwide.1,2,3 Magnetic resonance imaging (MRI), including diffusion-weighted imaging and spectroscopy performed between the fifth and seventh day after birth, has become the cornerstone for establishing the degree of brain injury and predicting neurodevelopmental outcome.4 There is compelling evidence suggesting that perinatal asphyxia and the administration of supplemental oxygen may influence the brain at the molecular level by stimulating epigenetic modifications, including DNA methylation/demethylation, histone modifications, and miRNAs and epigenetic mechanisms, that may be involved in the development of a hypoxic/ischemic-sensitive phenotype.5,6
It has been observed that the combination of decreased cerebral perfusion and subsequent hypoxia triggers an evolving sequence of biochemical alterations that, lasting days, weeks, or even months, lead to anatomical and physiological injuries characterizing NE.7,8 Therefore, a longitudinal biochemical assessment of NE is needed from birth until beyond the duration of therapeutic hypothermia (TH). NE has been associated with damage to the integrity of the blood–brain barrier, and therefore brain injury could be reflected in peripheral blood samples or other biofluids9 theoretically enabling the implementation of noninvasive monitoring approaches. Metabolomics is a systems biology approach aiming at the holistic study of the complete pool of small molecules forming the metabolome of a biological sample. While metabolites are downstream products of the genome, transcriptome, and proteome, they reflect their interactions and the interaction with the environment, thereby providing a direct, dynamic, and meaningful read out of the biochemical status of a system closely related to its actual phenotype.10 Literature reports on the use of metabolomics for the study of perinatal asphyxia in newborns with NE are highly encouraging, although the main scope has been the early diagnosis of NE based on the comparison of asphyxiated/NE infants to healthy control infants,11,12,13,14,15,16 and metabolomics has only been scarcely used for progressing our understanding of the multilayer effects of NE and TH on the newborn.16,17 Recently, the analysis of metabolic profiles of plasma samples collected between birth and until completion of TH enabled the identification of a dynamic perturbation of 11 metabolic pathways, including amino acid and purine metabolism, and the steroid hormone biosynthesis, in addition to pyruvate and lactate, in newborns with pathologic MRI outcomes.18
In this study, we hypothesized that the urinary metabolome of newborns with NE recorded during the first 96 h of life encompasses the evolution of the injury pattern of the brain that is evidenced by MRI outcomes. Hence, the goal of this study is to provide insight into metabolic alterations secondary to brain injury in a clinical cohort of newborn infants with NE undergoing TH, with the aim of generating a knowledge base for the future development of a noninvasive monitoring approach, supporting the prediction of long-term clinical outcomes.
Methods
Study approval and population
The study was approved by the Ethics Committee for Biomedical Research of the Health Research Institute La Fe (Valencia, Spain) and registered under the acronym HYPOTOP (EudraCT 2011-005696-17). The HYPOTOP trial is a randomized, controlled, multicenter, double-blinded clinical trial for assessing the efficacy of topiramate vs. placebo in newborns, with NE undergoing TH. All methods were performed in accordance with relevant guidelines and regulations, and informed consent was obtained from legal representatives of enrolled infants. For a detailed description of the HYPOTOP trial, including the study design and the established inclusion and exclusion criteria, the reader is referred to the literature.19 In this study, a sub-cohort of 55 newborns enrolled in the HYPOTOP trial was included.
Samples were collected from urinary catheters or using sterile cotton pads placed in the diaper following a standard operating procedure to avoid bias and systematic errors. Urine samples were collected before initiation of TH at 6 h, as well as 12, 24, 48, 72, and 96 h after the initiation of TH. Samples were aliquoted into three dry microcentrifuge tubes and stored immediately at −80 °C until analysis.
Magnetic resonance imaging
MRI was carried out using different high-field magnet systems between days 4 and 8 after birth. In all cases, 3D Gradient Echo T1-weighted MR images, axial Fast Spin Echo T2-weighted MR images, and diffusion-weighted images were acquired. The interpretation of MRI results was carried out by an experienced, blinded pediatric neuroradiologist, rating the extent of the injury in posterior limb internal capsule, basal ganglia, thalami, white matter, and cortex20 and using a standardized score according to the National Institute of Child Health and Human Development (NICHD) Neonatal Research Network (NRN), discerning between injury patterns as described by Shankaran et al.21.
