Abstract
Background
Genetic defects account for a substantial proportion of pediatric cholestasis. This study explored the molecular findings in a large cohort of Chinese patients with inherited cholestasis.
Methods
Between January 2012 and June 2016, 809 Chinese pediatric patients with suspected inherited intrahepatic cholestasis were evaluated by Sanger sequencing and/or panel sequencing.
Results
Of the 809 patients, 273 (33.7%) obtained a genetic diagnosis. The rate of positive genetic diagnosis in patients with disease onset at 0–3 month of age was higher than that in patients with disease onset at 4 month of age or later. There were 17 distinct genetic defects diagnosed. The top 4 resulted from mutations in SLC25A13 (44.3%), JAG1 (24.5%), ABCB11 (11.0%), and ATP8B1 (5.9%). All 17 genetic disorders were diagnosed in patients with disease onset at 0–3 months of age; but only 5 were diagnosed in patients with disease onset beyond 4 months of age. A total of 217 distinct pathogenic variants, including 41 novel variants, were identified. Ten recurrent mutations were detected in SLC25A13, ATP8B1, and CYP27A1. They accounted for 48.2% of the total 477 mutant alleles.
Conclusions
There were 17 distinct genetic disorders diagnosed in Chinese pediatric patients with inherited cholestasis.
Similar content being viewed by others
Introduction
Cholestasis, one of the most common manifestations of liver diseases in infancy, affects about 1 in 2500–5000 live births. It results from the impairment of bile acid biosynthesis, modification, secretion, or excretion.1 The etiologies include obstructive, infectious, metabolic, toxic as well as others.2 Due to the advancement in genetic tests, more and more cholestatic patients who failed to obtain a definite diagnosis by routine tests have been found to have specific genetic defects,3,4 especially in pediatric group.5 A range of cholestatic disease-causing genes has been identified,6,7 and the top 4 common disease-causing genes are SERPINA1, JAG1, ATP8B1, and ABCB11 in Western Countries.8,9 Using a phenotype-based candidate genes approach, majority of confirmed patients were diagnosed by Sanger sequencing. Quite often, obtaining a genetic diagnosis is complex by Sanger sequencing because the differential diagnoses of cholestasis are numerous.2 Multi-gene panel can simultaneously test most known genes associated with cholestasis,2,7,10 and can greatly facilitate genetic diagnosis. It also offers an opportunity to fully clarify the disease spectrum of pediatric inherited intrahepatic cholestasis.
As the largest center for pediatric liver disease in China, our patients are referred from all parts of China.11 Several genetic disorders have been reported in Chinese pediatric cholestatic patients.10,11,12,13,14,15,16,17 Most diagnosed cases are also diagnosed by Sanger sequencing. However, no study focuses on a comprehensive description of Chinese pediatric inherited intrahepatic cholestasis. Recently, we have established a multi-gene panel which enrolls most known cholestatic disease-causing genes.10 In this study, we report the molecular findings of Sanger sequencing and panel sequencing in a large cohort of pediatric patients with cholestatic diseases. It takes a step forward the full disease spectrum of pediatric inherited cholestatic disease in China.
Patients and methods
Patients
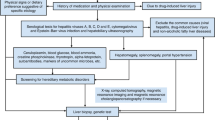
Between January 2012 and June 2016, a total of 809 pediatric cholestatic patients were tested by Sanger sequencing and/or panel sequencing. Some were reported previously.10,17,18,19,20 Cholestasis was defined as: direct bilirubin (DB) >20% of the total bilirubin (TB) if TB >5 mg/dL; or DB >1 mg/dL if TB <5 mg/dL.7,10 The patients were referred to the Center for Pediatric Liver Disease, Children’s Hospital of Fudan University and the Department of Pediatrics, Jinshan Hospital of Fudan University. Following an extensive workup as described previously,12,21 other obvious causes of liver diseases had been excluded, including infectious, drug-induced, metabolic and surgical causes. Cytomegalovirus infection was diagnosed as reported previously,10,18 and not excluded due to its high prevalence in Chinese population.22 The age ranged from 1 month to 17 years old when genetic tests were ordered.
This study was approved by the ethics committees of the Children’s Hospital of Fudan University and of the Jinshan Hospital of Fudan University. All the procedures were in accordance with the 2013 Helsinki Declaration. Written informed consent had been obtained from the parents/guardian when genetic analysis was assigned. Further informed consent was dispensed for this study.
