Abstract
As human skin hosts a diverse microbiota in health and disease, there is an emerging consensus that dysregulated interactions between host and microbiome may contribute to chronic inflammatory disease of the skin. Neonatal skin is a unique habitat, structurally similar to the adult but with a different profile of metabolic substrates, environmental stressors, and immune activity. The surface is colonized within moments of birth with a bias toward maternal strains. Initial colonists are outcompeted as environmental exposures increase and host skin matures. Nonetheless, early life microbial acquisitions may have long-lasting effects on health through modulation of host immunity and competitive interactions between bacteria. Microbial ecology and its influence on health have been of interest to dermatologists for >50 years, and an explosion of recent interest in the microbiome has prompted ongoing investigations of several microbial therapeutics for dermatological disease. In this review, we consider how recent insight into the host and microbial factors driving development of the skin microbiome in early life offers new opportunities for therapeutic intervention.
Impact
-
Advancement in understanding molecular mechanisms of bacterial competition opens new avenues of investigation into dermatological disease.
-
Primary development of the skin microbiome is determined by immunological features of the cutaneous habitat.
-
Understanding coordinated microbial and immunological development in the pediatric patient requires a multidisciplinary synthesis of primary literature.
Similar content being viewed by others
Introduction
The complex microbial community that inhabits barrier tissues, called the microbiome, is essential to understanding human health. Resident microbes support many functions of the human body, including metabolism,1 synthesis of vitamins,2 protection against pathogen invasion,3 and immune development.4,5 “Germ-free” gnotobiotic animals, born of sterile parents in a permanently sterile environment, demonstrate numerous physiological derangements and disease susceptibilities not seen in genetically identical animals enveloped by their microbiota.6,7,8 Thus microbes found on the healthy host include commensals and symbiotes, colonization with which is mutually beneficial.
With the follicular surface included, skin is the largest epithelial surface of the human body for interaction with microbes.9 This surface is a dynamic interface rather than an impermeable barrier, as the microbiome extends into the dermis and dermal adipose.10 The skin microbiome is characterized by diversity, skin site specificity, and stability. Human skin is a unique habitat with a microbiota distinct from other primates’, showing a greater preponderance of skin specialists and lower diversity overall.11 Nonetheless, healthy skin hosts a diverse microbial community with >200 genera from 19 different phyla.12,13,14 Different parts of the body display markedly different microbial communities on the skin.15 Shared features of the skin microbiome between sites reflect shared skin physiology.13,16 For example, Cutibacterium acnes (formerly Propionibacterium acnes) is a frequent colonist of sebaceous skin, while the harsher environment of dry skin is dominated by staphylococci and streptococci.13,16 Much like a fingerprint, the adult skin microbiome is highly personalized and stable in strain composition over years.16,17
Cohabitation with trillions of microorganisms is not without risk. Breakdown of the symbiotic relationship has been linked to chronic diseases of the skin, including atopic dermatitis18 and psoriasis,19 consistent with a role for microbiota in chronic diseases of other organs, such as the lung20 and the gut.21,22,23,24,25 An enduring question is how and why symbiosis with microbes either fails to develop or devolves into chronic disease.
Microbiologists have long appreciated that the presence of bacteria alone is insufficient to predict illness.26,27 Microscopic observations of abundant colonies of bacteria in skin abscesses,27 acne comedones,28 and eczematous dermatoses29 led early authors to propose that disease followed from excessive bacterial proliferation. Sabouraud proposed that bacterial overgrowth was prevented in health by constant epithelial desquamation,30 while later authors described the skin as an “acid mantle” inhospitable to bacterial growth due to low pH,31 osmotic stress,32 and desiccation.33 However, by the 1970s, improved culture methods had revealed that acne folliculitis, and its response to treatment, had no association with bacterial burden.34,35,36 This finding led to the hypothesis that host maturation induced native C. acnes toward an inflammatory phenotype, with colonizing strains showing variable propensity for inflammatory conversion.37 A key insight of this hypothesis is that genetic differences between different strains of the same species of bacterium can have drastically different and long-lasting effects on the host.
Interest in the strain specificity of disease has been renewed by increasingly intractable resistance to antimicrobial therapy and the technical ability to examine composition of the whole microbiome at strain-level resolution using metagenomics.38 In the past decade, culture-independent surveys of the indoor environment have revealed myriad commensal and pathogenic strains absent in individual microbiota despite persistent exposure.39,40,41,42,43,44 These observations suggest that understanding the developmental ecology of the microbiome, such as the host and microbial factors influencing strain acquisition from the environment, may add considerable insight into chronic skin diseases associated with dysbiosis. Therefore, the mechanisms by which infants acquire and retain specific microbes is of great interest. This review will focus on skin microbiome composition in early life and the host and microbial mechanisms governing its strain-specific development.
Skin microbiome assembly in early life
The order and timing of colonization events determines how strains subsequently interact with one another, an effect known as priority.45,46 Indeed, in mouse models of enteric disease, prior colonization of a naive host with a benign strain can limit subsequent engraftment of a pathogenic strain of the same species and prevent mucosal injury.47 Thus an infant’s first microbial encounters may have long-term consequences for microbiome composition and skin health.