Metabolomic fingerprinting of urine samples
Three complementary analytical approaches were employed for the analysis of metabolic fingerprints of urine samples to enhance metabolome coverage, i.e., (i) a semi-targeted capillary electrophoresis–time-of-flight mass spectrometry (CE–TOFMS) assay, (ii) an untargeted ultra-performance liquid chromatography–quadrupole time-of-flight mass spectrometry (UPLC–QTOFMS) assay, and (iii) a targeted UPLC-tandem MS (MS/MS)-based method for the quantification of amino acids. Results were normalized to creatinine concentrations and are made available as Supplementary Table 1.
Semi-targeted CE–TOFMS analysis
A total of 279 urine samples were thawed on ice and homogenized on a Vortex® mixer for 1 min. A total of 50 μL of the sample were mixed with 50 μL of 0.2 M formic acid containing 0.4 mM methionine sulfone (internal standard, IS). After homogenization on a Vortex® mixer for 5 min and centrifugation at 13,200 × g for 10 min at 4 °C, 60 μL of the supernatant were collected and mixed with 180 μL of cold methanol for 5 min. Samples were centrifuged at 13,200 × g for 15 min at 4 °C, and 200 μL of supernatant were transferred into microcentrifuge tubes and evaporated to dryness using a Speedvac concentrator. Finally, sample extracts were redissolved in 60 μL of 0.2 M formic acid, shaken for 5 min on a Vortex® mixer, and centrifuged at 2000 × g at 4 °C for 5 min. A total of 50 μL of supernatant were transferred to CE vials for analysis. Quality control (QC) samples were prepared by pooling 5 μL from each sample and processed applying the same procedure, as described for samples.
The analysis of urine samples was carried out by capillary electrophoresis–electrospray–time-of-flight MS (CE-ESI-TOFMS), using instrumental conditions described elsewhere.22 Briefly, CE–ESI–TOFMS analysis was performed on an Agilent 7100 Capillary Electrophoresis system (Agilent Technologies, CA) coupled to an Agilent 6224 TOFMS instrument. The separation was performed on a fused silica capillary (Agilent; total length, 100 cm; i.d., 50 μm) in normal polarity with a background electrolyte composed of 1.0 M HCOOH solution in 10% CH3OH (v/v). Samples were hydrodynamically injected at 50 mbar for 17 s followed by the injection of background electrolyte at 100 mbar for 10 s. The separation voltage applied was 30 kV with 25 mbar of internal pressure and the run time was 40 min. The sheath liquid (6 µL min−1) composition was CH3OH:H2O (1:1, v/v) containing two reference masses (purine and HP-0921). The MS parameters used were: fragmentor 125 V, skimmer 65 V, octopole 750 V, nebulizer pressure 10 p.s.i., drying gas temperature at 200 °C, and flow rate 10 mL min−1. The capillary voltage was 3500 V. Full-scan MS spectra from m/z 70 to 1000 were acquired in positive dual-ESI mode at a scan rate of 1.02 scan s−1. MassHunter Workstation version B.06.01 controlled the CE–TOFMS system. The analysis of the samples was carried out in four batches of randomized samples. At the beginning of each batch, six QCs were injected for pre-sequence system conditioning, and a QC was injected every ninth sample to monitor the system performance. CE–TOFMS data processing using the Profinder software (B.06.00, Agilent) enabled the identification of 102 metabolites (Supplementary Table 2) in the urine sample set.
Untargeted UPLC–QTOFMS analysis
A total of 293 urine samples were prepared and analyzed by UPLC–QTOFMS assay, as described elsewhere.23 Urine samples were randomly split into three analytical batches. Briefly, urine samples were thawed on ice, homogenized on a Vortex® mixer during 30 s, and centrifuged (16,000 × g during 15 min at 4 °C). A total of 50 µL of supernatant were withdrawn and spiked with 50 µL of IS solution in 96-well plates containing phenylalanine-D5 (Cambridge Isotopes Laboratory Inc., Andover, MA), caffeine-D9 (Toronto Research Chemicals, Toronto, Ontario, Canada), leukine enkephalin (Sigma-Aldrich Química SA, Madrid, Spain), and reserpine (Sigma-Aldrich Química SA) in H2O:CH3OH (1:1, 0.1% v/v HCOOH) at concentrations ranging between 2 and 4 µM. Blanks were prepared by replacing urine with H2O and a QC was prepared by mixing 5 µL of each final sample extract. The QC was split into three aliquots, each being measured in a single analytical batch to avoid additional freeze–thaw cycles. Sample extracts, blanks, and QCs were stored at −80 °C until analysis. Before analysis of each batch, sample extracts, blanks, and a QC aliquot were thawed on ice.
