Abstract
Background
Human induced pluripotent stem cells (iPSCs) are a promising tool to investigate pathogenic mechanisms underlying human genetic conditions, such as congenital anomalies of the kidney and urinary tract (CAKUT). Currently, iPSC-based research in pediatrics is limited by the invasiveness of cell collection.
Methods
Urine cells (UCs) were isolated from pediatric urine specimens, including bag collections, and reprogrammed using episomal vectors into urinary iPSCs (UiPSCs). Following iPSC-quality assessment, human kidney organoids were generated.
Results
UCs were isolated from 71% (12/17) of single, remnant urine samples obtained in an outpatient setting (patients 1 month–17 years, volumes 10–75 ml). Three independent UCs were reprogrammed to UiPSCs with early episome loss, confirmed pluripotency and normal karyotyping. Subsequently, these UiPSCs were successfully differentiated into kidney organoids, closely resembling organoids generated from control fibroblast-derived iPSCs. Importantly, under research conditions with immediate sample processing, UC isolation was successful 100% for target pediatric CAKUT patients and controls (11/11) after at most two urine collections.
Conclusions
Urine in small volumes or collected in bags is a reliable source for reprogrammable somatic cells that can be utilized to generate kidney organoids. This constitutes an attractive approach for patient-specific iPSC research involving infants and children with wide applicability and a low threshold for participation.
Similar content being viewed by others
Introduction
Reprogramming of terminally differentiated somatic cells into induced pluripotent stem cells (iPSCs)1,2 has provided the biomedical research community with an important new tool for mechanistic and translational research. iPSCs provide a virtually unlimited supply of human cells that can be re-differentiated into a multitude of cell types with potential therapeutic use. iPSCs can also be used to generate disease-specific stem cells that, in combination with the growing arsenal of protocols to generate different types of organoids, can be used to model a broad spectrum of human conditions.3,4 The latter holds the potential to play an important complementary role in disease modeling limited by interspecies differences. In the context of modeling human genetic diseases, iPSC-based research depends on gene-editing tools (e.g., clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein 9 (Cas9)) either for the generation of mutated control iPSC lines or for the correction of (putative) pathogenic mutations in patient-derived iPSCs.5 The latter, which allows for the analysis of mutations in the genetic background of actual patients, is of particular interest for modeling of genetic diseases with variable penetrance/expressivity.
Congenital anomalies of the kidney and urinary tract (CAKUT), the leading cause of childhood renal failure worldwide, display a high degree of phenotypic variability.6 With the introduction of iPSC-derived kidney organoids7,8,9 and the validation of their use as a tool in CAKUT research with CRISPR/Cas9-mutated control iPSCs,10 there will be a demand for the generation of patient-specific CAKUT iPSCs. Owing to the high proportion of infants and young children among CAKUT patients with a confirmed genetic diagnosis, a reliable and acceptable somatic cell source is required to ensure both successful cell isolation and willingness of patients or caregivers to consent to participation.
Commonly used iPSC-generating protocols are based on the invasive collection of somatic cells, including skin fibroblasts and peripheral blood cells,1,2,11,12,13 which raises the threshold for research study participation. In contrast, urinary cells (UCs) can be obtained non-invasively, and these cells, which were shown to be of epithelial origin by a combination of immunofluorescence imaging and gene expression analysis, have previously been shown to be reprogrammable.14 Patient-specific, UC-derived iPSCs (UiPSCs) have been differentiated into several cell types, including cardiomyocytes,15 primary neurons,16 hepatocyte-like cells,17 and retinal organoids.18 Although these studies involved pediatric patients, the feasibility of using urine as a somatic cell source for iPSC-based research projects involving infants and children remains to be demonstrated. In this age group, urine collection itself may be more challenging as patients are often not toilet-trained and alternative collection methods, such as urine bags, are notoriously prone to microbial contamination. Furthermore, patients may not provide quantities of urine required to generate UC cultures and/or may not be conducive to measures, including increasing fluid intake prior to urine collection, aimed at enhancing the success rate.
Here we determined the feasibility of using urine as a reliable source of reprogrammable cells to generate UiPSCs from infants and children and subsequently generate kidney organoids. Results demonstrate that UCs could be isolated from 71% of remnant urine samples collected routinely in outpatient Pediatric Nephrology clinic, which would otherwise have been discarded. Three UCs, obtained from small-volume, bag-collected infant urine specimens, were successfully reprogrammed and kidney organoids were generated with characteristics similar to those generated from fibroblast-derived control iPSC. Moreover, under research conditions with immediate urine sample processing, UC isolation was successful in 100% of target patients while requiring at most two urine collections.
Methods
Study population
Phase 1: participants were recruited from an outpatient clinic dedicated to patients with urinary tract malformations and/or urinary tract infections (UTIs). Patients with known genetic mutations or suspicion of concurrent UTI based on history, physical exam, and urinalysis, were excluded. Urine specimens were obtained in routine fashion using either urine collection bags for non-toilet-trained children or by parentally supervised collection of mid-stream urine (MSU). Timing of the latter relative to completion of clinic visit was not documented. Urine was not immediately cooled. After completion of their clinic visit, eligible patients and/or caregivers were approached by investigators and informed consent was obtained to use remnant urine.