Genetic tests
Genetic tests were done in the Translational Center of Children’s Hospital of Fudan University. In this center, only Sanger sequencing was available before 2015. The candidate genes were ordered by experienced pediatric physician basing on patients’ clinical information and were tested by Sanger sequencing. We established a cholestatic panel in 2014, which was applied to clinic in 2015.10 In 2015, both Sanger and panel sequencing methods were available. Patients with negative Sanger sequencing results were further evaluated by panel sequencing. From January 2016, panel sequencing became the first-line method. The patients were directly evaluated by panel sequencing. Sanger sequencing was only used to confirm variants of interest.
Sanger sequencing
Ten known cholestatic disease-causing genes were evaluated by Sanger sequencing, including SLC25A13, ABCB11, ABCB4, ATP8B1, CYP27A1, HSD3B7, AKR1D1, CYP7B1, JAG1, and AMACR. All coding exons and about 50bp of adjacent intronic sequence were amplified by polymerase chain reaction (PCR) and directly sequenced. PCR conditions and primer lists are available upon request. Given a high frequency of large insertions/deletions (indels) in SLC25A13,23 IVS16ins3kb were amplified by long and accurate-PCR and confirmed by electrophoresis as reported previously.19
Panel sequencing
Panel sequencing was done as described previously.10 Briefly, 25 known cholestasis disease-causing genes were enrolled, including ABCB11, ABCB4, ABCC2, AKR1D1, AMACR, ATP8B1, BAAT, CFTR, CIRH1A, CLDN1, CYP27A1, CYP7B1, FAH, GALT, HSD3B7, JAG1, NOTCH2, NPC1, NPC2, LIPA, SERPINA1, SLC25A13, TJP2, VIPAS39, and VPS33B. Library construction, emulsion PCR, and sequencing were done according to the manufacturer’s instructions (Life Technologies, USA). The average depth of coverage was more than 250x, which resulted in a desired coverage of target regions. Variation calling was carried out using NextGene software (version 2.3.3). Following an optimized protocol, variants beyond 5bp from exon boundaries and variants with poor coverage or poor allele balance were discarded. Benign variants, common variants and sequencing errors were filtered by applying 1000 Genomes Project (TGP), Exome Aggregation Consortium (ExAC), dbSNP, and an internal database. About 5 variants per sample were retained for further analysis. Variants of interest were validated by Sanger sequencing.
Parental studies
If parental blood samples were available, the mutation origin was tested by Sanger sequencing.
Bioinformatics analysis
The pathogenicity was assessed according to the standards for interpretation of sequence variants.24 Known disease mutations, canonical splice site, nonsense, and frameshift variants were considered pathogenic. Novel pathogenic missense variants must satisfy the following:1 minor allele frequency (MAF) <0.01% in both TGP and ExAC;2 predicted to be damaging by Pholyphen2, MutationTaster, and SIFT.
Molecular diagnosis
For autosomal recessive diseases, a definite genetic diagnosis was assigned if pathogenic variants were detected on both alleles of the same gene. If parental data were unavailable, the homozygotes could also receive a definite genetic diagnosis, while the patients with two different pathogenic variants in the same gene obtained a possible genetic diagnosis. For autosomal dominant diseases, the patients received a definite genetic diagnosis if heterozygous pathogenic variants were identified in a dominant gene.
Patients with one heterozygous pathogenic variant in a recessive gene and patients without pathogenic variant were treated as negative whether or not a clinical diagnosis was reached basing on their clinical presentations.
Statistical analysis
Statistic analysis was done using SPSS Inc. version 17.0 software (University of Chicago, Chicago). Chi-square test was used to test the difference among ratios. P < 0.05 was considered significant.
Results
Clinical data and general information
Of the 809 pediatric patients with suspected intrahepatic cholestasis, 779 (96.3%) had first symptoms with jaundice (Table 1); only 30 cases first presented for other reasons, including bleeding, pruritus, failure to thrive, liver failure, portal hypertension, and hepatomegaly and/or splenomegaly. Cholestasis was confirmed in all cases by liver function tests: the highest values of DB in the disease course ranged from 1.2 mg/dL to 27.3 mg/dL, while the ratio of DB/TB ranged from 32.7 to 98.8%. A total of 273 (33.7%, 273/809) patients obtained a potential genetic diagnosis (Supplemental Table S1 (online)), including 105 reported previously.10,17,18,19,20
Of the 664 patients with disease onset at 0–3 months of age, 247 (37.2%; 247/664) obtained a molecular diagnosis. The positive rate was higher than that in patient with disease onset at 4 month of age or later (17.9%; 26/145) (37.2 vs. 17.9%, χ2 = 19.761, P < 0.001), while the positive rates were similar between patients with disease onset at 4–12 month of age (22.0%; 13/59) and those beyond one year old (15.1%; 13/86) (22.0 vs. 15.1%, χ2 = 1.138, P = 0.286).