UPLC–QTOFMS analysis was performed on an Agilent 1290 Infinity UPLC chromatograph equipped with a UPLC BEH C18 (100 × 2.1 mm, 1.7 µm, Waters, Wexford, Ireland) column running a binary mobile phase gradient (mobile phase A: H2O, 0.1% v/v HCOOH and mobile phase B: CH3CN, 0.1% v/v HCOOHH), and acquiring full-scan MS data from 100 to 1700 m/z with a scan frequency of 6 Hz on an iFunnel QTOF Agilent 6550 spectrometer in the positive electrospray (ESI+) mode. The QC sample was injected ten times at the beginning of each batch for system conditioning, as well as after every five urine samples for the monitoring and control of the instrumental performance. The blank extract was injected twice, at the beginning and end of each batch.
The peak table was generated for each batch separately using XCMS (version 3.4.2) (https://bioconductor.org/packages/release/bioc/html/xcms.html) running in R (version 3.6.1), using the centWave method.24 Intensity weighted m/z values of each feature were calculated using the wMean function and peak limits used for integration were found through descent on the Mexican hat filtered data. Peak grouping was carried out using the nearest method, and the fillPeaks method was applied to fill missing peak data.
All further calculations were carried out in MATLAB R2019b (Mathworks Inc., Natick, MA) using in-built, as well as in-house written scripts and functions and the PLS Toolbox 8.7 (Eigenvector Research Inc., Wenatchee). Blank samples were used to identify and remove features arising from contaminants and carryover with a ratio of median intensities in QCs and blanks <9.25 The three data sets were aligned using m/z and RT tolerances of 50 mDa and 0.1 min, and a mzVsRT = 1. Within-batch effects were corrected for each batch separately employing the Quality Control-Support Vector Regression algorithm23 and the LIBSVM library26 method with an ε-range from 2.5 to 5, a γ-range from 1 to 105, and a C-interval of 50.27,28 Between-batch effects were linearly corrected using the median values in QCs as reference.23 UPLC-MS features with a relative intensity change across batches >5, and those with a D-ratio ≥ 20% in QCs after within and between-batch effect correction were classified as unreliable29 and removed.
Quantitative amino acid profiling
A total of 237 urine samples were thawed on ice and homogenized on a Vortex S0200 mixer (LabNet, Edison, NJ) during 10 s. A total of 70 µL of water (Optima LC/MS grade, Fisher Scientific) were added to 5 µL of urine. Amino acid levels were quantified using a ultraderivation kit (AccQ TagTM Ultra Derivatization Kit, Waters), following the manufacturer’s protocol. Blanks were prepared by replacing the urine sample with water (Optima LC/MS grade). A QC was prepared by mixing 5 µL of each sample. Quantitative analysis of amino acids was performed employing a 1290 Infinity UPLC system from Agilent equipped with a UPLC CORTECS C18 column (150 × 2.1 mm, 1.6 µm) from Waters coupled to an Agilent 6460 triple quadrupole MS system operating in ESI+ mode. The flow rate was set to 500 µL min−1 running a binary gradient with 0.1% v/v HCOOH in H2O and 0.1% v/v HCOOH in CH3CN as mobile phase components. Column and autosampler were kept at 55 and 4 °C, respectively and the injection volume was 3 µL. Raw data were acquired and processed employing MassHunter Workstation (version B.07.00, Agilent). Missing values were imputed as 0.5 × LOQ of each compound. In Supplementary Table 3 a list of the parameters employed for amino acid quantification can be found.
Creatinine determination
Creatinine was quantified following the protocol of the Urinary Creatinine Detection Kit (Arbor Assays™, Ann Arbor, MI), employing a dilution factor of 1:4 and measuring the absorbance at 490 nm.