Phase 2: patients and first-degree relatives with known mutations in either CAKUT-causing genes (WT1, HNF1β, GLI3) or genes encoding structural components of the nephron (COL4A3) were approached by investigators during regular clinic visits and asked to donate fresh urine specimens. Informed consent was obtained.
All procedures were performed in accordance with the protocol approved by the Hospital for Sick Children Research Ethics Board. For Phase 1, this required sample anonymization and limited data collection at recruitment to age, sex, specimen volume, and collection method.
UC isolation
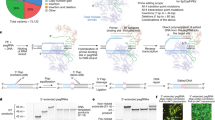
For isolation of UCs, the protocol of Zhou et al.19 was adapted (Fig. 1a). On Day 0, urine was placed on ice upon obtaining consent (Phase 1) or immediately upon collection (Phase 2) and processed within 1 h. Urine was centrifuged at 400 × g for 10 min at room temperature. Cell pellets (with 0.5–1 ml remnant supernatant) were washed with 10 ml Hank’s buffered saline without magnesium/calcium (HBSS, Gibco, Gaithersburg, MD) with penicillin/streptomycin (Gibco) and re-spun at 200 × g for 10 min. After careful aspiration, pellets (0.2–0.3 ml remnant buffer) were resuspended in 1 ml primary UC medium (PM: Dulbecco’s modified Eagle’s medium (DMEM)/F12 medium with 10% (vol/vol) fetal calf serum (FCS; Gibco), penicillin/streptomycin, Normocin (InvivoGen, San Diego, CA), and SingleQuot Kit (Lonza, Basel, Switzerland)) and transferred to a 0.1% gelatin-coated well (Millipore, Burlington, MA) of a 12-well culture plate and incubated under standard cell culture conditions. On Days 1–3, new PM was added to the seeded cells (1 ml/day) without replacing spent medium. On Day 4, PM (total volume 4 ml) was replaced with 1 ml renal epithelial proliferation medium (REPM (Lonza) with SingleQuot kit, penicillin/streptomycin, and Normocin), which was replaced 50% every 48 h. Once single colonies were approximately 2–4 mm in diameter (1–2 mm for wells with multiple colonies) or colonies ceased to expand, UCs were washed with HBSS and passaged 1:1 using TrypLE (Gibco, 8–10 min, followed by inactivation with two volumes of neutralization medium (NM: DMEM:F12 supplemented with 10% (vol/vol) FCS)) into a new single well of a 12-well culture plate (gelatin coated as all vessels were for culturing UCs). As passaged UCs were spread evenly over the well, timing of next passages for expansion (subsequently to a single 6-well plate well and a 10-cm culture dish (all gelatin coated)) was based on reaching 70–90% confluence. In case of minute, initial colonies, UCs were passaged 1:1 back into a single well of a 24-well-plate first. Once 75–90% confluence in 10-cm dishes was reached (approximately 1 × 106 cells), UC isolation was considered successful. UCs were frozen and stored on liquid nitrogen until reprogramming was performed. Mycoplasma-negative status of UCs was confirmed by routinely performed PCR test.
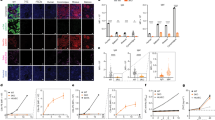
Urine cell isolation and reprogramming. a Schematic of UC isolation and UiPSC generation protocols: Gray arrows indicate cell passages; PM primary urine cell medium, R REPM, N2B27 N2B27/bFGF-medium; b Proliferation of urine cells during days 3–10 of initial culture. (Rectangle shows further magnification.) Debris, as seen in upper part of Day 3 image, is typically present until the first medium change (Day 4) (scale bar = 1 mm). c, d Representative images of UiPSC colonies surrounded by confluent non-reprogrammed UCs (c) and after selective passaging (d) (scale bars = 300 μm); e PCR check for episomal reprogramming vectors at passage 5 (passage 4 for clone #8): Gray arrows indicate episome-free clones; UC urine cells, neg negative control (Sendai-virus reprogrammed iPSCs), Pos positive control (whole-cell lysate of early passage Epi5-reprogrammed adult urine cells); 2× Pos double quantity of positive control
UC reprogramming
UCs were reprogrammed to UiPSCs using a non-integrating, episomal reprogramming kit (Epi5, Invitrogen; Fig. 1a). Thawed UCs were cultured in a gelatin-coated 10-cm dish until 75–90% confluence at Day 0. For the last 24 h prior to transfection, UCs were cultured in REPM without penicillin/streptomycin or Normocin.
