Abstract
Pancreatic cancer has an extremely terrible prognosis and is a common cause of cancer death. In this study, the clinic value, biological function and underlying mechanisms of Zinc finger protein 655 (ZNF655) in human pancreatic cancer were evaluated. The expression level of ZNF655 in pancreatic cancer was determined by immunohistochemistry (IHC) staining. The biological effects of ZNF655 in pancreatic cancer cells was investigated by loss/gain-of-function assays in vitro and in vivo. The downstream molecular mechanism of ZNF655 was explored using co-immunoprecipitation (Co-IP), dual-luciferase reporter and chromatin immunoprecipitation (Ch-IP). ZNF655 expression was significantly elevated in human pancreatic cancer and possessed clinical value in predicting poor prognosis. Functionally, ZNF655 knockdown inhibited the biological progression of pancreatic cancer cells, which was characterized by weaken proliferation, enhanced apoptosis, arrested cell cycle in G2, impeded migration, and suppressed tumor growth. Mechanistically, ZNF655 played an important role in promoting the binding of E2F transcription factor 1 (E2F1) to the cyclin-dependent kinase 1 (CDK1) promoter. Furthermore, knockdown of CDK1 alleviated the promoting effects of ZNF655 overexpression in pancreatic cancer cells. The promotive role of ZNF655 in pancreatic cancer via CDK1 was determined, which drew further interest regarding its clinical application as a promising therapeutic target.
Similar content being viewed by others
Introduction
Pancreatic cancer is a malignancy of the digestive tract that originating from the glandular epithelium, responsible for substantial morbidity and mortality worldwide [1]. The sobering reality is that pancreatic cancer has an extremely poor prognosis, with a 5-year survival rate of only about 10%, and it is becoming a common cause of cancer death [2]. Pancreatic cancer is generally classified into four types according to the degree of disease: resectable, marginally resectable, locally advanced and metastatic [3]. For a small number of patients diagnosed with resectable tumors, surgical resection is a great opportunity of curing the disease [4]. Unfortunately, only 20% of patients can survive 5 years after surgery [5]. Poor prognosis and widespread drug resistance continue to plague pancreatic cancer patients. Recently, with the development of genomics and proteomics, the exploration of targeted drugs for precision medicine has become a hot spot. For instance, targeted medicine that overexpress or mutation-related pathway dysfunction of epidermal growth factor receptor (EGFR) [6], vascular endothelial growth factor (VEGF) [7, 8], Notch [9], Hedgehog [10], mitogen-activated protein extracellular signal-regulated kinase (MEK) [11], and KRAS [12] common in pancreatic tumors are being extensively investigated. Although trials of these novel combination therapies have been modestly successful in the clinic, the overall survival of pancreatic cancer has been little improved [13]. To that end, further exploration on pancreatic cancer is extremely important for the identification of novel targets.
Zinc finger (ZNF) proteins are the largest family of transcription factors in the human genome and have a variety of molecular functions [14]. The different combinations of ZNF motifs lead to diverse functions of ZNF proteins in biological processes such as development, differentiation, metabolism and autophagy [15, 16]. Over the past few decades, accumulating evidence has been shed on the potential role of ZNF proteins in cancer progression [17]. However, the underlying mechanism of zinc finger protein in cancer varies among different cancer types [18]. For the most part, Cys2His2 ZNF proteins act as trans regulator of gene expression, which exert key role in cellular processes such as cell development, differentiation and inhibition of malignant cell transformation [19]. Numerous reports have shown a direct relationship between apoptosis resistance, chemotherapy resistance and increased expression of ZNF703. Moreover, ZNF703 has oncogene effects in breast cancer [20]. In addition, reduced expression of ZNF233 in hepatocellular carcinoma inhibits cell proliferation and tumorigenesis [21]. In contrast, ZNF554 is regarded as a potential tumor suppressor in malignant gliomas, and its reduced expression may lead to the loss of oncogene suppression and the activation of tumor pathways [22]. As a transcription factor of Cys2His2 type ZNF, the role of ZNF655 in pancreatic cancer has not been reported.
At present study, we evaluated the expression level, biological function and underlying mechanisms of ZNF655 in human pancreatic cancer. ZNF655 was abundantly expressed in pancreatic cancer, and its high expression was significantly correlated with poor prognosis. Reduced expression of ZNF655 resulted in reduced malignant phenotypes in pancreatic cancer cells. Mechanistically, ZNF655 played an important role in promoting the binding of E2F1 to the CDK1 promoter. Furthermore, knockdown of CDK1 alleviated the promoting effects of ZNF655 overexpression in pancreatic cancer cells. These findings suggested that ZNF655 had a stimulative effect on pancreatic cancer via CDK1 and was a candidate target for molecular therapy.