There were 562 patients only tested by Sanger sequencing: 226 (40.2%; 226/562) cases received a genetic diagnosis (Table 1). Other 247 patients had been evaluated by panel sequencing, including 38 cases with negative Sanger sequencing results, 47 (19.0%) received a genetic diagnosis (40.2% vs. 19.0%, χ2 = 34.444, P < 0.001). Of the 536 patients without a genetic diagnosis, 74 (13.8%; 74/536) were found to harbor heterozygous pathogenic variants (Supplemental Table S2 (online)).
Molecular diagnoses
For the 273 diagnosed patients, 17 distinct genetic defects were diagnosed (Table 2). The top 4 resulted from mutations in SLC25A13 (44.3%; 121/273), JAG1 (24.5%, 67/273), ABCB11 (11.0%, 30/273), and ATP8B1 (5.9%, 16/273). They together accounted for 85.7% of the diagnosed cases. Only 4 (1.5%; 4/273) patients were diagnosed with progressive familiar intrahepatic cholestasis type 3 (PFIC3) resulting from mutations in ABCB4. It was less common than Cerebrotendinous xanthomatosis (CTX) (n = 8; 2.9%, 8/273) and Dubin-Johnson syndrome (DJS) (n = 12; 4.4%, 12/273). Nine genetic disorders were diagnosed only in one or two cases.
All the 17 genetic disorders could develop cholestasis within 0–3 months of age. For the 247 diagnosed cases with disease onset at 0–3 months of age, SLC25A13 (47.0%, 116/247) was the most common causal gene, followed by JAG1 (23.9%, 59/247), ABCB11 (10.5%, 26/247), and ATP8B1 (4.0%, 10/247). However, only 5 genetic disorders were diagnosed in patients with disease onset beyond 3 months of age: JAG1 (30.8%, 8/26), ATP8B1 (23.1%, 6/26), SLC25A13 (19.2%, 5/26), ABCB11 (15.4%, 4/26), and ABCB4 (11.5%, 3/26).
By Sanger sequencing, nine distinct genetic disorders were diagnosed: the top 4 resulted from mutations in SLC25A13, JAG1, ABCB11, and ATP8B1. However, 13 distinct genetic diseases were diagnosed by panel sequencing: neonatal intrahepatic cholestasis caused by citrin deficiency (NICCD) still was the most common; 8 genetic disorders were first diagnosed in our laboratory; mutations in ABCC2 and CYP27A1 were commonly detected.
Mutation Spectrum
In the 273 diagnosed patients, 217 distinct pathogenic or likely pathogenic variants were detected, including 176 known and 41 novel variants. The 41 novel variants included 11 nonsense, 14 frameshift indels leading to truncated protein, 4 canonical splice site variants known to affect splicing, and 12 missense variants predicted to be damaging by MutationTaster, Polyphen-2, and SIFT (Supplemental Table S3 (online)). All the 12 missense pathogenic variants were identified in recessive disease genes and had MAF < 0.01% in both TGP and ExAC. Of them, 10 were compounded with other pathogenic variants; 2 were homozygous with mutant alleles inheriting from each carried parent.
A full range of mutation types were identified: 48 frameshift indels, 4 in-frame indels, 46 nonsense, 25 canonical splice-site, 7 intronic and 87 missense pathogenic variants (Table 3). Missense pathogenic variants accounted for the majority of mutant allele of ABCC2 (62.5%, 15/24), ABCB11 (61.7%, 37/60), and ATP8B1 (59.4%, 19/32); but only a few of SLC25A13 (4.5%). Frameshift indels and nonsense variants accounted for 77.6% (52/67) of mutant allele of JAG1.
Ten distinct pathogenic variants detected in >3 families (Table 4), and accounted for 48.2% of total 477 mutant alleles. Eight were detected in SLC25A13.
Parental Study
Heterozygous pathogenic variants were identified in 2 dominant genes of 69 patients, including JAG1 and NOTCH2. Parental studies were done in 37 patients, revealing that only 8 (21.6%; 8/37) patients had mutant alleles inherited from their symptomatic parents; the remaining 29 (78.4%; 29/37) mutant allele arose from de novo.