Day 0: UCs were washed, dissociated, and centrifuged as described for passaging. The resulting pellet was resuspended in 120 μl Buffer R with Epi5 vectors (1.0–1.5 μl each). UCs were transfected using the Neon system (Invitrogen, 100 μl tip, pulse voltage 1400 V, pulse width 20 ms, two pulses). Transfected cells were transferred to 10 ml REPM. An additional 100 μl Buffer R was added to the remnant cell suspension; again 100 μl suspension was transfected and added to the 10 ml REPM cell suspension. This suspension was transferred to a Geltrex-coated 10-cm dish (Gibco) and incubated for 24 h. On Day 1, medium was switched to N2B27/basic fibroblast growth factor (bFGF) medium (DMEM/F12 w/ HEPES, containing (final concentrations) N2 supplement (1×), B27 supplement (1×), MEM-non-essential amino acids (MEM-NEAA) (1×), GlutaMAX-1 (1/2×), β-mercaptoethanol (55 μM) (all Gibco) and bFGF (Peprotech, Rocky Hill, NJ, 100 ng/ml). N2B27/bFGF medium was replaced 50% every 48 h. On Day 9, medium was switched to Nutristem hPSC XF medium (Corning, New York, NY), which was replaced 50% every 48 h. By Days 21–23, UiPSC colonies were manually picked and transferred to Matrigel-coated (Corning, batch-specific recommended concentration) 24-well culture plates with mTeSR-1 medium (StemCell Technologies, Vancouver, Canada). Medium was fully replaced daily with at most one double-feeding per week. Proliferating UiPSC colonies were passaged using ReLeSR (StemCell Technologies, 5 min). Once UiPSC clones were cultured in 6-well plates, cells were passaged weekly (1:10–1:15).
UiPSC characterization
Loss of episomal reprogramming vectors
UiPSC DNA was isolated using DNeasy (Qiagen, Hilden, Germany). PCR (Platinum Taq, Invitrogen) was performed using manufacturer-supplied primer sequences for OriP and EBNA1 (Invitrogen).
Pluripotency
UiPSC pluripotency was confirmed by: (1) Expression analysis of marker genes NANOG, OCT4, SOX2, and TERT using quantitative reverse transcriptase–PCR (QRT-PCR). (2) Immunostaining of UiPSC colonies probing for the pluripotency marker proteins NANOG, OCT4, SSEA-4, and TRA1–60. (3) Embryoid body (EB) spontaneous differentiation assays. For the latter, nearly confluent UiPSC colonies in 6-well plates were treated with collagenase (1 ml Knock-Out DMEM (Gibco) with 2 mg/ml collagenase type IV (Gibco, 17104–019)) on Day 0. After complete detachment (30–45 min at 37 °C), EB-KnockOut (EB-KO, 1 ml) medium was added (250 ml KnockOut DMEM with 45 ml KnockOut serum replacement (Gibco), MEM-NEAA (1×), GlutaMAX (1×), penicillin/streptomycin (1×), and 540 μl β-mercaptoethanol (55 mM stock)). Cell suspensions were transferred to 15 ml conical to precipitate. Supernatant was aspirated, and cells were washed once with EB-KO and transferred to low-adherence plates (Corning). EB-KO was replaced every 48 h. On Day 8, EBs were transferred to a 0.1% gelatin-coated 24-well plate (4–8 EBs/well) containing EB-FCS medium (components identical to EB-KO except with FCS instead of KnockOut serum replacement). EB-FCS medium was fully replaced every 48 h. On Day 16, differentiated cells were fixed and stained for markers of the three germ layers: smooth muscle actin (SMA, mesoderm), α-fetoprotein (AFP, endoderm), and β3-tubulin (b3-Tub, ectoderm).
Euploidy
Karyotyping was performed by the Toronto Center for Applied Genomics (The Hospital for Sick Children).
Generation of human kidney organoids
Kidney organoids were generated according to Takasato et al.20 with several modifications similar to subsequent reports,21,22 i.e., feeder-free iPSC culturing and the use of dissociation agent Accutase (Innovative Cell Technologies, San Diego, CA) and ROCK-inhibitor Y-27632 (Millipore) for iPSC seeding. The fibroblast-derived, episomally reprogrammed iPSC line WTC11 (WTC)23 served as control iPSC line.
On Day −1, iPSCs were dissociated using Accutase and seeded at 7500–12,000 cells/cm2 on Matrigel-coated cell culture plates (Matrigel concentration 1.43× of batch-specific recommendation) in mTeSR-1 with 10 μM Y-27632. On Day 0, directed differentiation was started by changing seeding medium to APEL-2 (StemCell Technologies, containing Antibiotic/Antimycotic (1% v/v, Gibco)) supplemented with 8 μM CHIR99021 (Tocris Bioscience, Bristol, UK). Medium was fully replaced every 48 h, and on Day 5, CHIR medium was switched to APEL-2 supplemented with 200 ng/ml human FGF 9 (hFGF9) (R&D Systems) and 1 μg/ml heparin (Sigma-Aldrich). On Day 7, differentiated cells were dissociated using Trypsin-EDTA (0.05%, Gibco) for 1 min at 37 °C and, after addition of two volumes mTeSR-1, centrifuged at 400 × g for 3 min. Cell pellets were resuspended in APEL-2 medium and aggregated by centrifugation at 500 × g for 3–4 min (500,000 cells per 1.5 ml microcentrifuge tube). Aggregates were transferred to 6-well culture plate Transwell-inserts (0.4-μm pore membrane, Corning) using wide-bore p200 pipette tips. Aggregates were pulsed with APEL-2 with 5 μM CHIR for 1 h. Medium subsequently was switched back to APEL-2 (with previously used concentrations of hFGF9 and heparin). Medium was replaced every 48 h thereafter. From Day 7 + 5 onward, APEL-2 was supplemented with heparin only.