Materials and methods
Pancreatic cancer tissue collection and cell culture
Human survival pancreatic cancer tissues (n = 97) and paired pancreatic tissues (n = 69) were collected during surgical resection at Beijing Chaoyang hospital. The experimental procedures of this study were approved by the ethics committee of Beijing Chaoyang hospital (No. 2019-S-243) and all patients signed an informed consent form. Pancreatic cancer cell lines (PANC-1, SW1990 and BXPC-3) and normal pancreatic cell line HPDE6-C7 were purchased from Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The cells were cultured in DMEM supplemented with 10% fetal bovine serum (Invitrogen Gibco) and maintained in a 37 °C, 5% CO2 incubator.
Immunohistochemistry (IHC) staining
Formalin-immobilized tissues were dewaxed in xylene, rehydrated in ethanol solution, incubated with 3% hydrogen peroxide to block endogenous peroxidase and nonspecific binding sites. The tissues were incubated with the primary anti-ZNF655 (1:200, Invitrogen, PA5-56183), anti-CDK1 (1:50, Sigma, HPA003387) antibody at 4 °C overnight and then with the secondary antibody HRP goat anti-rabbit IgG (1:200, Beyotime, A0208) at room temperature for 1 h. After incubation with peroxidase conjugated streptavidin and diaminobenzidine, hematoxylin was stained. The staining intensity was scored according to the criteria described in the literature [23]. The result greater than or equal to median values of IHC was defined as ZNF655 high expression, otherwise low expression.
Lentivirus transfection and cell infection
Three shRNAs against ZNF655 or CDK1 interference sequences were synthesized (shZNF655-1, 5'-GCCCAGGAAGCAGCAGGGTCA-3'; shZNF655-2, 5'-CACCGACATGGAACAGGGACT-3'; shZNF655-3, 5'-TCCAGTTTCAGTCTTTGGAGA-3'; shCDK1-1, 5'-TTCCATGGATCTGAAGAAATA-3'; shCDK1-2, 5'-AGACTAGAAAGTGAAGAGGAA-3'; shCDK1-3, 5'-ATGGAGTTGTGTATAAGGGTA-3') and the lowest expression interference sequence of ZNF655 or CDK1 was selected, respectively. The fragment was inserted into lentiviral vector BR-V108 with green fluorescent protein (GFP) (BIOSCIRES). The 293 T cells were co-transfected with 10 μg recombinant BR-V108 vector and virus packaging plasmid (7.5 μg pMD2.G and 5 μg pSPAX2) (BIOSCIRES) for 72 h. Meanwhile, the amplified sequence of ZNF655 (ZNF655) was synthesized and inserted into lentiviral vector LV-003 (BIOSCIRES) to construct overexpression of ZNF655. After that, 2 × 105 PANC-1 and SW1990 cells were cultured for 24 h and infected with lentivirus shZNF655, shCDK1 and ZNF655 (1 × 108 TU/mL) at a MOI (multiplicity of infection) of 10. Finally, the expression of GFP was observed under fluorescence microscope (OLYMPUS) 72 h after lentivirus infection.
RNA extraction and qPCR
PANC-1 and SW1990 cells RNA was purified with Trizol [24] and reverse transcribed into cDNA using the Maxima First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). Quantification of mRNA expression levels of ZNF655 and CDK1 was accomplished by SYBR Green master mix (Thermo Fisher Scientific) by ABI Prism 7500 sequence detection system (Applied Biosystems) with normalization to the expression of GAPDH. The primer sequences were listed in Table S1.
Western blotting (WB) and Co-immunoprecipitation (Co-IP) assay
PANC-1 and SW1990 cells protein was purified with RIPA (Beyotime) and the concentration was detected using BCA Protein Assay Kit (Beyotime). The 20 μg/well total protein was subjected to 10% SDS–PAGE, transferred to PVDF membrane (Millipore), hybridized with corresponding primary antibody (Table S2) overnight at 4 °C, incubated with secondary antibody at room temperature for 2 h. After that, protein signal was visualized through chemiluminescence ECL kit (Thermo Fisher Scientific) and GAPDH as load control. ZNF655 and CDK1 protein–protein interaction was analyzed by Co-IP assay and the experimental procedures were performed as previously described [25].
MTT and cell counting assay
PANC-1 and SW1990 cells were cultured in 96-well plates at a density of 2000 cells/well. Cell viability was detected by MTT assay for 5 days [26] and cell proliferation ability was analyzed by drawing growth curve. The cells with GFP were identified with Celigo, photographed, counted, and the cell growth curve was plotted for 5 days.