The sterility of newborns at birth has been questioned. Several groups have reported bacterial DNA in the placenta, amnion, and fetus,48,49,50,51,52,53 including viable bacteria visualized and cultured from fetal mice during the second trimester,53 while others have failed to find evidence of microbial colonization before birth except in cases of clinically significant infection.54,55 Some authors have proposed that maternal microbiota are selectively transported to the placenta in order to colonize the fetus.48,49 Indeed, in a recent report, human infants were found to have oral and meconium microbiota at the time of Caesarean delivery predicted to originate from the placenta.53
The most extensive microbial colonization begins at birth. Immediately postpartum, the microbiota is homogeneously distributed across the human body regardless of delivery method or gestational age.56,57,58,59 Culture-dependent surveys of the skin within 5 min of vaginal delivery showed that culturable microbiota were overwhelmingly staphylococci (phylum Firmicutes) at every body site, with a minority of diptheroids (phylum Actinobacteria, including cutibacteria and corynebacteria).59 Neonates born by Caesarean section had no detectable bacteria,59 consistent with a sterile uterine environment. More recent culture-independent surveys of newborn infants showed vaginally delivered neonates were preferentially colonized by vaginal Prevotella and Lactobacillus, while the skin of neonates born by Caesarean section was found to have a diverse community of cutibacteria, corynebacteria, and micrococcae presumably contaminating the operative field from maternal skin.57,60
The infant’s first bath may alter the process of microbiome assembly. The newborn infant is coated with vernix caseosa, an unevenly distributed waxy layer derived from sebaceous glands with a unique profile of lipids, ceramides, and antimicrobial peptides (AMPs).61 Bathing in the first 24 h of life is known to disrupt this layer and is no longer recommended due to increased risk of hypothermia.62 Culture-dependent studies have shown that the first bath is also associated with immediate changes in the composition of the microbiome that are not yet fully characterized.63,64 These effects may depend on gestational age, as the distribution of vernix caseosa is different between preterm, term, and post-term infants.65 Moreover, preterm infants admitted to the neonatal intensive care unit (NICU) are more likely to be washed with antimicrobial soap.66
The long-term consequences of these initial perturbations are not known. The early life microbiome undergoes frequent strain replacements over time.67,68,69 Thus the association between birth environment and microbiome fades within weeks to months.56,70,71 In the absence of perturbation, a significant fraction of the skin microbiota of hospitalized infants at any given time originates from their hospital rooms.72 Bacterial communities at different skin sites begin to diverge into distinct functional communities like those found in the adult as soon as 2 days after delivery.73 Six weeks after delivery, the skin microbiota of mother and infant are more similar than their microbiota at other body sites, like the gut and oropharynx, which diverge more rapidly.56
The similarity observed between maternal and infant microbiota may be due to vertical transmission of maternal strains. Maternal microbiota are crucial for developing the microbiome of the skin, as of the gut.74,75,76 Postnatal vertical transmission of the mother’s bacteria may provide early colonists that can reside in the child’s gastrointestinal tract and influence its composition for many years while it develops into a distinct community.77 Indeed, maternal strains are more likely to persist in the infant gut than non-maternally acquired strains due to shared environment and common genetic and immunological factors.78,79 This may explain why probiotic supplementation during pregnancy is associated with lower incidence of childhood atopic dermatitis,80 a disease with significant microbial involvement.18 Vertical transmission of strains on the skin has not yet been directly demonstrated, however, and any mechanistic basis for mother–infant strain specificity remains unclear.
Identification of environmental reservoirs of bacteria, their contribution to skin microbiota, and the mechanisms of transmission in early life is currently underway. Initial colonists are those available in the immediate environment, such as the NICU or hospital room.72,81 As a result, seeding of the birthing room with maternal microbiota may be an important determinant of initial bacterial exposure, as the microbiota found throughout an indoor space can match their occupants within hours.41,82 The similarity between mother and infant microbiota decreases over the first year of life,83 suggesting successful invasion of environmental strains. These strains may be dispersed from the skin of close contacts, as has been shown for gut microbiota.46 A significant fraction of bacterial transmission may be indirect. A large study of pediatric patients around the age of 3 years, their homes, and their household contacts found that frequent handwashing could prevent household colonization of new Staphylococcus aureus strains, while strain transmission within households was associated with high household burden of the strain, as well sharing bedrooms, cosmetics, and bathroom towels.44 This work suggests that microbial transmission can be controlled by modifiable behaviors.
Cutaneous determinants of microbial ecology
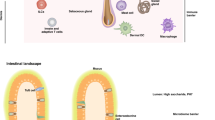
The restricted composition of the skin microbiome was first attributed to an active “degerming” process of normal skin.30 Growing appreciation of bacterial physiology led to the hypothesis that skin microbiota was constrained by environmental limitations. The epidermis is structurally mature by 34 weeks of gestation.84 However, the cutaneous environment of early life differs from the adult by having higher water content, higher pH, fewer lipids, and more rapid epidermal turnover.85 Infant skin has higher water content than the adult within 3 months,86 which change corresponds to eccrine sweat gland maturation.87 This may be significant for microbial colonization, as surface bacteria in the adult experience severe water restriction.33 Humans produce much more sebum than other mammals, which in part explains the greater abundance of C. acnes.88 Secretion is reduced in early life,89 which may explain why children have proportionally fewer corynebacteria and cutibacteria.90 Desquamation, originally proposed as the skin’s principal “degerming” mechanism,30 is elevated in infants, as sebum production is inversely related to epidermal turnover.85,86 Melanin production also changes with age due to sun exposure.91 While melanin utilization is widespread among bacteria,92 its role in skin ecology is unknown.
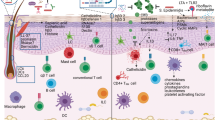
Recent attention has turned to host selection of bacteria through the targeted action of the immune system, including innate cells like keratinocytes. In healthy skin, keratinocytes are the primary source of AMPs93; each AMP with its own antimicrobial profile, including cathelicidins, beta-defensins, and S100A peptides.94 AMP accumulation in epidermal and dermal compartments restricts tissue invasion95 and peaks in early life. Keratinocytes express microbial pattern recognition receptors and upregulate AMP secretion in response to microbial ligands,96 thus balancing microbial ligand density with AMP secretion. Surprisingly, a population of AMP-secreting keratinocytes were recently shown in the mouse to express the antigen presentation complex major histocompatibility complex class II in response to the barrier-protective cytokine interleukin (IL)-22.97 These specialized cells are physically associated with cutaneous CD4+ T cells that express interferon (IFN)-γ after stimulation with commensal antigens.97 IFN-γ signaling induced by commensal microbiota in the murine gut has been shown to inhibit colonization with a pathogenic Salmonella strain.98 Similarly, recent work shows that IL-4Rα blockade, which potentiates IFN-γ signaling, is associated with reduced S. aureus colonization and increased microbial diversity in adult atopic patients,99 which may in part explain its efficacy in ameliorating the symptoms of atopic dermatitis.100 Whether tonic IFN-γ activity modifies skin ecology to select certain microbiota over others, and whether this selection protects the host or predisposes to disease, remains to be shown.