Immunostaining
For UiPSC colonies and EB assays, cells were fixed using 4% paraformaldehyde (in phosphate-buffered saline (PBS)) at room temperature for 10 min, washed (PBS, 3×), permeabilized with 0.1% Triton X-100 (Sigma) in PBS for 10 min, washed, blocked with 5% goat serum (in PBS) for 1 h, and incubated with primary antibodies for 1 h at room temperature. Next, cells were washed (3×) and incubated with secondary antibodies for 45 min in the presence of 4,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich, 1:1000). For kidney organoids, Day 7 + 18 organoids were fixed as described previously20 and sectioned (6 μm) after paraffin embedding. The immunostaining protocol used for organoid sections and that for imaging used was described previously.24 Images were processed using Fiji25 or Volocity (Perkin Elmer, Waltham, MA) for background subtraction and range expansion.
Antibodies
Primary antibodies used for immunostaining are listed in Supplementary Table S1. Secondary antibodies used were Alexa-fluor-labeled species-specific antibodies (ThermoFisher Scientific, dilution 1:500–1000. Fluorescein-labeled Lotus Tetragonolobus Lectin (FL-1321, Vector Laboratories, Burlingame, CA, USA, dilution 1:200) was used to stain proximal tubules and DAPI for nuclei.
Gene expression analysis
Total RNA was isolated using the RNeasy Mini kit (Qiagen) and quantified using Nanodrop 2000C (Thermo Scientific). RNA was reverse transcribed using the SuperScript First-Strand Kit (Invitrogen). QRT-PCR was performed using the ViiA7 and PowerSYBR Green PCR reagent (Applied Biosystems, Foster City, CA). Primer sequences are listed in Supplementary Table S2. Quantitation was performed using the standard curve method. mRNA levels were normalized to ribosomal 18S levels and expressed relative to the reference group as indicated.
Statistics
Categorical data were analyzed using Fisher exact test and gene expression data using Student’s T test using Prism8 (GraphPad, San Diego, CA). A p value < 0.05 was considered significant.
Results
UC isolation from pediatric urine specimens (Phase 1)
Seventeen participants (13 boys, 4 girls) were recruited within our outpatient clinic focused on urinary tract malformations and UTI (Table 1). Ages ranged from 1 month to 17 years. Urine bags were used for all children aged <1 year. Volume of available remnant urine varied from 8 to 75 ml. Adherent UCs were first identified on Days 2–10. For some samples, colonies were easily detected early (Fig. 1b). For others, minute but viable colonies consisting of only a few cells were identified much later, often at the side of the culture wells (Supplementary Fig. S1). These colonies frequently expanded despite an initial impression that isolation of UCs failed. A potential factor contributing to the highly variable time to first UC identification was the presence of more or less debris until the first full medium change (Day 4) (compare Day 3 vs. Day 6 in Fig. 1b). UCs were successfully isolated and cultured from 12 samples (71%) with the lowest urine volume being 10 ml. No statistically significant differences were found in success rates between sexes and collection methods. No evidence of microbial contamination was observed in any culture, including those initiated from bag urine collections. The time between urine collection and obtaining sufficient UCs for reprogramming (≈1 × 106 cells) ranged between 16 and 25 days in Phase 1 requiring a maximum of four passages. UCs, which were routinely frozen at that point, could later be thawed without limitation to subsequent proliferation and reprogramming.
Reprogramming of UCs
Three UC cultures underwent episomal reprogramming. As a “proof-of-principle”, we chose to reprogram UCs from the three smallest urine specimens all from infants (first participant G (labeled PedG), followed by participants A and O). Twenty-three days post-reprogramming, the first PedG UiPSC colonies (Fig. 1c) of a total of 32 were picked. After the first two passages, nine, well-proliferating clones were selected based on typical iPSC morphology and being devoid of non-reprogrammed UCs or differentiated UiPSCs (Fig. 1d). The use of the non-enzymatic dissociation agent ReLeSR greatly aided in selective passaging of UiPSCs and obtaining pure cultures. After passages 4–5 (approximately 60 days after reprogramming), PCR screening for episomal vectors revealed that five clones were episome-free (Fig. 1e). Analysis of previous passages showed that these clones had become episome-free as early as passage 3 (data not shown). To confirm our findings with PedG, we subsequently reprogrammed UCs from participants PedO and PedA. A total of 35 (PedO) and 36 (PedA) colonies were picked after 21–24 days and episome screening was performed after passage 3. For PedO, 4 of the 17 clones were episome-free after this passage and, for PedA, 1 of the 12 clones with a second clone episome-free after passage 4 (data not shown). Taken together, these results demonstrate that UCs from small volume, bag-collected urine specimens can be reprogrammed successfully and that episome-free clones can be identified in an early stage, which allows for early selection of UiPSC clones to expand.