Detection of cell cycle and apoptosis
PANC-1 and SW1990 cells were inoculated into 6-well plates (2 mL/well) for 5 days.
For apoptosis analysis, the cell precipitates were successively washed by precooled D-hanks (pH = 7.2~7.4) and 1×binding buffer (eBioscience), resuspended by 200 μL 1× binding buffer, stained with 10 μL Annexin V-APC (eBioscience) at room temperature in the dark for 15 min and detected by flow cytometry (Millipore). For cell cycle analysis, the cells were centrifuged for 5 min, the cell precipitates were eluted with precooled PBS (pH = 7.2~7.4), fixed with 70% ethanol for at least 1 h, stained with PI (Sigma) and monitored by flow cytometry.
Human apoptosis antibody array assay
PANC-1 cells protein was purified with RIPA (Beyotime) and the concentration was detected using BCA Protein Assay Kit (Beyotime). The concentrations of 43 kinds of human apoptotic markers in cell lysate were simultaneously detected in strict accordance with the instructions of the kit (Human Apoptosis Antibody Array–Membrane, Abcam). After that, the signal of apoptotic markers was visualized through chemiluminescence ECL kit (Thermo Fisher Scientific) and quantified by Image J software (National Institutes of Health).
Wound-healing assay
PANC-1 and SW1990 cells were cultured in 6-well plates (100 μL/well) at a density of 4000 cells/well. The specific experimental procedures followed the description in the literature [27]. In order to avoid the influence of cell proliferation on migration, the cells were placed in a serum-free medium and treated with mitomycin for 1 h before performing wound-healing experiments. A line wound was drawn by a pipette tip across the cell layer. The cells were washed with PBS, fixed with 3.7% paraformaldehyde (Corning) for 15 min, stained with 1% crystal violet (Corning) for 10 min. The cells were viewed under a microscope for image acquisition and Image J software (National Institutes of Health) was used to quantify the distance (μm) between the scratches at 0 h, 8 h and 72 h.
Transwell assay
PANC-1 and SW1990 cells at a density of 80,000 cells/well were cultured into Transwell chambers (24-well, 8-mm pore, Corning) for 24 h at 37 °C, of which 100 μL cell suspension in the inner compartment and 500 μL DMEM medium containing 30% FBS in the outer compartment. The non-invading cells on the upper chamber were removed, while the cells adhering to the Polycarbonate membrane were fixed with 4% precooled paraformaldehyde for 30 min and stained with 0.1% crystal violet for 20 min at room temperature. Finally, the cells were photographed from five randomly selected fields under a 200× microscope.
Gene microarray
After PANC-1 cells infected with lentivirus shZNF655 and shCtrl, RNA was purified and sequenced through Affymetrix human Gene Microarray Prime View (Affymetrix Scanner 3000 scan) to recognize differentially expressed genes (DEGs). DEGs was selected with criterion of |Fold Change | ≥ 1.3 and false discovery rate (FDR) ≤ 0.05 and presented as a volcano plot and hierarchical clustering. Significant enrichment of DEGs in classical pathways, disease and function, and interaction networks was explored based on Ingenuity Pathway Analysis (IPA).
Dual-luciferase reporter assay
The CDK1 promoter region was amplified and the fragment was cloned into the luciferase reporter vector GL002 (Promega Madison, USA), named as GL002- CDK1. Mutant construct GL002-CDK1-Mut was generated by site-directed mutagenesis. Luciferase assay was performed as described previously [28]. Each experimental analysis was repeated three times.
Chromatin immunoprecipitation (Ch-IP) assay
According to the manufacturer’s instructions, Ch-IP assay was performed using the Simple ChIP® Enzymatic Chromatin IP Kit (Cat No, 9002 S, CST, USA). In brief, the cells were transfected with ZNF655-overexpressing vector or its empty vector and incubated for 48 h at 37 °C with 5% CO2. Subsequently, the cells were crosslinked with 37% formaldehyde followed by lysed in SDS buffer and sheared sonication to fragment the DNA. Afterwards, the sonicated chromatin was precipitated by incubating it with according antibody overnight at 4 °C. The Protein–DNA complexes were then purified and the purified DNA was dissolved in nuclease-free water followed by qPCR analysis using the primers of CDK1 promoter and SYBR Green I Master (Roche, USA).
