Abstract
Background
We previously demonstrated an association between placental leptin (LEP) methylation levels and macrosomia without gestational diabetes mellitus (non-GDM). This study further explored the association between LEP methylation in cord blood and non-GDM macrosomia.
Method
We carried out a case–control study of 61 newborns with macrosomia (birth weight ≥4000 g) and 69 newborns with normal birth weight (2500–3999 g). Methylation in the LEP promoter region was mapped by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry.
Results
Average cord blood LEP methylation levels were lower in macrosomia newborns than in control newborns (P < 0.001). Eleven CpG sites were associated with macrosomia. Multivariate logistic regression revealed that low LEP methylation levels [adjusted odds ratio (AOR) = 2.84, 95% confidence interval (CI): 1.72–4.17], high pre-pregnancy body mass index (AOR = 7.44, 95% CI: 1.99–27.75), long gestational age (AOR = 3.18, 95% CI: 1.74–5.79), high cord blood LEP concentration (AOR = 2.25, 95% CI: 1.34–3.77), and male newborn gender (AOR = 3.91, 95% CI: 1.31–11.69) significantly increased the risk of macrosomia.
Conclusions
Lower cord blood LEP methylation levels and certain maternal and fetal factors are associated with non-GDM macrosomia.
Similar content being viewed by others
Introduction
Macrosomia, typically defined as a birth weight ≥4000 g or above the 90th percentile for gestational age,1 is a common obstetric phenomenon. The incidence of macrosomia is rising in many regions and countries. Over the past two decades, the global incidence of macrosomia has increased by 15–25%.2 In China, the incidence of macrosomia is similarly trending upward.3,4 Maternal factors during pregnancy, such as gestational weight gain (GWG) and pre-pregnancy body mass index (pre-BMI), are positively correlated with the incidence of macrosomia.5,6 Although many studies have focused on macrosomia in gestational diabetes mellitus (GDM),7,8,9 the molecular mechanism of non-GDM macrosomia remains unclear.
Leptin (LEP), a 16-kD peptide encoded by the obesity gene, is a neuroendocrine hormone involved in food intake, energy expenditure, reproduction, and metabolic diseases.10,11,12,13 As an intermediate molecule linking the neuroendocrine system with adipose tissue, LEP participates in the regulation of fetal growth and development.14,15 A meta-analysis confirmed that birth weight is associated with cord blood LEP concentration and that small for gestational age (SGA) babies have lower LEP concentrations than appropriate for gestational age (AGA) babies.16 In addition, a recent study by Lesseur et al.17 has found that SGA newborns have higher cord blood LEP methylation levels than AGA or large for gestational age newborns. Evidence from a cohort study indicates that high maternal pre-BMI is associated with downregulation of neonatal cord blood LEP methylation.18 Another study of breastfed 17-month-old children and their mothers reports that higher maternal pre-BMI and LEP concentration are associated with lower LEP methylation in fetal serum.19 Furthermore, our previous research shows greater hypermethylation of individual CpG dinucleotides of the LEP promoter in the placenta in non-GDM macrosomia pregnancies than in control pregnancies at 39 weeks gestation.20 Nonetheless, the association between non-GDM macrosomia and cord blood LEP methylation levels remains unclear.
Several common single-nucleotide polymorphisms (SNPs) in the LEP gene are associated with obesity-related diseases.21,22,23 Specifically, rs7799039 and rs2167270 SNPs, which are located in the LEP promoter, have been associated with birth weight.17,24 However, whether these two SNPs are associated with macrosomia is unknown. Therefore, the aims of our study were to investigate the association between cord blood LEP methylation levels and non-GDM macrosomia and to determine whether the occurrence of macrosomia is related to rs7799039 or rs2167270 mutations.