Data processing and statistics
Metabolic fingerprints were normalized to creatinine, log10 transformed, and pareto scaled. Between group comparisons of clinical and demographic variables were carried out employing the Student’s t test for unequal variances (α = 0.05), Wilcoxon rank-sum test (α = 0.05), or χ2-test (α = 0.05), according to the underlying distribution. For determining differentially regulated metabolites or UPLC–QTOFMS features, as well as for comparing MRI outcomes at each time point, Student’s t tests for unequal variances (α = 0.05) were applied and fold change (FC) was calculated as the ratio of means between groups.
The standardized mean difference using the simplified Cohen equation to determine the pooled standard deviation30 was calculated for each metabolite/metabolic feature at each of the six sample collection time points. Then, the Pearson correlation coefficient was calculated between the standardized mean difference and time and obtained FDR-adjusted P values were calculated.31
MetaboAnalyst (version 5.0)32 was used for the generation of heatmaps from autoscaled data employing Euclidean distance and Ward’s method. Pathway analysis was carried out using the mummichog algorithm (P value cutoff set to 0.05) with a m/z accuracy = 10 p.p.m. and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway library (Homo sapiens). As an input, t test P values estimated assuming unequal variances between distributions, and FCs were computed for each metabolic feature comparing normal and pathologic outcomes. Pathways with gamma P values <0.05 were considered as significantly altered. The PLS Toolbox 8.7 (Eigenvector Research Inc., Wenatchee) was used for partial least squares (PLS) and PLS discriminant analysis (PLSDA) of autoscaled data. For k-fold cross validation, samples were distributed into folds by subject to prevent person-specific information crossover between the training folds and the test fold. Receiver operating characteristic (ROC) curves were constructed based on PLSDA predictions and cross validation results.
Results
Patient characteristics
A total of 55 infants from the HYPOTOP cohort were included in this study. The characteristics of the studied population, including 55 newborns with NE undergoing TH are shown in Table 1. No significant differences between the control (i.e., normal neurological outcome) and pathologic groups were found in any of the collected anthropometric, clinical, or biochemical parameters. Furthermore, both groups were balanced in the proportion of infants receiving topiramate as adjuvant therapy.
Dynamics of the urinary metabolome
The urinary metabolome of newborns with NE was studied employing three complementary analytical methods, i.e., untargeted UPLC–QTOFMS, semi-targeted CE–TOFMS, and a targeted method for the determination of amino acids. Using UPLC–QTOFMS, a total of 293 urine samples were analyzed with N = 37 collected before the initiation of TH and N = 45, N = 54, N = 54, N = 54, and N = 49 collected 12, 24, 48, 72, and 96 h, respectively, after the initiation of TH. After initial data preprocessing and filtering, 11,561 features were retained and used for further analysis. In case of CE–TOFMS, a total of 279 urine samples were analyzed with N = 35 collected before the initiation of TH and N = 44, N = 52, N = 53, N = 53, and N = 42 collected 12, 24, 48, 72, and 96 h, respectively, after the initiation of TH. A list of metabolites that were consistently detected in urine samples with the CE–TOFMS target assay can be found in Supplementary Table 2. Amino acids were quantified in 237 samples, N = 27 collected before the initiation of TH and N = 40, N = 41, N = 47, N = 43, and N = 39 collected 12, 24, 48, 72, and 96 h, respectively, after the initiation of TH (see Supplementary Table 3).
The longitudinal analysis of the metabolic fingerprint of infants with NE showed a dynamic behavior. Figure 1 (left) depicts the percentage of differentially expressed features and metabolites (t test P value < 0.05) after the initiation of TH, using the first urine samples collected as reference. The majority of detected signals showed a decreasing trend with time ranging between 16 and 59% at 12 and 96 h after the initiation of TH, respectively. Simultaneously, only between 0.08 and 1% were upregulated. Changes are most pronounced during the first 48 h after the initiation of TH, whereas rewarming (96 h) did not seem to have a strong impact. The effect of time is further represented in Fig. 2, where metabolite concentrations with a significant correlation with time are represented. In total, 43% of the features of the urinary metabolic fingerprint showed a significant correlation with time, and out of the correlated features, 53% tended to increase with time.
Left: % of significantly changing features (P value < 0.05) and % of upregulated and downregulated features at each time point during 96 h after initiating TH with respect to the first urine sample collected before the initiation of TH. Right: % of significantly changing features (P value < 0.05) and % of upregulated and downregulated features in urine samples from infants with vs. without brain injury, as detected by MRI.