In vivo xenograft model of mice
The experimental procedures performed on mice were approved by the Ethics Committee of Beijing Chaoyang Hospital and the Animal Protection Association. The total of 20 BALB/c nude SPF mice (18–22 g, 4–6 weeks) were obtained from Viton Lihua Laboratory Animal Technology Co., Ltd. The 500 μL SW1990 cells infected with lentivirus shZNF655 (n = 5) and shCtrl (n = 5) were subcutaneously injected into the right armpit of mice (1 × 107 cells/mouse), respectively. Tumor volumes of mice was monitored 7 days after injection and then collected once or twice a week. Notably, the tumor volume was calculated using the formula π/6 × L × W × W (L: longest dimension, W: dimension perpendicular to length). After 26 days, mice were sacrificed by intraperitoneal injection of 0.7% pentobarbital sodium at a dose of 10 μL/g, and the tumors were removed for taking photos and weighting. Finally, the signal intensity of ZNF655 and CDK1 in mice tumor tissues was determined by IHC staining.
Statistical analysis
All data were presented as means ± SD and P < 0.05 were considered statistically significant. Comparisons between different groups were conducted by Student’s t-test or ANOVA as appropriate. The clinical relevance between ZNF655 and clinicopathological characteristics of pancreatic cancer patients was analyzed by univariate and multivariate analyses and Kaplan–Meier method.
Results
ZNF655 is highly expressed in human pancreatic cancer and predicts poor prognosis
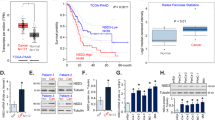
To clarified the clinical relevance of ZNF655 in the progression of human pancreatic cancer, we first examined expression level of ZNF655 in primary pancreatic cancer samples by IHC staining. The signal expression intensity of ZNF655 in pancreatic cancer tissues was significantly higher than that in adjacent normal pancreatic tissues (Fig. 1A, Table 1). According to the eighth edition of the American Joint Committee on Cancer (AJCC) and the Union for International Cancer Control (UICC), the clinical relevance between ZNF655 and clinicopathological characteristics of pancreatic cancer patients was analyzed. High expression of ZNF655 was observed in 21 of 66 low-grade pancreatic cancer (WHO II; 13.13%) and in 22 of 31 high-grade pancreatic cancer (WHO III–IV; 70.97%) (Table 2), suggesting that increased ZNF655 expression was found in higher tumor grade. Furthermore, correlation analysis between the expression level of ZNF655 (high and low) and the overall survival of patients with pancreatic cancer (Grade II, III and IV) was performed using Kaplan–Meier survival. As the expression of ZNF655 increased, the survival time was shortened, suggesting that higher expression of ZNF655 predicted a worse prognosis in patients with pancreatic cancer (Fig. 1B). Collectively, ZNF655 expression was significantly elevated in human pancreatic cancer and had clinical value in predicting poor prognosis.
A The expression level of ZNF655 in pancreatic cancer was determined by IHC staining and representative images were shown. Magnification is 200 and 400. B The clinical relevance between ZNF655 and Overall survival of pancreatic cancer patients was analyzed by Kaplan–Meier method. *P = 0.032. C The mRNA expression level of ZNF655 in pancreatic cancer cell lines (PANC-1, SW1990 and BXPC-3) and normal pancreatic cell line HPDE6-C7 was identified.
Knockdown of ZNF655 inhibits proliferation, promotes apoptosis and impedes migration of pancreatic cancer cells
Additionally, the mRNA expression level of ZNF655 in pancreatic cancer cell lines PANC-1 and SW1990 was significantly higher than that of normal pancreatic cell line HPDE6-C7 (Fig. 1C). In order to further determine the biological functions of ZNF655 in human pancreatic cancer, a series of cell functional experiments were carried out in vitro. The sequences of shRNA targeting ZNF655 (shZNF655-1/2/3) were synthesized and the two sequences with reduced expression of ZNF655 (shZNF655-1/2) were screened out (Fig. S1A). Compared with shCtrl, the protein expression of ZNF655 in PANC-1 and SW1990 cells was significantly reduced after the lentivirus shZNF655-1 and shZNF655-2 were transfected (Fig. S1B). Subsequently, knockdown of ZNF655 in PANC-1 and SW1990 cells was used to investigate the role of ZNF655 in pancreatic cancer. Cell growth curves based on the MTT assay demonstrated that ZNF655 knockdown significantly decreased cell viability and proliferation in PANC-1 and SW1990 cells (Fig. 2A). Furthermore, we examined the cell apoptosis and cycle progression in response to downregulation of ZNF655. The apoptotic index of PANC-1 and SW1990 was significantly increased after ZNF655 expression was reduced, as detected by flow cytometry (Fig. 2B). At the same time, we observed that knockdown of ZNF655 in PANC-1 and SW1990 cells with an accumulation of cells in the G2 phase and a decrease in the S phase (Fig. 2C). In wound-healing assays, the PANC-1 and SW1990 cells transfected with shZNF655-1 and shZNF655-2 lentivirus displayed impairment of migration ability when compared with cells transfected with shCtrl (Fig. 2D). These findings demonstrated that the proliferation and migration were inhibited, cell cycle was arrested in G2, while the apoptosis was enhanced in ZNF655-knockdown PANC-1 and SW1990 cells.