Skin is also home to innate lymphoid cells (ILCs) and lymphocytes with innate-like functions, including γδ T cells, natural killer T (NKT) cells, and mucosal-associated invariant T (MAIT) cells. Skin-resident ILCs, recruited to pilosebaceous units via C-C chemokine motif receptor 6, have been described to nurture Gram-positive commensalism in the mouse by expressing tumor necrosis factor receptor ligands that downregulate Notch signaling in sebocytes and restrain antimicrobial fatty acid secretion.101 Interestingly, ILCs in different tissue layers express significantly different genetic programs, indicating selective compartmentalization of immune functions.101 Most human epidermal T cells are γδ T cells.102 These cells may respond to microbial cues103 and are thought to be important in regulating insulin-like growth factor-1-induced keratinocyte turnover.104,105 NKT cells alter the intestinal microbiome in mouse models.106 In the skin, the NKT cell population expresses a pro-inflammatory phenotype107 that may, for example, contribute to alopecia areata.108 MAIT cells depend on riboflavin derivatives generated by skin microbiota and do not develop in germ-free animals.109 They have been described to control bacterial translocation109,110 and direct tissue repair.111,112,113 As MAIT cell development in mice is restricted to early life,111 dysregulated immune–microbe crosstalk during childhood may predispose an individual to skin dysbiosis throughout life.
Early life is also critical for the development of adaptive lymphocytes. Anti-inflammatory regulatory T cells (Tregs) occur at higher density in the neonatal skin of humans and mice, which biases the neonate toward an anti-inflammatory response.114,115 For example, neonatal germ-free mice exposed to Staphylococcus epidermidis develop antigen-specific Tregs that dampen inflammation against this benign commensal later in life.115 Colonization of hair follicles, an important niche for coagulase-negative staphylococci such as S. epidermidis, stimulates Treg recruitment to the hair follicles via chemokine C-C chemokine motif ligand 20.116 In the gut, Treg recruitment may promote selective bacterial colonization by facilitating B cell class switch to immunoglobulin A (IgA).117 Indeed, Bacteroides fragilis promotes mucosal Treg polarization and invites IgA binding to associate intimately with host epithelium and exclude exogenous competitors.118,119,120 Like the gut microbiome, the skin microbiome is highly stable after infancy.77,121,122 It is tempting to speculate that skin commensals are similarly adapted to direct host immunity for species- or strain-specific tolerance.
Mechanisms of microbial competition
Competition between microbiota for the hospitable early life environment may be especially important in structuring the microbiome to favor long-term skin health.123 Medicinal use of antagonism between bacteria has been discussed by every generation of microbiologists since Louis Pasteur in the nineteenth century.124,125,126 Several therapeutic approaches have been identified. In the early twentieth century, a Danish physician named Schiotz observed that a young patient with S. aureus pharyngitis, mistakenly placed in the diphtheria ward, was resistant to acquiring the disease. Schiotz cultured benign staphylococci from a surgical patient and reported success in protecting Corynebacterium diphtheriae carriers against diphtheria by spraying the staphylococci culture into the throat.127,128 A number of physicians in the United States reported employing this “overriding” therapy for pediatric diphtheria before widespread use of antitoxin superseded the practice.129,130,131,132 Variability in engraftment of S. aureus and displacement of C. diphtheriae was frequently observed but could not be explained.131 Similarly, the variability of exogenously introduced bacteria in successfully colonizing the enteric niche may explain the frequent failure of oral probiotic therapy in randomized trials.133,134 Recent attempts to displace S. aureus in atopic dermatitis patients with daily administration of competing strains of streptococci, coagulase-negative staphylococci, and the Gram-negative Roseomonas mucosa have shown efficacy in limited studies.135,136 Successful niche displacement, as measured by durable colonization of the probiotic strain, may significantly enhance the therapeutic effect.
An alternative approach for the pediatric population is to preempt initial pathogen invasion of the cutaneous niche. In the 1960s, a strain of S. aureus defined by phage type 80/81 that resisted existing antibiotics became epidemic to hospital nurseries worldwide.137,138,139 After identifying a benign S. aureus strain 502A common in infants that resisted strain 80/81, Henry Shinefield and colleagues showed that prophylactic inoculation of 502A lowered the rate of strain 80/81 engraftment into the neonatal microbiome and decreased the incidence of staphylococcal disease.140,141,142,143,144 The mechanism of antagonism was never defined, however, and 502A proved susceptible to displacement from environmental strains over time.145
Increasing awareness of the role of dysbiosis in chronic disease has renewed interest in how such niche competition structures the microbiome. An elaborate system of strain competition among Gram-negative bacteria has recently been described involving contact-dependent injection into neighboring bacteria of bacteriocidal toxins through the type VI secretion system (T6SS).146,147 Effectors are classically paired with immunity proteins to prevent self-intoxication; dozens of such pairings, specific to different strains, have been reported.68 The T6SS is a large protein complex that requires significant metabolic commitment to operate.148 Nonetheless, there appears to be strong evolutionary pressure among enteric commensals to accumulate genetic elements that neutralize T6S toxins,149 indicating their central place in structuring the enteric microbial community.
Many Gram-positive bacteria of the skin share a similar type VII secretion system (T7SS). S. aureus secretes four ESS proteins through the T7SS, which promote persistent infection in murine hosts.150 While one study reported association between S. aureus T7S expression and bacterial antagonism,151 efforts to identify antibacterial activity in analogy to T6S have not been successful.152 A T7SS-dependent system of LXG proteins with bacteriocidal properties, expressed together with an antidote against self-intoxication, was recently described in Streptococcus intermedius.153 As in the gut, these proteins could mediate single strain stability by preventing cutaneous engraftment of competing strains. However, the prevalence of T7SS and the specificity of effectors among strains is unknown.
The inhibitory effect of S. aureus against corynebacteria like C. diphtheriae was attributed to an antibiotic peptide isolated in 1947,130 one of an enormous class of peptides called bacteriocins first identified in Escherichia coli in the 1920s.154 While bacteriocins have been described in all lineages of prokaryotes,155 Gram-positive bacteria show bacteriocin-specific regulation and encode dedicated transport machinery that prevents self-intoxication.156 The toxicity of bacteriocins is generally restricted to other Gram-positive bacteria,157 with narrow activity often limited to cells of the same genus, species, or strain, although bacteriocins with broad activity have also been described.158
Colicin from E. coli has been used as a model bacteriocin for investigating microbial community assembly for >50 years.159,160,161 Bacteriocins are predicted to be most relevant for physically structured habitats,162,163 and in models of skin biology, their efficacy depends on tissue context and structure.164 A recent survey of human skin identified many novel bacteriocins with activity against C. acnes, S. epidermidis, and S. aureus.165 Success has been reported in using specific bacteriocin-secreting strains S. epidermidis and Streptococcus hominis to reduce S. aureus colonization of the skin in adult atopic dermatitis patients.136 Such antagonistic relationships show strain and microenvironment specificity. For example, phenol-soluble modulins secreted by a Staphylococcus capitis strain E12 were found to selectively inhibit C. acnes proliferation on the skin surface of mice and pigs,166 while a bacteriocin produced by C. acnes named cutimycin found widely distributed across individuals was associated with exclusion of S. epidermidis from human hair follicles.167 Consistent with their importance in microbial ecology of the skin, like T6SS genes in the gut, bacteriocins are often carried by mobile genetic elements, vary in composition between different strains of the same species, and show extensive history of gene transfer.168 How the sum of these diverse competitive interactions shapes assembly of a durable and long-lasting skin microbiome in early life remains to be understood.