Two different pathogenic variants were detected in the same recessive gene of 151 patients. Parental studies were performed in 51 patients: 49 (96.1%; 49/51) were compound heterozygous; other 2 (3.9%; 2/51) had both likely pathogenic variants located on the same allele and were treated as negative.
There were 55 patients harboring homozygous pathogenic variants in recessives genes. Parental data from 13 families revealed that all mutant alleles were inherited from respective carrier parents.
Discussion
Genetic defects account for a substantial proportion of pediatric intrahepatic cholestasis. Several inherited cholestatic diseases have been reported in China,10,11,12,13,14,15,16,17 but the disease spectrum is still not fully clarified. In this study, we reported the molecular findings in a large cohort of pediatric cholestatic patients from all parts of China, who ruled out other obvious causes of liver diseases, including infective, drug-induced, metabolic and surgical causes. A molecular diagnosis was assigned to 273 patients. Beside a small number of cases with atypical clinical presentations, most of them could obtain a possible clinical diagnosis. Their clinical diagnoses were confirmed by sequencing the candidate genes and/or multi-gene panel sequencing. There were 17 distinct genetic disorders diagnosed. The top 4 were resulted from mutations in SLC25A13, JAG1, ABCB11, and ATP8B1. It took a step forward the full spectrum of pediatric inherited cholestatic disease in China.
By Sanger sequencing and panel sequencing, 33.7% of pediatric cholestatic patients without obvious causes of liver disease were assigned a potential genetic diagnosis. The positive rate might be underestimated because of the following reasons:1 not all cholestatic genes were enrolled in our panel, especially those reported recently;2 only a part of enrolled patients was tested by panel sequencing;3 limitations of sequencing methods, such as poor coverage, insensitivity to large indels, leading to false negatives. The positive rate of Sanger sequencing was found to be higher than panel sequencing. It could attribute to that Sanger sequencing was frequently done in patients with distinguishable clinical features, such as Alagille syndrome and Citrin deficiency,17,25 who got the highest yields. Therefore, Sanger sequencing could be the first lines sequencing method for those patients. Panel sequencing was proposed for patients with negative Sanger sequencing results and patients without distinguishable features, even patients with distinguishable features if the sequencing cost of one panel test was comparable to that of one single gene test.
Nine distinct genetic disorders were diagnosed by Sanger sequencing. Additional eight genetic disorders were diagnosed by panel sequencing. The disease spectrum might be further expanded if more patients were tested by panel or whole exome sequencing.26,27 However, only a small number of patients with negative Sanger sequencing results underwent panel sequencing. It could be attributed to the following: reasons:1 most cholestatic children manifested transient neonatal cholestasis (TNC) and had a good prognosis;28,2 a significant number of patients were lost in follow-up. NICCD was the most common disease, followed by Alagille syndrome, PFIC, bile acid synthesis defects (BASD). Therefore, we proposed that serum amino acid could be routine, and that spine x-ray and opthalmology examination could be routine in patients with high GGT. Liver biopsy and immune-stain should be done if PFIC was considered. It would establish a genetic diagnosis before genetic reports were available.20 Urine bile acid profiles was proposed in cholestatic patients with low TBA level. Recently, we found the atypical bile acid profile in infantile CTX make it easily neglected.29 None was diagnosed with α1-antitrypsin deficiency, which was the most common cause of infant inherited cholestasis in Western Countries.8,9 Meanwhile, NICCD was less in countries outside Asia.7,30 Hence, we inferred that the disease spectrum of pediatric inherited cholestasis in China was different from that in Western Countries.
A large number of pathogenic variants were identified in this study. It expanded the mutation spectrums greatly. Ten distinct recurrent pathogenic variants were detected in recessive genes, but not dominant genes. E297G and D482G were often detected in ABCB11 of European,31 but not our patients. It indicated that the mutation spectrum might be different between China and Western Countries. For recessive disease genes, most mutant alleles were inherited from carrier parents. By parental studies, we found that 96.1% of patients with two different pathogenic variants in the same recessive genes were confirmed to be compound heterozygotes. Hence, it was reasonable to treat the patients with two different pathogenic variants as compound heterozygotes if parental data were unavailable.20,32 Different from recessive genes, only a fraction of mutant alleles were inherited from symptomatic parents for dominant disease genes; most mutant allele arose from de novo. Variants arising from de novo were more likely to be pathogenic.17 However, it was needed to note that some mutant alleles inherited from parents with mutations in germ cells might be treated as de novo.