Methods
Participants
This hospital-based case–control study was conducted in the Second Affiliated Hospital of Wenzhou Medical University (Zhejiang, China). One hundred and thirty healthy maternal–neonatal pairs were recruited, including 61 newborns with macrosomia and 69 newborns with normal birth weight as controls. Macrosomia was defined as a birth weight ≥4000 g. Controls were newborns with birth weights between 2500 and 3999 g who were born within 3 days of macrosomia newborns. Basic information about the mothers and newborns was collected from self-report questionnaires administered after delivery, and other maternal medical information was obtained from the departmental perinatal database. Participants were included if they had normal pregnancies with normal maternal glucose tolerance and singleton newborns born full-term (gestational age 37–42 weeks) by normal delivery or cesarean. A normal pregnancy was defined as no hypertension, gestational diabetes, heart disease, hepatitis, or psychological disorder.25 Participants were excluded if they had pregnancies involving preeclampsia, uteroplacental insufficiency, post-term pregnancy, pregnancy complications (e.g., diabetes, hypertension, or cardiovascular disease), or a newborn with congenital malformations.26 Each newborn was weighed immediately after delivery. The Wenzhou Medical University Ethics Committee reviewed and approved the study, and each mother provided informed consent.
Cord blood sampling and LEP concentration
Cord blood samples were collected from all participants immediately after delivery but before delivery of the placenta. The cord was clamped and cut, and 10 ml cord blood was collected in anti-freezing and enzyme-free tubes with a new syringe. A 5-ml aliquot was maintained at room temperature for 1 h and centrifuged at 3000 rpm for 20 min. The serum fraction was transferred to a 5-ml tube and immediately stored at −80 °C to determine LEP concentration. The remaining 5 ml was transferred to an EDTA-coated anticoagulant tube, mixed well, and stored at −80 °C for methylation and SNP analysis. LEP concentration in the cord blood was measured using a double antibody sandwich Avidin-Biotin Complex enzyme-linked immunosorbent assay (ABC-ELISA, Sigma, USA).
LEP DNA methylation
The LEP gene sequence was obtained from GenBank, and the position of the LEP promoter was predicted using Eukaryotic Promoter Prediction sites (http://epd.vital-it.ch/). Genomic DNA was extracted from cord blood using a BioTek DNA Purification Kit (BioTek, Beijing, China) and subsequently treated with a EZ DNA Methylation-Gold Kit (Zymo Research, Irvine, CA, USA) according to the manufacturers’ protocol. The resulting genomic DNA was then treated with bisulfite to convert unmethylated cytosine (C) bases to uracil (U). DNA methylation was performed using the gold standard Sequenom MassARRAY platform (CapitalBio, Beijing, China), which combines base-specific cleavage (molecular cleavage) and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. DNA was subjected to polymerase chain reaction (PCR) amplification using primer sets previously designed27 by EpiDesigner. Data were analyzed using the EpiTYPER software version 1.0 (SEQUENOM, San Diego, CA, USA) to determine the methylation results for each CpG site.
SNP genotyping of rs7799039 and rs2167670
Genotyping of rs7799039 and rs2167670 LEP SNPs were performed using PCR/restriction fragment length polymorphism as previously described.27 PCR was performed with a Bio-Rad PCR instrument (Bio-Rad, USA) using a 20-μl reaction volume containing a final primer concentration of 0.2 mM in Ampliqon master mix (1.5 mM MgCl2). PCR products were digested using HhaI (rs7799039) and MspAIL (rs2167270) restriction enzymes at 37 °C for 24 h.
Statistical analysis
Questionnaire and experimental data were entered into an EpiData3.1 database, and statistical analyses were performed using SPSS version 14.0 (SPSS, Inc., Chicago, IL, USA). The normality of the distributions of all variables was assessed using Kolmogorov–Smirnov tests. Normally distributed data are presented as mean ± standard deviation (SD), and non-normally distributed data are presented as median and range. Pre-BMI and GWG were grouped by World Health Organization (WHO)28 and American Institute of Medicine (IOM) criteria.29 Differences between groups were assessed using independent-sample t tests or Mann–Whitney U tests. Analysis of variance was used to determine differences in the methylation of CpG sites between groups. Categorical and rank data are presented as frequencies and percentages, and differences between groups were determined using Chi-square (χ2) or rank-sum tests. Differences in genotype and allele frequencies between groups were determined using χ2 or Fisher’s exact tests. The Hardy–Weinberg equilibrium (HWE) in the two groups was evaluated and haplotype tests were performed using the SHEsis software (http://analysis.bio-x.cn/myAnalysis.php).30 Differences in cord blood methylation levels among G/G, G/A, and A/A genotypes were analyzed by Tukey’s post hoc tests. Correlations were computed using Spearman correlation analysis. Multivariate logistic regression analysis was used to evaluate the effects of LEP methylation and gestational factors on macrosomia, and adjusted odds ratio (AOR) and 95% confidence interval (CI) were used to estimate the relative risk of macrosomia. Two-tailed P values <0.05 were considered statistically significant.