Conclusion
The human microbiome is an attractive therapeutic target in chronic disease. Unlike the genome, it is modifiable. Neonates born by Caesarean section and artificially exposed to vaginal microbiota readily accepted maternal microbes.60 However, artificial inoculation with entire microbial communities carries considerable risks.169 Strain replacement is a superior approach where disease-causing strains, or an inappropriate immune reaction to benign strains, can be identified. Advances in metagenomics and gnotobiotic animals have been used to define minimal consortia of healthy bacteria that resist pathogen invasion of the gut.170 In concert with this approach, benign variants of disease-associated bacteria could be genetically engineered for optimum bacterial antagonism such that engraftment on the skin of the engineered strain would preclude acquisition of pathogenic variants encountered in the environment for durable and long-term health. Gaps remain in understanding which strains are beneficial to the skin, from where they originate, the immunological determinants of successful engraftment, and the molecular mechanisms by which successful colonists exclude competitors. The effect on the skin microbiome of existing interventions, such as bathing and formula feeding, is also poorly understood. The developing microbiome is dynamic and vulnerable, and microbial exposures must be carefully curated. As understanding improves, however, early life may offer a rich opportunity for strain-level engineering of the microbiome.
References
Goodman, A. L. & Gordon, J. I. Our unindicted coconspirators: human metabolism from a microbial perspective. Cell Metab. 12, 111–116 (2010).
Degnan, P. H., Taga, M. E. & Goodman, A. L. Vitamin B12 as a modulator of gut microbial ecology. Cell Metab. 20, 769–778 (2014).
Buffie, C. G. & Pamer, E. G. Microbiota-mediated colonization resistance against intestinal pathogens. Nat. Rev. Immunol. 13, 790–801 (2013).
Kau, A. L., Ahern, P. P., Griffin, N. W., Goodman, A. L. & Gordon, J. I. Human nutrition, the gut microbiome and the immune system. Nature 474, 327–336 (2011).
Thaiss, C. A., Zmora, N., Levy, M. & Elinav, E. The microbiome and innate immunity. Nature 535, 65–74 (2016).
Belkaid, Y. & Naik, S. Compartmentalized and systemic control of tissue immunity by commensals. Nat. Immunol. 14, 646–653 (2013).
Gordon, H. A. & Pesti, L. The gnotobiotic animal as a tool in the study of host microbial relationships. Bacteriol. Rev. 35, 390–429 (1971).
Naik, S. et al. Compartmentalized control of skin immunity by resident commensals. Science 337, 1115–1119 (2012).
Gallo, R. L. Human skin is the largest epithelial surface for interaction with microbes. J. Investig. Dermatol. 137, 1213–1214 (2017).
Nakatsuji, T. et al. The microbiome extends to subepidermal compartments of normal skin. Nat. Commun. 4, 1431 (2013).
Ross, A. A., Müller, K. M., Weese, J. S. & Neufeld, J. D. Comprehensive skin microbiome analysis reveals the uniqueness of human skin and evidence for phylosymbiosis within the class Mammalia. Proc. Natl Acad. Sci. USA 115, E5786–E5795 (2018).
Grice, E. A. et al. A diversity profile of the human skin microbiota. Genome Res. 18, 1043–1050 (2008).
Grice, E. A. et al. Topographical and temporal diversity of the human skin microbiome. Science 324, 1190–1192 (2009).
Kong, H. H. & Segre, J. A. The molecular revolution in cutaneous biology: investigating the skin microbiome. J. Investig. Dermatol. 137, e119–e122 (2017).
Costello, E. K. et al. Bacterial community variation in human body habitats across space and time. Science 326, 1694–1697 (2009).
Oh, J. et al. Biogeography and individuality shape function in the human skin metagenome. Nature 514, 59–64 (2014).
Oh, J. et al. Temporal stability of the human skin microbiome. Cell 165, 854–866 (2016).
Paller, A. S. et al. The microbiome in patients with atopic dermatitis. J. Allergy Clin. Immunol. 143, 26–35 (2019).
Fyhrquist, N. et al. Microbe-host interplay in atopic dermatitis and psoriasis. Nat. Commun. 10, 4703 (2019).
Stokholm, J. et al. Maturation of the gut microbiome and risk of asthma in childhood. Nat. Commun. 9, 141 (2018).
Morgan, X. C. et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 13, R79 (2012).
Parekh, P. J., Balart, L. A. & Johnson, D. A. The influence of the gut microbiome on obesity, metabolic syndrome and gastrointestinal disease. Clin. Transl. Gastroenterol. 6, e91 (2015).
Kostic, A. D. et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 22, 292–298 (2012).
Vogtmann, E. et al. Colorectal cancer and the human gut microbiome: reproducibility with whole-genome shotgun sequencing. PLoS ONE 11, e0155362 (2016).
Zeller, G. et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol. Syst. Biol. 10, 766 (2014).
Koch, R. Ueber den augenblicklichen stand der bakteriologischen choleradiagnose. Z. Hyg. Infekt. 14, 319–338 (1893).
Ogston, A. Report upon micro-organisms in surgical diseases. Br. Med. J. 1, 369.b2–369.b375 (1881).
Whitfield, A., Sabouraud, R. & MacKenna, R. W. Discussion on acne and seborrhoea, their causation and treatment. Br. Med. J. 2, 286–289 (1912).
Dr, Sabouraud et al. A discussion on the role of cocci in the pathology of the skin. Br. Med. J. 2, 794–797 (1901).
Marples, R. R., Richardson, J. F. & Newton, F. E. Staphylococci as part of the normal flora of human skin. Soc. Appl. Bacteriol. Symp. Ser. 19, 93S–99S (1990).