Note: color represents standardized mean difference between time points; blue—metabolite levels lower than average, and red—metabolite levels higher than average. P values from Pearson correlation with time were computed, and significant metabolites after multiple testing correction are shown; heatmap calculated using Euclidean distance and Ward algorithm representing autoscaled features.
Perturbation of the urinary metabolome in infants with pathologic MRI outcome
Differences in the urinary metabolic fingerprints of infants with normal and pathologic MRI outcome were assessed throughout the first 96 h of life. As shown in Fig. 1 (right), the number of significantly altered metabolic features reaches a minimum at 12 h and ranges between 4 and 8% at the remaining time points (6 ± 2%). The proportion of upregulated features ranges between 0 and 0.2%, whereas the proportion of downregulated features was rising from 0.8% at 12 h to 7% at 96 h.
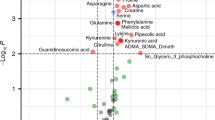
Figure 3 illustrates the metabolic pathways altered between newborn infants with pathologic and normal MRI outcomes. Significant alterations on the pathway level were found in urine samples collected at all studied time points. Pathways related to the amino acid and lipid metabolism were consistently identified to be affected in infants with pathologic MRI. Pathways related to the metabolism of cofactors and vitamins were altered during TH. Differences in pathways associated to xenobiotics biodegradation and metabolism were found repeatedly before, during and after TH. Carbohydrate metabolism and glycan synthesis and metabolism were altered during the first 24 h. Other pathways related to the metabolism of terpenoids and polyketides, and nucleotide and energy metabolism were affected transiently. Accordingly, Fig. 3 also shows the classes of changing metabolites, with “carboxylic acids and derivatives”, “organooxygen compounds”, “fatty acyls”, “steroids and steroid derivatives”, “indoles and derivatives”, and “tetrapyrroles and derivatives” being the most prominent metabolite classes affected. A detailed list of identified altered metabolites is provided in Supplementary Table 4.
With the aim of defining a disease pattern in urine metabolic fingerprints, pathway analysis was repeated for all samples collected between 24 and 96 h after the initiation of TH together, as the differences between newborns with NE and pathologic vs. normal MRI outcomes in the metabolome became more stable (see Fig. 1). Pathway analysis confirmed the consistent alteration of ten pathways associated to amino acid and lipid metabolism, metabolism of cofactors and vitamins, glycan biosynthesis and metabolism, and nucleotide metabolism, as shown in Fig. 3. Using the metabolic features corresponding to metabolites included in those pathways as annotated by the mummichog algorithm (i.e., 398 features), a PLSDA model with four latent variables was computed for discerning between metabolic fingerprints of newborns with pathologic (Scores 1, 2, or 3) and normal MRI outcomes (Score-0) retrieved between 24 and 96 h after the initiation of TH. The significance of the model was confirmed by a permutation test (500 permutations) and ROC curves are shown in Fig. 4.
ROC curves from a PLSDA model (four latent variables) for Score-0 (top, left), Score-1 (top, right), Score-2 (bottom, left), and Score-3 (bottom, right) calculated, using relative intensities of 398 metabolic features from altered metabolic pathways shown in Fig. 3. Note: black lines represent estimated PLSDA ROC curves (calibration set); gray lines represent estimated PLSDA ROC curves (cross validation); dashed lines represent 50% lines; and circles indicate model thresholds.
Discussion
A severe hypoxic insult suffered in the perinatal period causes an imbalance of the homeostasis manifesting at a systemic level in the newborn. This perturbation can be readily monitored employing urinary metabolomics allowing to study dynamic changes of the phenotype over time. In this study, a continuously increasing proportion of altered metabolic features with time was detected affecting up to 59% of features (Fig. 1, left) and 43% of features showed a significant correlation with time (Fig. 2). The number of downregulated features was steadily increasing and higher than the number of upregulated features at all studied time points (Fig. 1, left). This observation is in good agreement with earlier literature reports on metabolomics studies involving the analysis of the urinary metabolome of NE infants over time,15,17,33 reporting continuous changes during the neonatal period with some metabolites showing a relative increase in concentration, while others tended to decrease.