Most pediatric cholestatic patients had disease onset at first year of life, especially at 0–3 months of age. Patients with disease onset at 0–3 months of age had a higher possibility of genetic defects than patients with disease onset at 4 months old or later. Therefore, we proposed that genetic testing should be took into consideration if routine evaluation failed to reach a diagnosis, especially in infants with disease onset at 0–3 months. For cholestasis onset at 0–3 months of age, all the 17 genetic disorders could be the causes. As panel sequencing could test those causing-genes simultaneously, we proposed panel sequencing as the first-line sequencing method for cholestasis onset at 0–3 months of age. However, supplementary evaluation methods were needed for some patients, because large indels were frequently identified in SLC25A13 and JAG1.17,23 Recurrent large indels, including IVS4ins6kb and IVS16ins3kb, were quite often identified in SLC25A13.23 It was necessary to screen these recurrent variants if patients with high possibility of NICCD had negative results or only one mutant allele identified SLC25A13 by panel sequencing. Different from SLC25A13, recurrent large indels were not identified in JAG1. For patients with possibility of Alagille syndrome, multiplex ligation-dependent probe amplification (MLPA) should be done if negative results were got by panel sequencing.17
In summary, we here reported the molecular findings in a large cohort of Chinese pediatric inherited cholestatic patients. By Sanger and/or panel sequencing, 33.7% of patients obtained a molecular diagnosis. Patients with disease onset at 0–3 months of age were more likely to obtain a genetic diagnosis. Seventeen genetic disorders were diagnosed. The top 4 resulted from mutations in SLC25A13, JAG1, ABCB11, and ATP8B1. The spectrum was different from that in Western Countries.
References
Zollner, G. & Trauner, M. Mechanisms of cholestasis. Clin. Liver Dis. 12, 1–26 (2008).
Herbst, S. M. et al. Taking the next step forward- Diagnosing inherited infantile cholestatic disorders with next generation sequencing. Mol. Cell Probes 29, 291–298 (2015).
Feldman A. G., Sokol R. J. Neonatal cholestasis. Neoreviews 14, e63–e73 (2013).
Stormon, M. O. et al. The changing pattern of diagnosis of infantile cholestasis. J. Paediatr. Child Health 37, 47–50 (2001).
Mieli-Vergani, G., Howard, E. R. & Mowat, A. P. Liver disease in infancy: a 20 year perspective. Gut 32, S123–S128 (1991).
Hirschfield, G. M. et al. The genetics of complex cholestatic disorders. Gastroenterology 144, 1357–1374 (2013).
Togawa, T. et al. Molecular genetic dissection and neonatal/infantile intrahepatic cholestasis using targeted next-generation sequencing. J. Pediatr. 171, 171–177 (2016).
Fischler, B. & Lamireau, T. Cholestasis in the newborn and infant. Clin. Res Hepatol. Gastroenterol. 38, 263–267 (2014).
Liu, C. et al. Novel resequencing chip customized to diagnose mutations in patients with inherited syndromes of intrahepatic cholestasis. Gastroenterology 132, 119–126 (2007).
Wang, N. L. et al. A specially designed multi-gene panel facilitates genetic diagnosis in children with intrahepatic cholestasis: simultaneous test of known large insertions/deletions. PLoS ONE 11, e0164058 (2016).
Chen, R. et al. Different regional distribution of SLC25A13 mutations in Chinese patients with neonatal intrahepatic cholestasis. World J. Gastroenterol. 19, 4545–4551 (2013).
Fang, L. J. et al. Chinese children with chronic intrahepatic cholestasis and high γ-glutamyl transpeptidase: clinical features and association with ABCB4 mutations. J. Pediatr. Gastroenterol. Nutr. 55, 150–156 (2012).
Liu, L. Y. et al. ABCB11 gene mutations in Chinese children with progressive intrahepatic cholestasis and low gamma glutamyltransferase. Liver Int. 30, 809–815 (2010).
Liu, L. Y. et al. Characterization of ATP8B1 gene mutations and a hot-linked mutation found in Chinese children with progressive intrahepatic cholestasis and low GGT. J. Pediatr. Gastroenterol. Nutr. 50, 179–183 (2010).
Zhao, J. et al. Primary ∆4-3-oxosteroid 5β-reductase deficiency: two cases in China. World J. Gastroenterol. 18, 7113–7117 (2012).