Schade, H. & Marchionini, A. Der säuremantel der haut (nach gaskettenmessungen). Klin. Wochenschr. 7, 12–14 (1928).
Pillsbury, D. M. & Rebell, G. The bacterial flora of the skin; factors influencing the growth of resident and transient organisms. J. Investig. Dermatol. 18, 173–186 (1952).
Marples, R. R. in Skin Bacteria and their Role in Infection (eds Maibach, H. I. & Hildick-Smith, G.) 33–41 (McGraw-Hill, New York, 1965).
Cunliffe, W. J. et al. Tetracycline and acne vulgaris: a clinical and laboratory investigation. Br. Med. J. 4, 332–335 (1973).
Holland, K. T., Cunliffe, W. J. & Roberts, C. D. Acne vulgaris: an investigation into the number of anaerobic diphtheroids and members of the Micrococcaceae in normal and acne skin. Br. J. Dermatol. 96, 623–626 (1977).
Puhvel, S. M. & Amirian, D. A. Bacterial flora of comedones. Br. J. Dermatol. 101, 543–548 (1979).
Holland, K. T., Ingham, E. & Cunliffe, W. J. A review, the microbiology of acne. J. Appl. Bacteriol. 51, 195–215 (1981).
Planet, P. J., Parker, D., Ruff, N. L. & Shinefield, H. R. Revisiting bacterial interference in the age of methicillin-resistant Staphylococcus aureus: insights into Staphylococcus aureus carriage, pathogenicity and potential control. Pediatr. Infect. Dis. J. 38, 958–966 (2019).
Gibbons, S. M. et al. Ecological succession and viability of human-associated microbiota on restroom surfaces. Appl. Environ. Microbiol. 81, 765–773 (2015).
Hogan, P. G. et al. Interplay of personal, pet, and environmental colonization in households affected by community-associated methicillin-resistant Staphylococcus aureus. J. Infect. 78, 200–207 (2019).
Lax, S. et al. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science 345, 1048–1052 (2014).
Miller, M. et al. Staphylococcus aureus in the community: colonization versus infection. PLoS ONE 4, e6708 (2009).
Mork, R. L. et al. Comprehensive modeling reveals proximity, seasonality, and hygiene practices as key determinants of MRSA colonization in exposed households. Pediatr. Res. 84, 668–676 (2018).
Mork, R. L. et al. Longitudinal, strain-specific Staphylococcus aureus introduction and transmission events in households of children with community-associated meticillin-resistant S. aureus skin and soft tissue infection: a prospective cohort study. Lancet Infect. Dis. 20, 188–198 (2019).
Fukami, T. Historical contingency in community assembly: integrating niches, species pools, and priority effects. Annu. Rev. Ecol. Evol. Syst. 46, 1–23 (2015).
Sprockett, D., Fukami, T. & Relman, D. A. Role of priority effects in the early-life assembly of the gut microbiota. Nat. Rev. Gastroenterol. Hepatol. 15, 197–205 (2018).
Hecht, A. L. et al. Strain competition restricts colonization of an enteric pathogen and prevents colitis. EMBO Rep. 17, 1281–1291 (2016).
Aagaard, K. et al. The placenta harbors a unique microbiome. Sci. Transl. Med. 6, 237ra65 (2014).
Collado, M. C., Rautava, S., Aakko, J., Isolauri, E. & Salminen, S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci. Rep. 6, 23129 (2016).
Perez-Muñoz, M. E., Arrieta, M.-C., Ramer-Tait, A. E. & Walter, J. A critical assessment of the “sterile womb” and “in utero colonization” hypotheses: implications for research on the pioneer infant microbiome. Microbiome 5, 48 (2017).
Rodríguez, J. M. et al. The composition of the gut microbiota throughout life, with an emphasis on early life. Microb. Ecol. Health Dis. 26, 26050 (2015).
Tapiainen, T. et al. Maternal influence on the fetal microbiome in a population-based study of the first-pass meconium. Pediatr. Res. 84, 371–379 (2018).
Younge, N. et al. Fetal exposure to the maternal microbiota in humans and mice. JCI Insight 4, e127806 (2019).
de Goffau, M. C. et al. Human placenta has no microbiome but can contain potential pathogens. Nature 572, 329–334 (2019).
Theis, K. R. et al. Does the human placenta delivered at term have a microbiota? Results of cultivation, quantitative real-time PCR, 16S rRNA gene sequencing, and metagenomics. Am. J. Obstet. Gynecol. 220, 267.e1–267.e39 (2019).
Chu, D. M. et al. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat. Med. 23, 314–326 (2017).
Dominguez-Bello, M. G. et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl Acad. Sci. USA 107, 11971–11975 (2010).
Olm, M. R. et al. Identical bacterial populations colonize premature infant gut, skin, and oral microbiomes and exhibit different in situ growth rates. Genome Res. 27, 601–612 (2017).
Sarkany, I. & Gaylarde, C. C. Skin flora of the newborn. Lancet 1, 589–590 (1967).
Dominguez-Bello, M. G. et al. Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nat. Med. 22, 250–253 (2016).
Nishijima, K., Yoneda, M., Hirai, T., Takakuwa, K. & Enomoto, T. Biology of the vernix caseosa: a review. J. Obstet. Gynaecol. Res. 45, 2145–2149 (2019).
Bergström, A., Byaruhanga, R. & Okong, P. The impact of newborn bathing on the prevalence of neonatal hypothermia in Uganda: a randomized, controlled trial. Acta Paediatr. 94, 1462–1467 (2005).
Sarkany, I. & Gaylarde, C. C. Bacterial colonisation of the skin of the newborn. J. Pathol. Bacteriol. 95, 115–122 (1968).
Medves, J. M. & O’Brien, B. Does bathing newborns remove potentially harmful pathogens from the skin? Birth 28, 161–165 (2001).
Visscher, M. O. et al. Vernix caseosa in neonatal adaptation. J. Perinatol. 25, 440–446 (2005).
Lund, C. Bathing and beyond: current bathing controversies for newborn infants. Adv. Neonatal Care 16(Suppl 5S), S13–S20 (2016).
Lozupone, C. A., Stombaugh, J. I., Gordon, J. I., Jansson, J. K. & Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230 (2012).
Verster, A. J. et al. The landscape of type VI secretion across human gut microbiomes reveals its role in community composition. Cell Host Microbe 22, 411.e4–419.e4 (2017).