The main aim of this study was to search for a metabolic fingerprint able to discern urine samples from infants, which showed pathologic patterns during a subsequent MRI analysis from those with normal MRI results. This is the first study reporting distinct patterns in urinary metabolic fingerprints of newborns with NE and pathologic vs. normal MRI outcomes. The injury pattern showed a dynamic behavior. Initially detected differences between groups almost disappeared 12 h after the initiation of TH, which might reflect a partial recovery during the latent injury phase, and then became more evident again after 24 h of TH (Fig. 1, right).
To further elucidate the effect of brain injury on the metabolome, pathway analysis was employed. Pathway analysis aims to place the seemingly disjointed list of altered metabolites into a broader biological context by jointly assigning metabolites to relevant metabolic pathways, providing an interaction network that may identify centralized hubs, where metabolic pathways coincide or where bottlenecks may occur. Pathway alterations affecting between 3 and 11 pathways (Fig. 3) were detected across all studied time points, covering before and during TH, as well as after rewarming with some alterations being transient and others being detected consistently (e.g., tryptophan metabolism).
From the clinical perspective and with the aim of developing a monitoring approach the discovery of stable disease patterns is valuable. We found that a total of ten pathways were consistently altered in urine samples of infants with NE and pathologic MRI outcomes, when urine samples collected between 24 and 96 h after the initiation of TH were analyzed (Fig. 3). Seemingly, after a period of 24 h after the initiation of TH, biochemical alterations that evolve to anatomical lesions in the brain that are later detectable by MRI are mirrored in the urinary metabolome. Metabolites associated to the ten altered metabolic pathways were successfully employed to discern between newborns with different injury patterns (i.e., Scores 0, 1, 2, and 3) as shown by ROC curves in Fig. 4. Statistically highly significant models could be obtained for all comparisons, with exception of the model for the discrimination of Score-3, probably due to the limited number of samples, as only two infants showed the highest degree of brain injury. Hence, our data provides evidence of gradual changes detectable in urine samples, providing a knowledge base for the development of biomarkers that might support clinical monitoring compatible with TH in the future.
To the best of our knowledge, this study has analyzed and compared for the first time the evolution of the metabolome of newborns with NE and pathologic vs. normal MRI outcomes over time. The use of a representative control group of newborns with NE instead of healthy control newborns has facilitated the interpretation of the outcomes. Due to substantial differences in the experimental design between this study and previous results,15,17,33 a detailed comparison of the outcomes was not possible. However, alterations of amino acids and associated pathways, including tryptophan metabolism and aminoacyl-tRNA biosynthesis,17,33 had been reported. In umbilical cord blood samples from infants with NE vs. healthy control newborns, the terpenoids backbone biosynthesis pathway and, also a significant alteration of kynurenine, a metabolite from the tryptophan metabolism pathway, were identified.14 Interestingly, in a mouse model with induced excitotoxic lesion, changes in amino acid metabolism in brain tissue were sustaining acute and delayed responses to injury.34
In a previous study, we analyzed the plasma metabolic fingerprints of the newborns included in this study.18 Several metabolic pathways altered in urine (Fig. 3) were also found to be affected in plasma, i.e., arginine and proline metabolism (48 and 72 h in plasma vs. 72 h in urine), lysine degradation (72 h in plasma vs. 12 in urine), nitrogen metabolism (72 h in plasma and urine), and steroid hormone biosynthesis (0, 24, 48, and 72 h in plasma vs. 24 h in urine). Hence, the results from metabolic fingerprinting in plasma and urine samples are in good agreement, indicating that the development of brain injury secondary to asphyxia is broadly affecting metabolism.
This study has limitations. No newborns with asphyxia or classified as mild NE (Sarnat 1) were included. There is risk of adverse neurodevelopmental outcome of newborns with mild NE, but to date, there is no evidence to support cooling in these babies. Furthermore, as NE is frequently associated to a transient renal insufficiency secondary to proximal tubular necrosis with decreased urine output and creatinine clearance, hematuria, and proteinuria, analytical determinations might have been influenced by renal insufficiency especially in babies with severe NE.35
In summary, this is the first time that a dynamic perturbation of the urinary metabolome of newborns with NE undergoing TH, and showing pathologic and normal MRI outcomes is reported. Undoubtedly, TH significantly influences patient’s metabolism. However, since TH has become a standard of care, our results truly reflect the clinical situation undergone by patients with moderate to severe NE. From birth and until completing 24 h of TH, the metabolic fingerprint experienced a gradual shift and a distinct disease pattern evolved. This disease pattern is characterized by the alteration of amino acid and lipid metabolism, metabolism of cofactors and vitamins, glycan biosynthesis and metabolism, and nucleotide metabolism. Although follow-up studies are required for further confirming the observations from this study, this new insight might be meaningful for patient monitoring in the NICU and for supporting early outcome prediction.