UiPSC characterization
For each UC reprogramming, at least one UiPSC clone was fully characterized. Results for PedA-#16 are shown in Fig. 2a–d. Pluripotency was confirmed using gene expression analysis of pluripotency markers NANOG, OCT4, SOX2, and TERT (Fig. 2a) and immunostaining for NANOG, OCT4, SSEA-4, and TRA1-60 (Fig. 2b). EB assays confirmed that UiPSC clones could spontaneously differentiate into cell types corresponding to all three germ layers: mesoderm (SMA), endoderm (AFP), and ectoderm (b3-Tub) (Fig. 2c). Euploidy of clones was confirmed by karyotyping (Fig. 2d). Results for PedO-#30 and PedG-#8 are shown in Supplementary Fig. S2.
Confirmation of UiPSC pluripotency and euploidy. a Pluripotency marker gene expression (N = 4 replicates, horizontal line denoting mean): UC non-reprogrammed urine cells, WTC control fibroblast-derived iPSCs; b Immunostaining for pluripotency marker proteins (scale bars = 1 mm); c Immunostaining of spontaneously differentiated UiPSC (embryoid body assay): AFP α-fetoprotein, b3-Tub β3-tubulin, SMA smooth muscle actin (scale bars = 200 μm (SMA), 1 mm (AFP, b3-Tub)); d karyogram. (All data from PedA-#16.)
Kidney organoid generation from UiPSCs
To establish the feasibility of using UiPSC for renal development research, we next generated human kidney organoids using the protocol of Takasato et al.20 with minor modifications (Fig. 3a). This two-step protocol consists of a 7-day two-dimensional/monolayer iPSC differentiation phase followed by an 18-day three-dimensional self-reorganization phase during which aggregated differentiated cells are cultured at the air–liquid interface.20 UiPSC differentiation and organoid formation were performed at least twice per line (with 3–8 organoids per differentiation experiment but more (>20) for time course experiments) and compared to that of the fibroblast-derived iPSC line, WTC. As described previously,20,22 UiPSC seeding density (Day −1) had to be optimized for each of the three cell lines prior to successful organoid generation. To confirm proper differentiation in the 7-day monolayer phase, a limited screening of UiPSC gene expression profiles was performed (Fig. 3b). Loss of pluripotency marker NANOG, transient induction of primitive streak marker T, and the eventual induction of posterior intermediate mesoderm marker HOXD11, which is known to be very responsive to 5-day CHIR treatment,9 were observed and these followed patterns similar to those in WTC.
Generation of UiPSC-derived kidney organoids. a Schematic of differentiation protocol: CHIR CHIR99021, F FGF9, Hep heparin; b Gene expression profile during 2D differentiation (N = 3–4 replicates, horizontal line denoting mean); WTC control fibroblast-derived iPSCs; c Brightfield imaging of organoids at Day 7 + 7 (scale bar = 1 mm); Immunofluorescence imaging of organoid sections: d WT1 Wilm’s tumor-1, LTL Lotus Tetragonolobus Lectin, ECAD E-cadherin (scale bar = 200 μm) and e COL4 collagen IV, NPHS1 Nephrin (scale bar = 20μm); f Organoid gene expression at Day 7 + 0, 7 + 11, and 7 + 18 (N = 4–6 individual organoids, horizontal line denoting mean). *p < 0.05, **p < 0.01. (All data from PedA-#16.)
Upon aggregation of differentiated UiPSC, cellular reorganization and organoid development resembled that of differentiated WTC (Fig. 3c). Immunofluorescence imaging of Day 7 + 18 organoid sections showed staining of clusters of podocytes (WT1), proximal tubules (LTL), and (distal) tubules (ECAD) with patent lumina (Fig. 3d and Supplementary Fig. S3). In addition, clusters of WT1+ podocytes are surrounded by Collagen IV (COL4)-positive (basement) membrane and express Neprin (NPHS1) in close proximity to this membrane (Fig. 3e and Supplementary Fig. S3). Analysis of whole organoid gene expression at Days 7 + 0, 7 + 11, and 7 + 18 showed the induction of markers of different nephron segments (NPHS1, SLC3A1, SLC12A1), endothelial cells (PECAM), and stroma (FOXD1) (Fig. 3f). The differences in relative induction levels for SLC3A1 (higher for UiPSC compared to WTC) and PECAM (lower for UiPSC) and the earlier induction of NPHS1 and FOXD1 for UiPSC could be attributed not only to inherent differences between these two different iPSC lines but also to differences in nephron maturation and patterning or changes in relative abundances of renal and non-renal cell types in organoids. The latter were previously shown to account for remarkable interbatch variability in organoids generated from a single iPSC line.26
UC isolation from patients and first-degree relative controls (Phase 2)
The observed success rate of UC isolations in Phase 1 was similar to those reported for previous studies with adults despite the urine collections being subordinate to clinical care (e.g., delay in processing/cooling of specimens, additional handling for routine testing, and use of portions for other clinically indicated testing). However, these outpatient clinic patients could not be considered fully healthy. We could not rule out an effect of this on success rates, either positive or negative. To determine the efficiency of isolating UCs under research conditions, we studied a further five patients with known mutations and one (or two) first-degree relatives as controls (either with or without same mutation) for each of them. This group of patients involved both patients with a known renal phenotype and a patient carrying a mutation in GLI3, a gene involved in renal development.27 Complete urine collections were immediately cooled and processed solely for the purpose of the study within 1 h. For this group (N = 11), the success rate of the first attempt was 82% (9/11) but rose to 100% with a second attempt (Table 2). These results demonstrate that UCs can be isolated from a single urine collection under experimental conditions in the majority of patients and from all patients after a second collection in situations where the first collection failed.