Yatsunenko, T. et al. Human gut microbiome viewed across age and geography. Nature 486, 222–227 (2012).
Capone, K. A., Dowd, S. E., Stamatas, G. N. & Nikolovski, J. Diversity of the human skin microbiome early in life. J. Investig. Dermatol. 131, 2026–2032 (2011).
Costello, E. K., Carlisle, E. M., Bik, E. M., Morowitz, M. J. & Relman, D. A. Microbiome assembly across multiple body sites in low-birthweight infants. mBio 4, e00782–00713 (2013).
Younge, N. E., Araújo-Pérez, F., Brandon, D. & Seed, P. C. Early-life skin microbiota in hospitalized preterm and full-term infants. Microbiome 6, 98 (2018).
Kennedy, E. A. et al. Skin microbiome before development of atopic dermatitis: early colonization with commensal staphylococci at 2 months is associated with a lower risk of atopic dermatitis at 1 year. J. Allergy Clin. Immunol. 139, 166–172 (2017).
Asnicar, F. et al. Studying vertical microbiome transmission from mothers to infants by strain-level metagenomic profiling. mSystems https://doi.org/10.1128/mSystems.00164-16 (2017).
Korpela, K. et al. Selective maternal seeding and environment shape the human gut microbiome. Genome Res. 28, 561–568 (2018).
Zhu, T. et al. Age and mothers: potent influences of children’s skin microbiota. J. Investig. Dermatol. 139, 2497.e6–2505.e6 (2019).
Faith, J. J. et al. The long-term stability of the human gut microbiota. Science 341, 1237439 (2013).
Ferretti, P. et al. Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe 24, 133.e5–145.e5 (2018).
Yassour, M. et al. Strain-level analysis of mother-to-child bacterial transmission during the first few months of life. Cell Host Microbe 24, 146.e4–154.e4 (2018).
Doege, K. et al. Impact of maternal supplementation with probiotics during pregnancy on atopic eczema in childhood–a meta-analysis. Br. J. Nutr. 107, 1–6 (2012).
Shin, H. et al. The first microbial environment of infants born by C-section: the operating room microbes. Microbiome 3, 59 (2015).
Lax, S. et al. Colonization and succession of hospital-associated microbiota. Sci. Transl. Med. 9, eaah6500 (2017).
Gaitanis, G. et al. Variation of cultured skin microbiota in mothers and their infants during the first year postpartum. Pediatr. Dermatol. 36, 460–465 (2019).
Evans, N. J. & Rutter, N. Development of the epidermis in the newborn. Biol. Neonate 49, 74–80 (1986).
Oranges, T., Dini, V. & Romanelli, M. Skin physiology of the neonate and infant: clinical implications. Adv. Wound Care 4, 587–595 (2015).
Hoeger, P. H. & Enzmann, C. C. Skin physiology of the neonate and young infant: a prospective study of functional skin parameters during early infancy. Pediatr. Dermatol. 19, 256–262 (2002).
Saijo, S. & Tagami, H. Dry skin of newborn infants: functional analysis of the stratum corneum. Pediatr. Dermatol. 8, 155–159 (1991).
Webster, G. F., Ruggieri, M. R. & McGinley, K. J. Correlation of Propionibacterium acnes populations with the presence of triglycerides on nonhuman skin. Appl. Environ. Microbiol. 41, 1269–1270 (1981).
Agache, P., Blanc, D., Barrand, C. & Laurent, R. Sebum levels during the first year of life. Br. J. Dermatol. 103, 643–649 (1980).
Oh, J., Conlan, S., Polley, E. C., Segre, J. A. & Kong, H. H. Shifts in human skin and nares microbiota of healthy children and adults. Genome Med. 4, 77 (2012).
Mack, M. C. et al. Development of solar UVR-related pigmentation begins as early as the first summer of life. J. Investig. Dermatol. 130, 2335–2338 (2010).
Nosanchuk, J. D. & Casadevall, A. Impact of melanin on microbial virulence and clinical resistance to antimicrobial compounds. Antimicrob. Agents Chemother. 50, 3519–3528 (2006).
Gallo, R. L. & Nakatsuji, T. Microbial symbiosis with the innate immune defense system of the skin. J. Investig. Dermatol. 131, 1974–1980 (2011).
Lai, Y. & Gallo, R. L. AMPed up immunity: how antimicrobial peptides have multiple roles in immune defense. Trends Immunol. 30, 131–141 (2009).
Gläser, R. et al. Antimicrobial psoriasin (S100A7) protects human skin from Escherichia coli infection. Nat. Immunol. 6, 57–64 (2005).
Schauber, J. et al. Injury enhances TLR2 function and antimicrobial peptide expression through a vitamin D-dependent mechanism. J. Clin. Investig. 117, 803–811 (2007).
Tamoutounour, S. et al. Keratinocyte-intrinsic MHCII expression controls microbiota-induced Th1 cell responses. Proc. Natl Acad. Sci. USA 116, 23643–23652 (2019).
Thiemann, S. et al. Enhancement of IFNγ production by distinct commensals ameliorates Salmonella-induced disease. Cell Host Microbe 21, 682.e5–694.e5 (2017).
Callewaert, C. et al. IL-4Rα blockade by dupilumab decreases Staphylococcus aureus colonization and increases microbial diversity in atopic dermatitis. J. Investig. Dermatol. 140, 191.e7–202.e7 (2020).
Simpson, E. L. et al. Two phase 3 trials of dupilumab versus placebo in atopic dermatitis. N. Engl. J. Med. 375, 2335–2348 (2016).
Kobayashi, T. et al. Homeostatic control of sebaceous glands by innate lymphoid cells regulates commensal bacteria equilibrium. Cell 176, 982.e16–997.e16 (2019).
Davey, M. S. et al. Clonal selection in the human Vδ1 T cell repertoire indicates γδ TCR-dependent adaptive immune surveillance. Nat. Commun. 8, 14760 (2017).
Kabelitz, D. Function and specificity of human gamma/delta-positive T cells. Crit. Rev. Immunol. 11, 281–303 (1992).
Sharp, L. L., Jameson, J. M., Cauvi, G. & Havran, W. L. Dendritic epidermal T cells regulate skin homeostasis through local production of insulin-like growth factor 1. Nat. Immunol. 6, 73–79 (2005).
Toulon, A. et al. A role for human skin-resident T cells in wound healing. J. Exp. Med. 206, 743–750 (2009).