References
Kurinczuk, J. J., White-Koning, M. & Badawi, N. Epidemiology of neonatal encephalopathy and hypoxic-ischaemic encephalopathy. Early Hum. Dev. 86, 329–338 (2010).
Lee, A. C. C. et al. Intrapartum-related neonatal encephalopathy incidence and impairment at regional and global levels for 2010 with trends from 1990. Pediatr. Res. 74, 50–72 (2013).
Lehtonen, L., Gimeno, A., Parra-Llorca, A. & Vento, M. Early neonatal death: a challenge worldwide. Semin. Fetal Neonatal Med. 22, 153–160 (2017).
Groenendaal, F. & de Vries, L. S. Fifty years of brain imaging in neonatal encephalopathy following perinatal asphyxia. Pediatr. Res. 81, 150–155 (2017).
Ma, Q. & Zhang, L. Epigenetic programming of hypoxic-ischemic encephalopathy in response to fetal hypoxia. Prog. Neurobiol. 0, 28–48 (2015).
Lorente-Pozo, S. et al. Oxygen in the neonatal period: oxidative stress, oxygen load and epigenetic changes. Semin. Fetal Neonatal Med. 25, 101090 (2020).
Douglas-Escobar, M. & Weiss, M. D. Hypoxic-ischemic encephalopathy: a review for the clinician. JAMA Pediatr. 169, 397–403 (2015).
Edwards, A. D. & Azzopardi, D. V. Perinatal hypoxia-ischemia and brain injury. Pediatr. Res 47, 431–432 (2000).
Lee, W. L. A., Michael-Titus, A. T. & Shah, D. K. Hypoxic-ischaemic encephalopathy and the blood-brain barrier in neonates. Dev. Neurosci. 39, 49–58 (2017).
Nagana Gowda, G. A. & Raftery, D. Biomarker discovery and translation in metabolomics. Curr. Metabolomics 1, 227–240 (2013).
Walsh, B. H. et al. The metabolomic profile of umbilical cord blood in neonatal hypoxic ischaemic encephalopathy. PLoS ONE 7, e50520 (2012).
Reinke, S. N. et al. 1H NMR derived metabolomic profile of neonatal asphyxia in umbilical cord serum: implications for hypoxic ischemic encephalopathy. J. Proteome Res. 12, 4230–4239 (2013).
Ahearne, C. E. et al. Early cord metabolite index and outcome in perinatal asphyxia and hypoxic-ischaemic encephalopathy. Neonatology 110, 296–302 (2016).
Denihan N. M. et al. Untargeted metabolomic analysis and pathway discovery in perinatal asphyxia and hypoxic-ischaemic encephalopathy. J. Cereb. Blood Flow Metab. 39, 147–162 (2019).
Locci, E. et al. A longitudinal 1H-NMR metabolomics analysis of urine from newborns with hypoxic-ischemic encephalopathy undergoing hypothermia therapy. Clinical and medical legal insights. PLoS ONE 13, e0194267 (2018).
Efstathiou, N., Theodoridis, G. & Sarafidis, K. Understanding neonatal hypoxic-ischemic encephalopathy with metabolomics. Hippokratia 21, 115–123 (2017).
Noto, A. et al. Urinary gas chromatography mass spectrometry metabolomics in asphyxiated newborns undergoing hypothermia: from the birth to the first month of life. Ann. Transl. Med. 4, 417 (2016).
Piñeiro-Ramos, J. D. et al. Metabolic phenotypes of hypoxic-ischemic encephalopathy with normal vs. pathologic magnetic resonance imaging outcomes. Metabolites 10, 109 (2020).
Nuñez-Ramiro, A. et al. Topiramate plus cooling for hypoxic-ischemic encephalopathy: a randomized, controlled, multicenter, double-blinded trial. Neonatology 116, 76–84 (2019).
Rutherford, M. et al. Assessment of brain tissue injury after moderate hypothermia in neonates with hypoxic-ischaemic encephalopathy: a nested substudy of a randomised controlled trial. Lancet Neurol. 9, 39–45 (2010).