Discussion
Among the types of somatic cells that can be used to generate iPSCs,28 UCs are very attractive with respect to their availability and non-invasive collection, particularly for young children and indispensable healthy related controls (e.g., siblings, parents). In contrast to collection of peripheral blood, which requires skilled phlebotomists, urine collection can be performed by patients, caregivers, and non-medically trained research personnel. Our protocol can be incorporated easily into projects that aim to bank reprogrammable cells for rare diseases that may involve multiple, distant sites. UC isolation is relatively easy and inexpensive. Further, UCs can also be frozen and thus transported safely. We anticipate that this use of urine, which would otherwise be discarded as waste, will not only lower the threshold for their participation but will also lead to fewer concerns raised by institutional research ethics boards (REBs) carefully weighing the benefits of research projects and the burden on participants.
Our study demonstrates that it is feasible to obtain reprogrammable UCs from small volume urine specimens even when collected using urine bags. Previous studies mainly used MSU collections and generally had much larger volumes, ranging from 50 to 1000 ml.15,17,29,30,31,32,33,34 To obtain such large volumes, urine was pooled and/or participants were instructed to drink ample fluids prior to collection.34 The latter is clearly non-practical in young children. The 71% success rate of UC isolation from single, routinely collected specimens during Phase 1 is comparable to those reported for three studies with 35–57 participants19,34,35 but much higher than the rate of 32% in another study with 114 adults.30 Only Xue et al. reported a higher success rate (>90%, including eight children aged <10 years) with urine volumes of 50–200 ml, but it is not clear whether single or multiple urine collections had been performed.33 Only two studies specifically reported using urine volumes <50 ml, but no overall success rate was mentioned.15,36 By using whole urine specimens and immediate processing during Phase 2, we achieved an even higher success rate for single collection (82%). Zhou et al.19 showed previously that repeated collection only increased success rate to a maximum of 82% (N = 57 healthy participants), suggesting that there are participant-specific factors affecting success rates. We, however, found that with a second attempt UCs could be isolated for all patients and controls. It is unclear what causes this discrepancy. Using our single-specimen results (71–82%) as conservative estimates but assuming randomness of success, multiple urine collections would lift the likelihood of successful UC isolation to 91–96% with two and 97–99% with three attempts, which would most likely be acceptable for motivated research participants and caregivers.
The original UiPSC-generation protocol described by Zhou et al. involved retroviral delivery of reprogramming factors and used feeder cells.14 Subsequently, the protocol has been optimized rendering it “footprint-free” using non-integrating reprogramming strategies and “feeder-free.”34,37 Our protocol included both of these features making it suitable for research purposes by avoiding unpredictable genomic changes and practical. All our reprogrammings yielded sufficient numbers of UiPSC clones indicating that the successful UiPSC generation is limited by UC isolation and not by reprogramming efficiency as has been reported previously.34 Although iPSCs require a minimum number of passages to reach genomic stability,38 early detection of episome-free clones will reduce time to obtain usable iPSCs as multiple characterization steps can be performed in parallel, which also reduces costs substantially.
To generate kidney organoids, we used the protocol of Takasato et al.20 We validated the use of this modified protocol using control iPSC lines and determined optimal experimental seeding densities for the individual iPSC lines, similarly to van den Berg et al.22 Kidney organoids generated from UiPSCs closely resembled those obtained from control iPSC lines and those described elsewhere using the Takasato protocol.9,21,22 These results combined confirm that it is not only feasible to generate pediatric patient-specific UiPSCs within a time frame comparable to that reported previously34 but also that kidney organoids can be successfully generated shortly thereafter. This provides the direly needed tool for modeling human kidney disease (e.g., renal hypodysplasia) in the context of actual patients as has previously only been done using fibroblast-derived iPSCs.21,39
This study has several limitations. First, REB approval for Phase 1 only permitted us to use remnant urine samples from our outpatient clinic for children with mild renal malformations and UTIs. Since participants cannot be considered fully healthy individuals, we cannot rule out an effect of this on success rates. Yet, Phase 2 does demonstrate that UCs can be isolated for iPSC-based CAKUT research. Second, REB approval precluded analysis of potential factors determining success rate of UC isolation (e.g., underlying diagnosis, ethnic background, medication use) or changes in standard medical care (e.g., to give advice to drink more fluids or collection of repeat urine samples), which could not only increase success rate but also provide insight whether success rate is actually random. Third, there was a remarkably skewed gender distribution (76% boys) in Phase 1 of our study. Since we did not preferentially recruit boys, this most likely reflects the known higher incidence of (severe) CAKUT among boys.40 Fourth, in the context of demonstrating a proof-of-principle and the limited information on donors precluding their further use as either control or disease-specific line, we only generated kidney organoids from one fully characterized UiPSC clone per UC culture. However, it is accepted that multiple clones per patient should be used for iPSC-based disease modeling to account for the variability between clones from one donor (Hildebrandt et al., submitted). Based on the large number of pickable UiPSC colonies and the proportion of obtained episome-free clones per reprogramming, we are confident that this issue can be addressed easily by picking more UiPSC clones initially and screening for early episome loss (e.g., at passages 3–5).