Li, L. T. et al. Two novel VPS33B mutations in a patient with arthrogryposis, renal dysfunction and cholestasis syndrome in mainland China. World J. Gastroenterol. 20, 326–329 (2014).
Li, L. et al. JAG1 mutation spectrum and origin in Chinese children with clinical features of Alagille syndrome. PLoS ONE 10, e0130355 (2015).
Wang, N. L. et al. The features of GGT in patients with ATP8B1 or ABCB11 deficiency improve the diagnostic efficiency. PLoS ONE 11, e0153114 (2016).
Zhang, M. H., Gong, J. Y. & Wang, J. S. Citrin deficiency presenting as acute liver failure in an eight-month-old infant. World J. Gastroenterol. 21, 7331–7334 (2015).
Li, L. et al. Hypothyroidism Associated with ATP8B1 Deficiency. J. Pediatr. 167, 1334–1339 (2015).
Wang, J. S., Tan, N. & Dhawan, A. Significance of low or normal serum gamma glutamyl transferase level in infants with idiopathic neonatal hepatitis. Eur. J. Pediatr. 165, 795–801 (2006).
Chang, M. H. et al. Neonatal hepatitis: a follow-up study. J. Pediatr. Gastroenterol. Nutr. 6, 203–207 (1987).
Lin, W. X. et al. Molecular diagnosis of pediatric patients with citrin deficiency in China: SLC25A13 mutation spectrum and the geographic distribution. Sci. Rep. 6, 29732 (2016).
Richards, S. et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 17, 405–424 (2015).
Wang, J. S. et al. Biochemical characteristics of neonatal cholestasis induced by citrin deficiency. World J. Gastroenterol. 18, 5601–5607 (2012).
Qiu, Y. L. et al. Defects in MYO5B are associated with a spectrum of previously undiagnosed low γ-glutamyltransferase cholestasis. Hepatology 65, 1655–1669 (2017).
Gomez-Ospina, N. et al. Mutations in the nuclear bile acid receptor FXR cause progressive familial intrahepatic cholestasis. Nat. Commun. 7, 10713 (2016).
Liu, L. Y. et al. Association of variants of ABCB11 with transient neonatal cholestasis. Pediatr. Int. 55, 138–144 (2013).
Gong, J. Y. et al. Severe neonatal cholestasis in cerebrotendinous xanthomatosis: genetics, immunostaining, mass Spectrometry. J. Pediatr. Gastroenterol. Nutr. 65, 561–568 (2017).
Tabata, A. et al. Identification of 13 novel mutations including a retrotransposal insertion in SLC25A13 gene and frequency of 30 mutations found in patients with citrin deficiency. J. Hum. Genet. 53, 534–545 (2008).
Strautnieks, S. S. et al. Severe bile salt export pump deficiency: 82 different ABCB11 mutations in 109 families. Gastroenterology 134, 1203–1214 (2008).
Davit-Spraul, A. et al. ATP8B1 and ABCB11 analysis in 62 children with normal gamma-glutamyl transferase progressive familial intrahepatic cholestasis (PFIC): phenotypic differences between PFIC1 and PFIC2 and natural history. Hepatology 51, 1645–1655 (2010).
Acknowledgements
Great thanks to Prof. L.-Z. Wang for the revision of the manuscript. This work was supported by the National Natural Science Foundation of China, grant numbers 81361128006 and 81570468.
Author information
Authors and Affiliations
Contributions
Study design: N.L.W., J.L., J.Y.G., J.S.W.; sample and data acquisition: all authors; data interpretation: Y.L., J.Y.G., X.B.X., M.H.Z.; draft manuscript: N.L.W.; statistics: N.L.W., K.A.; review and approve manuscript: all authors; accountability for work: all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
About this article
Cite this article
Wang, NL., Lu, Y., Gong, JY. et al. Molecular findings in children with inherited intrahepatic cholestasis. Pediatr Res 87, 112–117 (2020). https://doi.org/10.1038/s41390-019-0548-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41390-019-0548-8
This article is cited by
-
Diagnostic yield and novel candidate genes by next generation sequencing in 166 children with intrahepatic cholestasis
Hepatology International (2024)
-
Neonatal cholestasis is an early liver manifestation of children with acid sphingomyelinase deficiency
BMC Gastroenterology (2022)
-
Genetic spectrum and clinical characteristics of 3β-hydroxy-Δ5-C27-steroid oxidoreductase (HSD3B7) deficiency in China
Orphanet Journal of Rare Diseases (2021)