Results
Baseline data
Maternal and neonatal characteristics are displayed in Table 1. Birth weight (P < 0.001), the proportion of male newborns (P = 0.02), maternal GWG (P = 0.03), pre-BMI (P = 0.007), and gestational age (P = 0.001) were significantly higher in the macrosomia group than in the control group. No significant differences in maternal age, height, parity, or educational level were detected between groups.
LEP DNA methylation
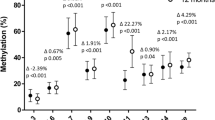
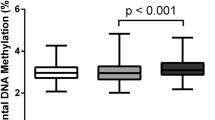
DNA methylation analysis focused on the 507-bp human LEP promoter region, which includes 36 CpG sites (Supplemental Figure S1). Three CpG sites (CpG15, CpG18, and CpG32) did not display a signal. Differences between the macrosomia and control groups in methylation levels of the 33 CpG sites are shown in Fig. 1. The average LEP DNA methylation levels among the 33 CpG sites were 24.38 ± 3.73% and 27.15 ± 3.17% for the macrosomia and control groups (P < 0.001), respectively. Average methylation levels of CpG sites were further compared between subgroups stratified by newborn gender, gestational age, pre-BMI, and GWG (Supplemental Table S1). Within newborn gender, gestational age, and GWG subgroups, LEP promoter methylation levels were significantly lower in the macrosomia group than in the control group (P < 0.05). In the subgroup with pre-BMI ≤24.9 kg/m2, LEP promoter methylation levels were significantly lower in the macrosomia group than in the control group, whereas no significant differences were found in the subgroup with pre-BMI >24.9 kg/m2. There was no multicollinearity among the 33 CpG sites (variance inflation factors of each parameter were <10), and we further performed multivariate logistic regression analysis on these sites after controlling for the confounding factors of neonatal gender, pre-BMI, gestational age, and GWG. We found that 11 CpG sites (CpG1, 2, 3; CpG11; CpG23, 24, 25; CpG26, 27, 28; and CpG36) were associated with macrosomia (Fig. 2), suggesting that low methylation levels increase the risk of macrosomia.
Association between the methylation levels of CpG sites in the leptin (LEP) promoter region and the risk of macrosomia. The dotted line represents adjusted odds ratio (AOR) = 1. Triangles represent the AOR values of 16 CpG dinucleotides in the LEP promoter region. Bars represent 95% confidence interval
SNP genotypes
Two common SNPs, rs7799039 and rs2167270, which are located in the LEP promoter region, were chosen to explore whether the non-GDM macrosomia is associated with LEP genetic variation. Both the macrosomia and control groups were in HWE. Genotypic and allelic frequencies of the two groups are shown in Supplemental Table S2. In the macrosomia group, the frequencies of wild-type G/G, heterozygous G/A, and homozygous A/A genotypes were 41, 49, and 10% for rs2167270 and 52, 43, and 5% for rs7799039, respectively. In the control group, the frequencies of wild-type G/G, heterozygous G/A, and homozygous A/A genotypes were 52, 39, and 9% for rs2167270 and 61, 38, and 1% for rs7799039, respectively. There were no significant group differences in genotypic or allelic frequencies for either SNP.