In conclusion, we present an optimized protocol to isolate UCs from pediatric urine specimens with volumes as low as 10 ml and collected using both urine bags and MSU, reprogram these UCs footprint- and feeder-free, and generate patient-specific human kidney organoids. This protocol is not only reliable as its success appears guaranteed when at least two urine samples are obtained but is also manageable in any laboratory interested in patient-specific iPSC research. All steps of the protocol can be implemented in any laboratory with general cell culture facilities. The ability to use bag-collected urines allows for inclusion of infants and non-toilet-trained children. Thus this protocol not only brings the use of patient-specific iPSCs and kidney organoids to model CAKUT within practical arm’s reach but its first part can also be employed for iPSC-based modeling of a broad spectrum of diseases.
References
Takahashi, K. et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 (2007).
Yu, J. et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920 (2007).
Li, M. & Izpisua Belmonte, J. C. Organoids - preclinical models of human disease. N. Engl. J. Med. 380, 569–579 (2019).
Durbin, M. D., Cadar, A. G., Chun, Y. W. & Hong, C. C. Investigating pediatric disorders with induced pluripotent stem cells. Pediatr. Res. 84, 499–508 (2018).
Musunuru, K. Genome editing of human pluripotent stem cells to generate human cellular disease models. Dis. Model Mech. 6, 896–904 (2013).
Nicolaou, N., Renkema, K. Y., Bongers, E. M., Giles, R. H. & Knoers, N. V. Genetic, environmental, and epigenetic factors involved in CAKUT. Nat. Rev. Nephrol. 11, 720–731 (2015).
Morizane, R. et al. Nephron organoids derived from human pluripotent stem cells model kidney development and injury. Nat. Biotechnol. 33, 1193–1200 (2015).
Taguchi, A. et al. Redefining the in vivo origin of metanephric nephron progenitors enables generation of complex kidney structures from pluripotent stem cells. Cell Stem Cell 14, 53–67 (2014).
Takasato, M. et al. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature 526, 564–568 (2015).
Przepiorski, A. et al. A simple bioreactor-based method to generate kidney organoids from pluripotent stem cells. Stem Cell Rep. 11, 470–484 (2018).
Okita, K. et al. An efficient nonviral method to generate integration-free human-induced pluripotent stem cells from cord blood and peripheral blood cells. Stem Cells 31, 458–466 (2013).
Seki, T. et al. Generation of induced pluripotent stem cells from human terminally differentiated circulating T cells. Cell Stem Cell 7, 11–14 (2010).
van Mil, A. et al. Modelling inherited cardiac disease using human induced pluripotent stem cell-derived cardiomyocytes: progress, pitfalls, and potential. Cardiovasc. Res. 114, 1828–1842 (2018).
Zhou, T. et al. Generation of induced pluripotent stem cells from urine. J. Am. Soc. Nephrol. 22, 1221–1228 (2011).
Guan, X. et al. Dystrophin-deficient cardiomyocytes derived from human urine: new biologic reagents for drug discovery. Stem Cell Res 12, 467–480 (2014).
Massa, M. G. et al. Multiple sclerosis patient-specific primary neurons differentiated from urinary renal epithelial cells via induced pluripotent stem cells. PLoS ONE 11, e0155274 (2016).
Jia, B. et al. Modeling of hemophilia A using patient-specific induced pluripotent stem cells derived from urine cells. Life Sci. 108, 22–29 (2014).
Li, G. et al. Generation of retinal organoids with mature rods and cones from urine-derived human induced pluripotent stem cells. Stem Cells Int. 2018, 4968658 (2018).
Zhou, T. et al. Generation of human induced pluripotent stem cells from urine samples. Nat. Protoc. 7, 2080–2089 (2012).
Takasato, M., Er, P. X., Chiu, H. S. & Little, M. H. Generation of kidney organoids from human pluripotent stem cells. Nat. Protoc. 11, 1681–1692 (2016).
Forbes, T. A. et al. Patient-iPSC-derived kidney organoids show functional validation of a ciliopathic renal phenotype and reveal underlying pathogenetic mechanisms. Am. J. Hum. Genet. 102, 816–831 (2018).
van den Berg, C. W. et al. Renal subcapsular transplantation of PSC-derived kidney organoids induces neo-vasculogenesis and significant glomerular and tubular maturation in vivo. Stem Cell Rep. 10, 751–765 (2018).
Kreitzer, F. R. et al. A robust method to derive functional neural crest cells from human pluripotent stem cells. Am. J. Stem Cells 2, 119–131 (2013).