Selvanantham, T. et al. NKT cell-deficient mice harbor an altered microbiota that fuels intestinal inflammation during chemically induced colitis. J. Immunol. 197, 4464–4472 (2016).
Doisne, J.-M. et al. Skin and peripheral lymph node invariant NKT cells are mainly retinoic acid receptor-related orphan receptor (gamma)t+ and respond preferentially under inflammatory conditions. J. Immunol. 183, 2142–2149 (2009).
Ito, T. et al. Maintenance of hair follicle immune privilege is linked to prevention of NK cell attack. J. Investig. Dermatol. 128, 1196–1206 (2008).
Gold, M. C. et al. Human thymic MR1-restricted MAIT cells are innate pathogen-reactive effectors that adapt following thymic egress. Mucosal Immunol. 6, 35–44 (2013).
Le Bourhis, L. et al. Antimicrobial activity of mucosal-associated invariant T cells. Nat. Immunol. 11, 701–708 (2010).
Constantinides, M. G. et al. MAIT cells are imprinted by the microbiota in early life and promote tissue repair. Science 366, eaax6624 (2019).
Hinks, T. S. C. et al. Activation and in vivo evolution of the mait cell transcriptome in mice and humans reveals tissue repair functionality. Cell Rep. 28, 3249.e5–3262.e5 (2019).
Leng, T. et al. TCR and inflammatory signals tune human MAIT cells to exert specific tissue repair and effector functions. Cell Rep. 28, 3077.e5–3091.e5 (2019).
Cordoro, K. M. et al. Skin-infiltrating, interleukin-22-producing T cells differentiate pediatric psoriasis from adult psoriasis. J. Am. Acad. Dermatol. 77, 417–424 (2017).
Scharschmidt, T. C. et al. A wave of regulatory T cells into neonatal skin mediates tolerance to commensal microbes. Immunity 43, 1011–1021 (2015).
Scharschmidt, T. C. et al. Commensal microbes and hair follicle morphogenesis coordinately drive treg migration into neonatal skin. Cell Host Microbe 21, 467.e5–477.e5 (2017).
Russler-Germain, E. V., Rengarajan, S. & Hsieh, C.-S. Antigen-specific regulatory T cell responses to intestinal microbiota. Mucosal Immunol. 10, 1375–1386 (2017).
Donaldson, G. P. et al. Gut microbiota utilize immunoglobulin A for mucosal colonization. Science 360, 795–800 (2018).
Lee, S. M. et al. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature 501, 426–429 (2013).
Mazmanian, S. K., Liu, C. H., Tzianabos, A. O. & Kasper, D. L. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122, 107–118 (2005).
Mehta, R. S. et al. Stability of the human faecal microbiome in a cohort of adult men. Nat. Microbiol. 3, 347–355 (2018).
Truong, D. T., Tett, A., Pasolli, E., Huttenhower, C. & Segata, N. Microbial strain-level population structure and genetic diversity from metagenomes. Genome Res. 27, 626–638 (2017).
Torow, N. & Hornef, M. W. The neonatal window of opportunity: setting the stage for life-long host-microbial interaction and immune homeostasis. J. Immunol. 198, 557–563 (2017).
Florey, H. W. The use of micro-organisms for therapeutic purposes. Yale J. Biol. Med. 19, 101–117 (1946).
Henderson, D. W. Bacterial interference. Bacteriol. Rev. 24, 167–176 (1960).
Sprunt, K. & Redman, W. Evidence suggesting importance of role of interbacterial inhibition in maintaining balance of normal flora. Ann. Intern. Med. 68, 579–590 (1968).
Schiotz, A. Uskadeliggorelse af infektionsbaerere ved difteri. Ugeskr. Læg. 71, 1382–1384 (1909).
Books received. J. Am. Med. Assoc. LIV, 422–422 (1910).
Albert, H. The treatment of diphtheria carriers. J. Am. Med. Assoc. 61, 1027–1031 (1913).
Jennings, M. A. & Sharp, A. E. Antibacterial activity of the Staphylococcus. Nature 159, 133 (1947).
Lorenz, W. F. & Ravenel, M. P. The treatment of diphtheria-carriers by overriding with Staphylococcus aureus. J. Am. Med. Assoc. LIX, 690–693 (1912).
Womer, W. A. Results of staphylococcus spray treatment in forty-two cases of diphtheria carriers. J. Am. Med. Assoc. 61, 2293–2294 (1913).
Panigrahi, P. et al. Long-term colonization of a Lactobacillus plantarum synbiotic preparation in the neonatal gut. J. Pediatr. Gastroenterol. Nutr. 47, 45–53 (2008).
Panigrahi, P. et al. A randomized synbiotic trial to prevent sepsis among infants in rural India. Nature 548, 407–412 (2017).
Myles, I. A. et al. First-in-human topical microbiome transplantation with Roseomonas mucosa for atopic dermatitis. JCI Insight 3, e120608 (2018).
Nakatsuji, T. et al. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci. Transl. Med. 9, eaah4680 (2017).
Blair, J. E. & Carr, M. Staphylococci in hospital-acquired infections; types encountered in the United States. J. Am. Med. Assoc. 166, 1192–1196 (1958).
Blair, J. E. & Carr, M. Distribution of phage groups of Staphylococcus aureus in the years 1927 through 1947. Science 132, 1247–1248 (1960).
Rountree, P. M. & Freeman, B. M. Infections caused by a particular phage type of Staphylococcus aureus. Med. J. Aust. 42, 157–161 (1955).
Boris, M. et al. Bacterial interference: its effect on nursery-acquired infection with Staphylococcus aureus. IV. The Louisiana epidemic. Am. J. Dis. Child. 105, 674–682 (1963).
Shinefield, H. R., Ribble, J. C., Boris, M. & Eichenwald, H. F. Bacterial interference: its effect on nursery-acquired infection with Staphylococcus aureus. I. Preliminary observations on artificial colonzation of newborns. Am. J. Dis. Child. 105, 646–654 (1963).
Shinefield, H. R., Sutherland, J. M., Ribble, J. C. & Eichenwald, H. F. Bacterial interference: its effect on nursery-acquired infection with Staphylococcus aureus. II. The Ohio epidemic. Am. J. Dis. Child. 105, 655–662 (1963).