A The proliferation of PANC-1 and SW1990 cells after knockdown of ZNF655 was measured using MTT assay. B, C Cell apoptosis and cycle of PANC-1 and SW1990 cells after knockdown of ZNF655 was analyzed by flow cytometry. D The migration of PANC-1 and SW1990 cells after knockdown of ZNF655 was measured using wound-healing assay. E The expression of typical proteins related to apoptosis, cycle and migration was detected in pancreatic cancer cells. shCtrl indicates PANC-1 and SW1990 cells infected with a vector-expressing GFP; shZNF655 indicates ZNF655 knockdown in PANC-1 and SW1990 cells. The presented results were representative of experiments repeated at least three times. Data was represented as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
Knockdown of ZNF655 regulates the expression of typical proteins related to apoptosis, cycle and migration
In addition, the protein expression of human apoptosis signaling pathway related regulator was detected in PANC-1 cells with reduced ZNF655 expression. Compared with the control group, the expression of Bax (BCL2 associated X), BIM (BCL2 like 11), Caspase3, cytoC (cytochrome c), IGFBP-6 (insulin-like growth factor binding protein-6), p21, p27 and p53 was upregulated in shZNF655 group, while the expression of Survivin, IGFBP-2 (insulin-like growth factor binding protein-2) and STNF-R2 (soluble tumor necrosis factor-receptor 2) was downregulated (Fig. S2C). Moreover, the expression of typical proteins related to apoptosis, cycle and migration was detected in pancreatic cancer cells. The results showed that the knockdown of ZNF655 upregulated Bax, p21, p53, and downregulated CCND1, E-cadherin, and N-cadherin (Fig. 2E). Previous study reported that phosphatidylinositol 3-kinase (PI3K)/AKT signaling pathway is one of the most downregulated pathways in human cancers, which is involved in a variety of carcinogenic processes, including cell proliferation, growth, survival, apoptosis and metastasis [29]. In this study, WB results showed that ZNF655 knockdown resulted in downregulation of p-AKT and PIK3CA (Fig. 2E).
ZNF655 promotes the binding of E2F1 to CDK1 promoter in pancreatic cancer cells
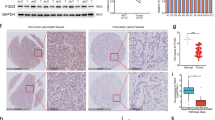
Furthermore, gene microarray results showed that ZNF655 knockdown resulted in 933 upregulated genes and 1430 downregulated genes (Fig. 3A). Additionally, significant enrichment of DEGs in disease and function was explored based on IPA (Fig. S2A), such as Cancer, Organismal Injury and Abnormalities, Cell Death and Survival. Fig. S2B showed the significant enrichment of DEGs in the classical pathway, such as TNFR2 signaling, Cell Cycle: G2/M DNA Damage Checkpoint Regulation. Moreover, the interaction network between ZNF655 and Cell Cycle: G2/M DNA Damage Checkpoint Regulation pathway was illustrated in the Fig. S2C. Accordingly, the expression levels of the first 19 DEGs most significantly enriched in these functions and pathways were further detected by qPCR and WB, respectively (Figs. S2D, 3B). We found that knockdown of ZNF655 downregulated CDK1. Considering that CDK1 is a typical cell cycle regulator and has promoted the progression of pancreatic cancer [30]. Consistently, the expression of CDK1 in pancreatic cancer tissues was higher than that in adjacent normal tissues, suggesting that CDK1 may exhibit an important role in pancreatic cancer. Additionally, the mRNA expression level of CDK1 in pancreatic cancer cell lines PANC-1 and SW1990 was significantly higher than that of normal pancreatic cell line HPDE6-C7 (Fig. 3D). Furthermore, we stably knocked down CDK1 in PANC-1 cells using lentiviral shCDK1 (Fig. S2E) and the loss-of-function results showed a significant inhibition in the proliferation and migration of CDK1-knocked-down PANC-1 cells (Fig. 3E–H).