We also analyzed methylation levels of the different genotypes. Because cord blood G/G and G/A genotype methylation levels were not statistically different (P = 0.77 for rs2167270, P = 0.63 for rs7799039), the wild-type G/G and heterozygous G/A genotypes were classified as a single group to test whether the two SNPs differed in average LEP methylation between groups. For both SNPs, the G/G+G/A genotypes had lower methylation levels in the macrosomia group than in the control group (P < 0.001; Supplemental Figure S2). For rs2167270, there was no group difference in LEP methylation levels of the homozygous A/A genotype. For rs7799039, there were only three instances of the homozygous A/A genotype in the macrosomia group and only one instance in the control group, making statistical comparison impossible.
LEP concentration in cord blood
LEP concentration in cord blood was significantly higher in the macrosomia group (mean: 11.39 ng/ml, range: 2.59–41.37 ng/ml) than in the control group (mean: 7.27 ng/ml, range: 1.52–46.19 ng/ml; Z = −3.522, P < 0.001; Fig. 3). Newborns were stratified by gender to further analyze cord blood LEP concentration. Among both males and females, cord blood LEP concentration was significantly higher in the macrosomia group (male mean: 9.71 ng/ml, range: 2.95–31.56 ng/ml; female mean: 15.08 ng/ml, range: 7.14–41.37 ng/ml) than in the control group (male mean: 5.22 ng/ml, range: 1.52–21.47 ng/ml, Z = −3.853, P < 0.001; female mean: 10.67 ng/ml, range: 2.28–46.19 ng/ml, Z = −2.555, P = 0.011). In addition, in the macrosomia group, cord blood LEP concentration was significantly lower in males than in females (Z = −2.731, P = 0.006). We further analyzed the correlation between LEP average methylation and LEP concentration but found no significant linear correlation (rs = −0.027, P = 0.766). Also, no significant correlations were found when data were stratified according to newborn gender, pre-BMI, gestational age, and GWG.
Comprehensive analysis of risk factors for non-GDM macrosomia
To further explore the association of LEP methylation with pregnancy-related factors in macrosomia, multivariate logistic regression analysis was performed with macrosomia as the dependent variable and newborn gender, gestational age, GWG, pre-BMI, average cord blood LEP methylation levels, and cord blood LEP concentration as independent variables. We found that low cord blood LEP DNA methylation levels (AOR = 2.84, 95% CI: 1.72–4.17), high pre-BMI (AOR = 7.44, 95% CI: 1.99–27.75), long gestational age (AOR = 3.18, 95% CI: 1.74–5.79), high cord blood LEP concentration (AOR = 2.25, 95% CI: 1.34–3.77), and male newborn gender (AOR = 3.91, 95% CI: 1.31–11.69) significantly increase the risk of macrosomia (Table 2).
Discussion
The results of our study suggest that low cord blood LEP methylation levels and high cord blood LEP concentration are associated with non-GDM macrosomia. Methylation levels at 11 CpG sites (CpG1, 2, 3; CpG11; CpG23, 24, 25; CpG26, 27, 28; and CpG36) were significantly lower in the macrosomia group than in the control group. Male newborn gender; lower LEP DNA methylation levels; and higher pre-BMI, gestational age, GWG, and LEP concentration synergistically increased the risk of non-GDM macrosomia. The SNPs rs2167270 and rs7799039 were not associated with the occurrence of non-GDM macrosomia.
Although no significant differences in average LEP methylation were found between macrosomia and normal birth weight pregnancies in our previous experiment using placental tissue, methylation levels at some CpG sites in the macrosomia group were significantly higher than those in the control group.20 Importantly, combined with the results of the present study, we found that methylation at some CpG sites in both placental tissue and cord blood (i.e., CpG23, 24, 25; CpG26, 27, 28) were associated with macrosomia. However, in contrast to methylation levels at these sites in the placenta, cord blood CpG site methylation levels were significantly lower in the macrosomia group than in the control group. One explanation may be the different sources of LEP, as LEP in the placenta is mainly secreted by the placenta to satisfy maternal and fetal growth needs during pregnancy, whereas LEP in cord blood originates from both placental and fetal adipose tissue.31 Thus LEP methylation differences between the placenta and cord blood could be because the LEP secreted by the fetus is more sensitive to its content of body fat. In addition, DNA methylation is tissue specific,32 and the placenta and cord blood have separate circulatory systems,33 in which LEP may have different roles.