Rowan, C. J. et al. Hedgehog-GLI signaling in Foxd1-positive stromal cells promotes nephrogenesis Via TGFbeta signaling. Development 145, dev159947 (2018).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Phipson, B. et al. Evaluation of variability in human kidney organoids. Nat. Methods 16, 79–87 (2019).
Cain, J. E. & Rosenblum, N. D. Control of mammalian kidney development by the Hedgehog signaling pathway. Pediatr. Nephrol. 26, 1365–1371 (2011).
Benda, C. et al. Urine as a source of stem cells. Adv. Biochem. Eng. Biotechnol. 129, 19–32 (2013).
Cheng, L. et al. Generation of urine cell-derived non-integrative human iPSCs and iNSCs: a step-by-step optimized protocol. Front. Mol. Neurosci. 10, 348 (2017).
Li, D. et al. Optimized approaches for generation of integration-free iPSCs from human urine-derived cells with small molecules and autologous feeder. Stem Cell Rep. 6, 717–728 (2016).
Sauer, V. et al. Human urinary epithelial cells as a source of engraftable hepatocyte-like cells using stem cell technology. Cell Transpl. 25, 2221–2243 (2016).
Wang, L. et al. Using low-risk factors to generate non-integrated human induced pluripotent stem cells from urine-derived cells. Stem Cell Res. Ther. 8, 245 (2017).
Xue, Y. et al. Generating a non-integrating human induced pluripotent stem cell bank from urine-derived cells. PLoS ONE 8, e70573 (2013).
Steichen, C. et al. Human induced pluripotent stem (hiPS) cells from urine samples: a non-integrative and feeder-free reprogramming strategy. Curr. Protoc. Hum. Genet. 92, 21.27.21–21.27.22 (2017).
Si-Tayeb, K. et al. Urine-sample-derived human induced pluripotent stem cells as a model to study PCSK9-mediated autosomal dominant hypercholesterolemia. Dis. Model Mech. 9, 81–90 (2016).
Afzal, M. Z. & Strande, J. L. Generation of induced pluripotent stem cells from muscular dystrophy patients: efficient integration-free reprogramming of urine derived cells. J. Vis. Exp. 52032 (2015).
Lee, K. I., Kim, H. T. & Hwang, D. Y. Footprint- and xeno-free human iPSCs derived from urine cells using extracellular matrix-based culture conditions. Biomaterials 35, 8330–8338 (2014).
Ronen, D. & Benvenisty, N. Genomic stability in reprogramming. Curr. Opin. Genet. Dev. 22, 444–449 (2012).
Tanigawa, S. et al. Organoids from nephrotic disease-derived iPSCs identify impaired NEPHRIN localization and slit diaphragm formation in kidney podocytes. Stem Cell Rep. 11, 727–740 (2018).
Smith, J. M., Stablein, D. M., Munoz, R., Hebert, D. & McDonald, R. A. Contributions of the Transplant Registry: The 2006 Annual Report of the North American Pediatric Renal Trials and Collaborative Studies (NAPRTCS). Pediatr. Transpl. 11, 366–373 (2007).
Acknowledgements
We thank Natasha Jawa, Wei Wei, Alina Piekna, Monica Piekut, Michele Reddon, and Josefina Brooks for their support. We thank Dr. Bruce Conklin (Gladstone Institute, San Francisco, CA) for providing the WTC11 iPSCs. This work was supported by Canadian Institute of Health Research and Tier I Canada Research Chair (to N.D.R.), The Hospital for Sick Children/Research Institute/RestraComp (to J.M. and S.S.), and Medicine by Design and McLaughlin Center grants (to J.E.).
Author information
Authors and Affiliations
Contributions
All authors met the Pediatric Research authorship requirements. Conception and design, acquisition of data, or analysis and interpretation of data: J.M., S.S., T.C., D.C.R., M.R.H., R.D.C., N.D.R. Drafting the article or revising it critically for important intellectual content: J.M., D.C.R., M.R.H., I.R., J.E., N.D.R. Final approval of the version to be published: J.M., N.D.R.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Mulder, J., Sharmin, S., Chow, T. et al. Generation of infant- and pediatric-derived urinary induced pluripotent stem cells competent to form kidney organoids. Pediatr Res 87, 647–655 (2020). https://doi.org/10.1038/s41390-019-0618-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41390-019-0618-y
This article is cited by
-
Advances and potential of regenerative medicine in pediatric nephrology
Pediatric Nephrology (2024)
-
Determining epigenetic memory in kidney proximal tubule cell derived induced pluripotent stem cells using a quadruple transgenic reprogrammable mouse
Scientific Reports (2022)
-
Organs-on-chip technology: a tool to tackle genetic kidney diseases
Pediatric Nephrology (2022)
-
Mechanisms of ion transport regulation by HNF1β in the kidney: beyond transcriptional regulation of channels and transporters
Pflügers Archiv - European Journal of Physiology (2022)
-
Identification of New Transcription Factors that Can Promote Pluripotent Reprogramming
Stem Cell Reviews and Reports (2021)