A The DEGs between shZNF655 and shCtrl groups of PANC-1 cells were identified. In the heat map of cluster analysis, each column represents a sample and each row represents a differential gene. The red indicates that the gene expression is upregulated, the green indicates that the gene expression is downregulated, the black indicates that the gene expression is not significantly changed, and the gray indicates that the signal strength of the gene is not detected. B The expression of several selected DEGs of PANC-1 cells after knockdown of ZNF655 was measured by WB. C The expression level of CDK1 in pancreatic cancer was detected by IHC staining and representative images were shown. Magnification is 200 and 400. D The mRNA expression level of CDK1 in pancreatic cancer cell lines (PANC-1, SW1990 and BXPC-3) and normal pancreatic cell line HPDE6-C7 was identified by qPCR. E–H Effects of knockdown of CDK1 on proliferation, apoptosis and migration of pancreatic cancer cells. shCtrl indicates PANC-1 cells infected with a vector-expressing GFP; shCDK1 indicates CDK1 knockdown in PANC-1 cells. I Cell lysates from PANC-1 cells were subjected to Co-IP, and anti-E2F1 antibody was used for IP (immunoprecipitation). IgG antibody, a negative control group, were designed to exclude nonspecific immune responses. Input, positive control, refers to using the lysate before IP as IB (immunoblotting) to exclude false positive interference. J We mutated the promoter region of CDK1 and performed a dual-luciferase reporter experiment to observe the effects of ZNF655 and E2F1 on the CDK1 promoter activity. K Chromatin immunoprecipitation (Ch-IP) assay confirmed that overexpression of ZNF655 induced E2F1 to be recruited to the CDK1 promoter region. The presented results were representative of experiments repeated at least three times. Data was represented as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
Combined with our data, we inferred that there may be a connection between ZNF655 and CDK1. Moreover, it is reported that E2F1 binds to the upstream region of CDK1 to accelerate transcription and upregulate its protein expression [31]. Here, cell lysates from PANC-1 cells were subjected to Co-IP, and anti-E2F1 antibody was used for IP. IgG antibody, a negative control group, were designed to exclude nonspecific immune responses. Interestingly, the present study indicated that there was interaction between ZNF655 and E2F1 (Fig. 3I). Therefore, we hypothesized that ZNF655 regulated CDK1 expression through E2F1. Subsequently, we mutated the promoter region of CDK1 and performed a dual-luciferase reporter experiment to observe the effects of ZNF655 and E2F1 on the CDK1 promoter activity. E2F1 could significantly enhance the luciferase activity of CDK1-WT but not CDK1-MUT, suggesting that presence of combination between E2F1 and CDK1. Moreover, overexpression of ZNF655 and E2F1 significantly enhance the activity of CDK1. The results indicated that overexpression of ZNF655 significantly promoted the combination of E2F1 and CDK1 (Fig. 3J). Ch-IP assay further confirmed that overexpression of ZNF655 induced E2F1 to be recruited to the CDK1 promoter region (Fig. 3K). Taken together, these data suggested that ZNF655 facilitated malignant behaviors of pancreatic cancer cells via promoting the binding of E2F1 to CDK1 promoter.
Knockdown of CDK1 alleviates the promoting effects of ZNF655 overexpression in pancreatic cancer cells
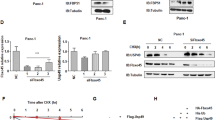
Functional recovery experiments were conducted to clarify whether there was a synergistic effect between ZNF655 and CDK1 in pancreatic cancer. PANC-1 cells overexpressing ZNF655 lead to a promotion in proliferation and migration, as well as an inhibition in apoptosis (Fig. 4A–D). Furthermore, the overexpression of ZNF655 and the knockdown of CDK1 (shCDK1 + ZNF655) were established in PANC-1 cells (Fig. S2F). The malignant progression of PANC-1 cells in shCDK1 + ZNF655 group was significantly inhibited compared with ZNF655, which was characterized by weaken proliferation (P < 0.001), impeded migration and enhanced apoptosis (P < 0.001) (Fig. 4A–D). Taken together, the loss/gain-of-function assays demonstrated that knockdown of CDK1 could alleviate the promoting effects of ZNF655 overexpression in PANC-1 cells.
Detection of alteration in proliferation (A), migration (B, C), apoptosis (D) after lentivirus ZNF655 and shCDK1 + ZNF655 infects PANC-1 cells. Control indicates PANC-1 cells infected with empty vector LV-003, as negative control; ZNF655 indicates ZNF655 overexpression in PANC-1 cells; NC(KD + OE) indicates PANC-1 cells infected with empty vector LV-003 and BR-V108, as negative control; shCDK1 + ZNF655 indicates simultaneously downregulated CDK1 and upregulated ZNF655 in PANC-1 cells. The presented results were representative of experiments repeated at least three times. Data was represented as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
Knockdown of ZNF655 suppresses tumor growth in the mouse xenograft model
To further verify the effects of ZNF655 in vivo, SW1990 cells were injected subcutaneously into the BALB/c nude mice and divided into shCtrl group and shZNF655 group. Monitoring of mice 26 days after the cell injection showed that that the tumor volume of shZNF655 group was significantly weaker than that of the control group (P < 0.01) (Fig. 5A). Consistently, the tumor weight of shZNF655 group was significantly lighter than that of the control group (Fig. 5B). Moreover, IHC staining performed on tumor sections from xenografts showed that ZNF655 and CDK1 expression level was markedly reduced in the ZNF655 knockdown group compared with the control group (Fig. 5C). Collectively, these results highlighted that reduced expression of ZNF655 suppressed the tumor growth in vivo.