Previous studies report that SGA newborns have higher cord blood LEP methylation levels than AGA newborns,17 although an association between the methylation of CpG sites and birth weight has not been described. In the present study, we found a low methylation level in the macrosomia group, consistent with the finding of a cross-sectional study that newborns whose blood cells had low LEP methylation levels had higher birth weights.19 We also examined 33 CpG sites in the LEP promoter region in cord blood and found that methylation at 11 CpG sites was associated with macrosomia, and this pattern was still observed after adjusting for confounding factors. Most CpG sites associated with macrosomia were located near the specificity protein 1 (SP1)- or CCAAT-enhancer binding protein (C/EPB)-binding motifs in the LEP promoter region. These results are in accord with the hypothesis by Hogg et al.34 that CpG sites proximal to SP1- or C/EBP-binding motifs are more sensitive to methylation changes. However, the mechanism underlying the influence of the CpG sites on the development of macrosomia requires additional investigation.
In our previous study, we found that cord blood brain-derived neurotrophic factor (BDNF) gene expression is downregulated in macrosomia pregnancies as compared with normal birth weight pregnancies.26 In the present study, we found that cord blood LEP concentration was higher in the macrosomia group and that an elevated LEP concentration was associated with macrosomia. However, LEP concentrations were not consistently increased. This might be due to LEP resistance,35 which might inhibit the expression of BDNF and in turn affect transmission of related signals in neurons, resulting in increased energy intake, reduced consumption, and obesity.36 According to the epigenetic regulation theory, decreased methylation in the LEP gene promoter region could promote LEP expression, resulting in appetite suppression, altered metabolism, and prevention of obesity. Although we did not find a negative correlation between LEP concentration and its methylation, it is possible that a non-linear correlation could exist. As the contribution of CpG locus methylation status in the gene promoter region to gene transcription is complicated, the specific mechanism of the epigenetic regulation of gene expression requires further study.
We found no differences in the frequency of the A/A genotype in the two LEP promoter SNPs (rs2167270 and rs7799039) between the macrosomia and control groups. Furthermore, we found no differences in the methylation levels of the A/A genotype. In a study of a Caucasian population, Lesseur et al.17 found elevated cord blood LEP DNA methylation levels in SGA newborns and in newborns with the A/A rs2167270 genotype, suggesting that LEP SNPs have different effects in different populations. Another possible explanation for the discrepancy between studies is that a larger sample size may be needed to detect LEP SNPs. Also, other LEP SNPs related to obesity need to be studied for the association with non-GDM macrosomia.
When we controlled for newborn gender, gestational age, GWG, and pre-BMI, there was still a significant difference in LEP methylation levels between macrosomia and normal birth weight newborns. Multivariate logistic regression indicated that LEP methylation, LEP concentration, and pregnancy-related factors were associated with non-GDM macrosomia, suggesting that epigenetic and maternal pre-pregnancy and pregnancy characteristics may influence fetal growth and development. Nonetheless, the specific molecular mechanisms of the link between epigenetic and pregnancy-related factors and macrosomia require further study.
This study has some limitations. As it was a hospital-based case–control study, not a cross-sectional study, it is difficult to determine whether cord blood LEP concentration and methylation occurs before or after macrosomia occurs, so we could not determine a causal relationship between LEP methylation and non-GDM macrosomia, and a prospective cohort study needs to be further investigated. In addition, inherent selection bias likely existed. However, samples were collected for an entire year to rule out the effect of time on macrosomia. All participants were enrolled from the Second Affiliated Hospital of Wenzhou Medical University. Although only one hospital was included, it was a comprehensive hospital that serves patients from all 11 counties in the Wenzhou region. Thus our sample provides a basic representation of the population of the Wenzhou region in China. In addition, the study sample size was small, especially for a SNP study; thus larger samples are required to more fully analyze associations with non-GDM macrosomia.