Shinefield, H. R., Boris, M., Ribble, J. C., Cale, E. F. & Eichenwald, H. F. Bacterial interference: its effect on nursery-acquired infection with Staphylococcus aureus. III. The Georgia epidemic. Am. J. Dis. Child. 105, 663–673 (1963).
Shinefield, H. R., Ribble, J. C., Eichenwald, H. F., Boris, M. & Sutherland, J. M. Bacterial interference: its effect on nursery-acquired infection with Staphylococcus aureus. V. An analysis and interpretation. Am. J. Dis. Child. 105, 683–688 (1963).
Light, I. J., Sutherland, J. M. & Schott, J. E. Control of a staphylococcal outbreak in a nursery: use of bacterial interference. JAMA 193, 699–704 (1965).
Russell, A. B. et al. Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475, 343–347 (2011).
Russell, A. B. et al. A type VI secretion-related pathway in Bacteroidetes mediates interbacterial antagonism. Cell Host Microbe 16, 227–236 (2014).
Wexler, A. G. et al. Human symbionts inject and neutralize antibacterial toxins to persist in the gut. Proc. Natl Acad. Sci. USA 113, 3639–3644 (2016).
Ross, B. D. et al. Human gut bacteria contain acquired interbacterial defence systems. Nature 575, 224–228 (2019).
Burts, M. L., DeDent, A. C. & Missiakas, D. M. EsaC substrate for the ESAT-6 secretion pathway and its role in persistent infections of Staphylococcus aureus. Mol. Microbiol. 69, 736–746 (2008).
Christensen, G. J. M. et al. Antagonism between Staphylococcus epidermidis and Propionibacterium acnes and its genomic basis. BMC Genomics 17, 152 (2016).
Ohr, R. J., Anderson, M., Shi, M., Schneewind, O. & Missiakas, D. EssD, a nuclease effector of the Staphylococcus aureus ESS pathway. J. Bacteriol. 199, e00528-16 (2017).
Whitney, J. C. et al. A broadly distributed toxin family mediates contact-dependent antagonism between gram-positive bacteria. eLife 6, e26938 (2017).
Gillor, O., Etzion, A. & Riley, M. A. The dual role of bacteriocins as anti- and probiotics. Appl. Microbiol. Biotechnol. 81, 591–606 (2008).
Klaenhammer, T. R. Bacteriocins of lactic acid bacteria. Biochimie 70, 337–349 (1988).
Nes, I. F. et al. Biosynthesis of bacteriocins in lactic acid bacteria. Antonie Van Leeuwenhoek 70, 113–128 (1996).
Riley, M. A. & Wertz, J. E. Bacteriocin diversity: ecological and evolutionary perspectives. Biochimie 84, 357–364 (2002).
Mota-Meira, M., LaPointe, G., Lacroix, C. & Lavoie, M. C. MICs of mutacin B-Ny266, nisin A, vancomycin, and oxacillin against bacterial pathogens. Antimicrob. Agents Chemother. 44, 24–29 (2000).
Ikari, N. S., Kenton, D. M. & Young, V. M. Interaction in the germfree mouse intestine of colicinogenic and colicin-sensitive microorganisms. Proc. Soc. Exp. Biol. Med. 130, 1280–1284 (1969).
Riley, M. A. & Gordon, D. M. The ecological role of bacteriocins in bacterial competition. Trends Microbiol. 7, 129–133 (1999).
Sassone-Corsi, M. et al. Microcins mediate competition among Enterobacteriaceae in the inflamed gut. Nature 540, 280–283 (2016).
Chao, L. & Levin, B. R. Structured habitats and the evolution of anticompetitor toxins in bacteria. Proc. Natl Acad. Sci. USA 78, 6324–6328 (1981).
Durrett, R. & Levin, S. Allelopathy in spatially distributed populations. J. Theor. Biol. 185, 165–171 (1997).
Noble, W. C. & Willie, J. A. Interactions between antibiotic-producing and non-producing staphylococci in skin surface and sub-surface models. Br. J. Exp. Pathol. 61, 339–343 (1980).
O’Sullivan, J. N., Rea, M. C., O’Connor, P. M., Hill, C. & Ross, R. P. Human skin microbiota is a rich source of bacteriocin-producing staphylococci that kill human pathogens. FEMS Microbiol. Ecol. 95, fiy241 (2019).
O’Neill, A. M. et al. Identification of a human skin commensal bacterium that selectively kills Cutibacterium acnes. J. Investig. Dermatol. 140, 1619.e2–1628.e2 (2020).
Claesen, J. et al. Cutibacterium acnes antibiotic production shapes niche competition in the human skin microbiome. Preprint at https://doi.org/10.1101/594010 (2019).
Janek, D., Zipperer, A., Kulik, A., Krismer, B. & Peschel, A. High frequency and diversity of antimicrobial activities produced by nasal staphylococcus strains against bacterial competitors. PLoS Pathog. 12, e1005812 (2016).
DeFilipp, Z. et al. Drug-resistant E. coli bacteremia transmitted by fecal microbiota transplant. N. Engl. J. Med. 381, 2043–2050 (2019).
Sorbara, M. T. & Pamer, E. G. Interbacterial mechanisms of colonization resistance and the strategies pathogens use to overcome them. Mucosal Immunol. 12, 1–9 (2019).
Acknowledgements
B.W.C. is supported by the National Institute of Allergy and Infectious Diseases (5 F30 AI126791).
Author information
Authors and Affiliations
Contributions
B.W.C. drafted the article, revised the article critically for important intellectual content, and approved the final version. A.S.P. revised the article critically for important intellectual content and approved the final version.
Corresponding author
Ethics declarations
Competing interests
B.W.C.: no competing interests. A.S.P.: consultant with honorarium for AbbVie, Amgen, Celgene, Dermavant, Dermira, Galderma, Eli Lilly, Forte, LEO Pharma Inc., Menlo, Novartis, Pfizer, Regeneron, and Sanofi-Genzyme.
Patient consent
None required.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Casterline, B.W., Paller, A.S. Early development of the skin microbiome: therapeutic opportunities. Pediatr Res 90, 731–737 (2021). https://doi.org/10.1038/s41390-020-01146-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41390-020-01146-2
This article is cited by
-
The role of Staphylococcus aureus quorum sensing in cutaneous and systemic infections
Inflammation and Regeneration (2024)
-
A genome catalog of the early-life human skin microbiome
Genome Biology (2023)
-
Mechanisms of microbe-immune system dialogue within the skin
Genes & Immunity (2021)