A Post injection of SW1990 cells for 7 days, the tumor volume in mice was measured. B Mice were sacrificed at day 26 post injection, and the tumor weight was measured and photographed. C The expression of ZNF655 and CDK1 in mice tumor tissues was detected by IHC staining. Data was represented as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
A significant finding of this study is the identification of a promoting role of ZNF655 in human pancreatic cancer. Previous evidence has shown that the development and progression of human pancreatic cancer is a complex, multifactorial process with multiple genetic abnormalities [3]. The characteristic of early pancreatic intraepithelial tumors is that mutations in the oncogene KRAS lead to the activation of the RAS/RAF and PI3K/AKT signaling pathways, which are intracellular signaling pathways that regulate the cell cycle [32]. In this study, we found that that malignant progression of ZNF655 knockdown pancreatic cancer cells was significantly inhibited, which was characterized by weaken proliferation, impeded migration, arrested cell cycle in G2 and enhanced apoptosis. In fact, ZNF655 knockdown by shRNA specifically enhanced the apoptotic sensitivity of pancreatic cancer cells, while ZNF655 overexpression resulted in reduced apoptotic capacity. In this way, why a single TF perturbs so many different tumor-promoting features still stands. we found that ZNF655, a TF, can affect a variety of functions in a variety of cancers. For example, Chen et al., indicated that ZNF655 promotes the progression of glioma through transcriptional regulation of AURKA [33]. Teng et al., demonstrated that the decrease of ZNF655 expression led to the inhibition of the malignant behaviors of non-small cell lung cancer, which was manifested by weakened proliferation, increased sensitivity to apoptosis, cycle repression at G2 and weakened migration [34]. Yang et al., identified role of ANKHD1/LINC00346/ZNF655 feedback loop in regulating the glioma angiogenesis via staufen1-mediated mRNA decay [35]. In general, ZNF655 may affect multiple functions of cancer by targeting certain genes.
Furthermore, ZNF655 knockdown resulted in downregulation of CCND1 expression and inhibition of PI3K/AKT signaling pathway in pancreatic cancer cells. Additionally, the characteristics of early lesions also include the inactivation of the tumor suppressor gene CDKN2A and the overexpression of the oncogene CDKN1A, which promote the transition of the cell cycle from the G1 phase to the S phase [36]. In later lesions, two key tumor suppressor genes, Smad4 and TP53, were inactivated. The inactivation of Smad4 and its coding protein blocks the typical downstream signaling of transforming growth factor β (TGFβ) receptor, while the inactivation of TP53 and its coding protein P53 promote the progression of cell cycle, survival and inhibition of apoptosis [37, 38]. Furthermore, ZNF655 knockdown upregulated the expression of Bax, BIM, Caspase3, cytoC, IGFBP-6, p21, p27 and p53, while downregulated the expression of Survivin, IGFBP-2 and STNF-R2 in PANC-1 cells. ZNF655 knockdown can promote the apoptosis of pancreatic cancer cells, which required the co-participation of a series of pro-apoptotic and anti-apoptotic factors. Indeed, ZNF655 may affect tumor progression through multiple pathways. Given that apoptosis is one of the biological phenotypes of cancer, this study focused on the effect of ZNF655 knockdown on apoptosis and the expression of apoptosis-related proteins. Of course, it is impossible to study all biological pathways related to cancer in one project. Therefore, we will delve into other biological processes related to cancer in future work.
The downstream mechanism of ZNF655 regulating pancreatic cancer progression was preliminarily investigated and found that knockdown of ZNF655 downregulated CDK1. To our knowledge, CDK1 is responsible for driving cell division and coordinating the transition from G2 phase to mitosis [39]. Uncontrolled proliferation signaling leads to dysregulation of the cell cycle, which is considered a hallmark of cancer [40]. Recently, it has been reported that that CDK1 is a typical cell cycle regulator and has promoted the progression of pancreatic cancer [30]. On the other hand, NCAPD2 could affect tumor cell proliferation, apoptosis, migration and invasion by targeting CDK1 in breast cancer [41]. Consistently, we stably knocked down CDK1 in PANC-1 cells using lentiviral shCDK1 and the loss-of-function results showed a significant inhibition in the proliferation and migration of CDK1-knocked-down PANC-1 cells. Additionally, knockdown of CDK1 alleviated the promoting effects of ZNF655 overexpression in pancreatic cancer cells. As a consequence, ZNF655 promoted the malignant behavior of pancreatic cancer cells through CDK1.