Conclusions
Our results suggest that cord blood LEP methylation levels and maternal and fetal factors are associated with the occurrence of non-GDM macrosomia. However, further research is required to elucidate the mechanisms involved in the development of non-GDM macrosomia, which is critical for a better understanding of the origins of health and disease.
References
Henriksen, T. The macrosomic fetus: a challenge in current obstetrics. Acta Obstet. Gynecol. Scand. 87, 134–145 (2008).
Filkaszova, A. et al. Ultrasound diagnosis of macrosomia. Bratisl. Lek. Listy 115, 30–33 (2014).
Li, G. et al. Prevalence of macrosomia and its risk factors in China: a multicentre survey based on birth data involving 101,723 singleton term infants. Paediatr. Perinat. Epidemiol. 28, 345–350 (2014).
Wang, D. et al. Risk factors and outcomes of macrosomia in China: a multicentric survey based on birth data. J. Matern. Fetal Neonatal Med. 30, 623–627 (2017).
Yang, W. et al. Relationship between gestational weight gain and pregnancy complications or delivery outcome. Sci. Rep. 7, 12531 (2017).
Zhao, R. et al. Maternal pre-pregnancy body mass index, gestational weight gain influence birth weight. Women Birth 31, e20–e25 (2017).
Gillman, M. W. et al. Maternal gestational diabetes, birth weight, and adolescent obesity. Pediatrics 111, e221–e226 (2003).
Suhonen, L. et al. Detection of pregnancies with high risk of fetal macrosomia among women with gestational diabetes mellitus. Acta Obstet. Gynecol. Scand. 87, 940–945 (2008).
Eriksson, U. J. & Swenne, I. Diabetes in pregnancy: fetal macrosomia, hyperinsulinism, and islet hyperplasia in the offspring of rats subjected to temporary protein-energy malnutrition early in life. Pediatr. Res. 34, 791–795 (1993).
Barash, I. A. et al. Leptin is a metabolic signal to the reproductive system. Endocrinology 137, 3144–3147 (1996).
Houseknecht, K. L. & Spurlock, M. E. Leptin regulation of lipid homeostasis: dietary and metabolic implications. Nutr. Res. Rev. 16, 83–96 (2003).
Meinders, A. E., Toornvliet, A. C. & Pijl, H. Leptin. Neth. J. Med. 49, 247–252 (1996).
Friedman, J. M. Leptin and the regulation of body weigh. Keio J. Med. 60, 1–9 (2011).
Ben, X., Qin, Y. & Wu, S. Placental leptin correlates with intrauterine fetal growth and development. Zhonghua Yi Xue Za Zhi 81, 489–492 (2001).
Hassink, S. G. et al. Placental leptin:an important new growth factor in intrauterine and neonatal development? Pediatrics 100, e1 (1997).
Ren, R. X. & Shen, Y. A meta-analysis of relationship between birth weight and cord blood leptin levels in newborns. World J. Pediatr. 6, 311–316 (2010).
Lesseur, C. et al. Tissue-specific leptin promoter DNA methylation is associated with maternal and infant perinatal factors. Mol. Cell Endocrinol. 381, 160–167 (2013).
Kadakia, R. et al. Maternal pre-pregnancy BMI downregulates neonatal cord blood LEP methylation. Pediatr. Obes. 12, 57–64 (2017).
Obermann-Borst, S. A. et al. Duration of breastfeeding and gender are associated with methylation of the LEPTIN gene in very young children. Pediatr. Res. 74, 344–349 (2013).
Xu, X. et al. Placental leptin gene methylation and macrosomia during normal pregnancy. Mol. Med. Rep. 9, 1013–1018 (2014).
Dasgupta, S. et al. Genetic variants in leptin: determinants of obesity and leptin levels in South Indian population. Adipocyte 4, 135–140 (2015).
Rodriguez-Arroyo, G. et al. LEP, LDLR and APOA4 gene polymorphisms and their relationship with the risk of overweight, obesity and chronic diseases in adults of the State of Sucre, Venezuela. Biomedica 36, 78–90 (2016).