Shankaran, S. et al. Neonatal magnetic resonance imaging pattern of brain injury as a biomarker of childhood outcomes following a trial of hypothermia for neonatal hypoxic-ischemic encephalopathy. J. Pediatr. 167, 987–993.e3 (2015).
López-Gonzálvez, Á., Godzien, J., García, A. & Barbas, C. Capillary electrophoresis mass spectrometry as a tool for untargeted metabolomics. Methods Mol. Biol. 1978, 55–77 (2019).
Sánchez-Illana, Á. et al. Evaluation of batch effect elimination using quality control replicates in LC-MS metabolite profiling. Anal. Chim. Acta 1019, 38–48 (2018).
Smith, C. A., Want, E. J., O’Maille, G., Abagyan, R. & Siuzdak, G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 78, 779–787 (2006).
Martínez-Sena, T. et al. Monitoring of system conditioning after blank injections in untargeted UPLC-MS metabolomic analysis. Sci. Rep. 9, 1–9 (2019).
Chang, C.-C. & Lin, C.-J. LIBSVM: a library for support vector machines. ACM Trans. Intell. Syst. Technol. 2, 27:1–27:27 (2011).
Sánchez-Illana, Á. et al. Model selection for within-batch effect correction in UPLC-MS metabolomics using quality control - Support vector regression. Anal. Chim. Acta 1026, 62–68 (2018).
Kuligowski, J., Sánchez-Illana, Á., Sanjuán-Herráez, D., Vento, M. & Quintás, G. Intra-batch effect correction in liquid chromatography-mass spectrometry using quality control samples and support vector regression (QC-SVRC). Analyst 140, 7810–7817 (2015).
Broadhurst, D. et al. Guidelines and considerations for the use of system suitability and quality control samples in mass spectrometry assays applied in untargeted clinical metabolomic studies. Metabolomics 14, 72 (2018).
Cohen, J. et al. Statistical Power Analysis for the Behaviorla Sciences (Lawrence Erlbaum Associates, 1988).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 57, 289–300 (1995).
Chong, J. et al. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 46, W486–W494 (2018).
Sarafidis, K. et al. Urine metabolomic profile in neonates with hypoxic-ischemic encephalopa-thy. Hippokratia 21, 80–84 (2017).
Blaise, B. J. et al. Persistently altered metabolic phenotype following perinatal excitotoxic brain injury. Dev. Neurosci. 39, 182–191 (2017).
Vento, M., Sastre, J., Asensi, M. A. & Viña, J. Room-air resuscitation causes less damage to heart and kidney than 100% oxygen. Am. J. Respir. Crit. Care Med. 172, 1393–1398 (2005).
Acknowledgements
The authors would like to express their gratitude to the parents and their newborns who participated in the study. This work was supported by the Instituto Carlos III, Ministry of Economy and Competitiveness, Spain [grant numbers CD19/00037, CP16/00034, EC11-244, and PI17/00127].
Author information
Authors and Affiliations
Consortia
Contributions
J.K., M.V., and C.B. were responsible for the conception and design of the study; J.D.P.-R., M.M.C., A.A.-D., A.S.G., J.M.-R., R.L.S., A.L.-G., D.S.-H., and G.Q. acquired the data; J.K., G.Q., J.D.P.-R., M.M.C., and A.N.-R. analyzed and interpreted the data; J.K. drafted the article; C.B., G.Q., and M.V. revised the manuscript critically and all authors approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Consent statement
Legal representatives of all participants signed an informed consent form.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Piñeiro-Ramos, J.D., Cascant, M.M., Núñez-Ramiro, A. et al. Noninvasive monitoring of evolving urinary metabolic patterns in neonatal encephalopathy. Pediatr Res 91, 598–605 (2022). https://doi.org/10.1038/s41390-021-01553-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41390-021-01553-z
This article is cited by
-
Longitudinal perturbations of plasma nuclear magnetic resonance profiles in neonatal encephalopathy
Pediatric Research (2023)
-
Neonatal encephalopathy plasma metabolites are associated with neurodevelopmental outcomes
Pediatric Research (2022)
-
Biomarkers in neonatal encephalopathy: new approaches and ongoing questions
Pediatric Research (2022)