Pawlik, A. et al. Adiponectin and leptin gene polymorphisms in women with gestational diabetes mellitus. J. Assist. Reprod. Genet. 34, 511–516 (2017).
Kroll, C., Mastroeni, S. S., Veugelers, P. J. & Mastroeni, M. F. Association of ADIPOQ, LEP, and FTO gene polymorphisms with large for gestational age infants. Am. J. Hum. Biol. https://doi.org/10.1002/ajhb.22893 (2017).
Mazaki-Tovi, S. et al. Could alterations in maternal plasma visfatin concentration participate in the phenotype definition of preeclampsia and SGA? J. Matern. Fetal Neonatal Med. 23, 857–868 (2010).
Cai, Q. Y. et al. Placental and cord blood brain derived neurotrophic factor levels are decreased in nondiabetic macrosomia. Arch. Gynecol. Obstet. 296, 205–213 (2017).
Okpechi, I. G. et al. Genetic variation at selected SNPs in the leptin gene and association of alleles with markers of kidney disease in a Xhosa population of South Africa. PLoS ONE 5, e9086 (2010).
WHO Expert Consultation. Appropriate body-mass index for Asian populations and its implications for policy and intervention strategies. Lancet 363, 157–163 (2004).
Rasmussen, K. M., Catalano, P. M. & Yaktine, A. L. New guidelines for weight gain during pregnancy: what obstetrician/gynecologists should know. Curr. Opin. Obstet. Gynecol. 21, 521–526 (2009).
Li, H. M. et al. Association of leptin and leptin receptor gene polymorphisms with systemic lupus erythematosus in a Chinese population. J. Cell Mol. Med. 21, 1732–1741 (2017).
Masuzaki, H. et al. Nonadipose tissue production of leptin: leptin as a novel placenta-derived hormone in humans. Nat. Med. 3, 1029–1033 (1997).
Stoger, R. In vivo methylation patterns of the leptin promoter in human and mouse. Epigenetics 1, 155–162 (2006).
Dotsch, J. et al. Leptin and neuropeptide Y gene expression in human placenta: ontogeny and evidence for similarities to hypothalamic regulation. J. Clin. Endocrinol. Metab. 84, 2755–2758 (1999).
Hogg, K., Blair, J. D., von Dadelszen, P. & Robinson, W. P. Hypomethylation of the LEP gene in placenta and elevated maternal leptin concentration in early onset pre-eclampsia. Mol. Cell Endocrinol. 367, 64–73 (2013).
Bjorbaek, C. et al. The role of SOCS-3 in leptin signaling and leptin resistance. J. Biol. Chem. 274, 30059–30065 (1999).
Liao, G. Y. et al. Dendritically targeted Bdnf mRNA is essential for energy balance and response to leptin. Nat. Med. 18, 564–571 (2012).
Acknowledgements
We thank Xin-Yun Xu for assistance with collecting cord blood samples and Zi-Wei Liu for reviewing and providing advice on the preparation of the manuscript. This work was supported by the National Natural Science Foundation of China (to X.-J.Y., No. 81072378) and the Natural Science Funds of Zhejiang (to X.-J.Y., No. Y2101185).
Author information
Authors and Affiliations
Contributions
H.-T.Y. and X.-J.Y. conceived and designed the study. Y.-H.W. was responsible for collecting the cord blood samples and data. Y.-C.W., Y.H. and Z.-F.L. conducted the statistical analyses. X.-X.X. and X.-J.Y. wrote the manuscript. All authors participated in discussions, made contributions to the manuscript, and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wang, YH., Xu, XX., Sun, H. et al. Cord blood leptin DNA methylation levels are associated with macrosomia during normal pregnancy. Pediatr Res 86, 305–310 (2019). https://doi.org/10.1038/s41390-019-0435-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41390-019-0435-3
This article is cited by
-
Genetic variation, intrauterine growth, and adverse pregnancy conditions predict leptin gene DNA methylation in blood at birth and 12 months of age
International Journal of Obesity (2020)