Combined with our data, we inferred that there may be a connection between ZNF655 and CDK1. Moreover, it was reported that E2F1 binds to the upstream region of CDK1 to accelerate transcription and upregulated its protein expression [31]. Interestingly, the present study indicated that there was interaction between ZNF655 and E2F1. In addition, dual-luciferase reporter experiment and Ch-IP assay further confirmed that overexpression of ZNF655 induced E2F1 to be recruited to the CDK1 promoter region. Mechanically, these data suggested that ZNF655 facilitated malignant behaviors of pancreatic cancer cells via promoting the binding of E2F1 to CDK1 promoter.
ZNF655 expression was significantly elevated in human pancreatic cancer and possessed clinical value in predicting poor prognosis. ZNF655 facilitated malignant behaviors of pancreatic cancer cells via promoting the binding of E2F1 to CDK1 promoter, which may contribute to the development of promising targets for tumor diagnosis and treatment.
Data availability
The data used and analyzed during the current study are available from the corresponding author on reasonable request.
References
Puckett Y, Garfield K. Pancreatic cancer. StatPearls. Treasure Island (FL): StatPearls; 2021.
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30.
Mizrahi JD, Surana R, Valle JW, Shroff RT. Pancreatic cancer. Lancet 2020;395:2008–20.
Masiak-Segit W, Rawicz-Pruszynski K, Skorzewska M, Polkowski WP. Surgical treatment of pancreatic cancer. Pol Przegl Chir. 2018;90:45–53.
Strobel O, Neoptolemos J, Jager D, Buchler MW. Optimizing the outcomes of pancreatic cancer surgery. Nat Rev Clin Oncol. 2019;16:11–26.
Grapa CM, Mocan T, Gonciar D, Zdrehus C, Mosteanu O, Pop T, et al. Epidermal growth factor receptor and its role in pancreatic cancer treatment mediated by nanoparticles. Int J Nanomed. 2019;14:9693–706.
Castro BA, Aghi MK. Bevacizumab for glioblastoma: current indications, surgical implications, and future directions. Neurosurg Focus. 2014;37:E9.
Masuda C, Yanagisawa M, Yorozu K, Kurasawa M, Furugaki K, Ishikura N, et al. Bevacizumab counteracts VEGF-dependent resistance to erlotinib in an EGFR-mutated NSCLC xenograft model. Int J Oncol. 2017;51:425–34.
Ristorcelli E, Lombardo D. Targeting Notch signaling in pancreatic cancer. Expert Opin Ther Targets. 2010;14:541–52.
Skoda AM, Simovic D, Karin V, Kardum V, Vranic S, Serman L. The role of the Hedgehog signaling pathway in cancer: a comprehensive review. Bosn J Basic Med Sci. 2018;18:8–20.
Lee S, Heinrich EL, Lu J, Lee W, Choi AH, Luu C, et al. Epidermal growth factor receptor signaling to the mitogen activated protein kinase pathway bypasses Ras in pancreatic cancer cells. Pancreas 2016;45:286–92.
Waters AM, Der CJ. KRAS: The critical driver and therapeutic target for pancreatic cancer. Cold Spring Harb Perspect Med. 2018;8:a031435.
Miller AL, Garcia PL, Yoon KJ. Developing effective combination therapy for pancreatic cancer: an overview. Pharm Res. 2020;155:104740.
Cassandri M, Smirnov A, Novelli F, Pitolli C, Agostini M, Malewicz M, et al. Zinc-finger proteins in health and disease. Cell Death Disco. 2017;3:17071.
Brayer KJ, Kulshreshtha S, Segal DJ. The protein-binding potential of C2H2 zinc finger domains. Cell Biochem Biophys. 2008;51:9–19.
Font J, Mackay JP. Beyond DNA: zinc finger domains as RNA-binding modules. Methods Mol Biol. 2010;649:479–91.
Ye Q, Liu J, Xie K. Zinc finger proteins and regulation of the hallmarks of cancer. Histol Histopathol. 2019;34:1097–109.
Jen J, Wang YC. Zinc finger proteins in cancer progression. J Biomed Sci. 2016;23:53.
Razin SV, Borunova VV, Maksimenko OG, Kantidze OL. Cys2His2 zinc finger protein family: classification, functions, and major members. Biochem (Mosc). 2012;77:217–26.
Marzbany M, Bishayee A, Rasekhian M. Increased expression of ZNF 703 in breast cancer tissue: an opportunity for RNAi-NSAID combinatorial therapy. Biotechnol Appl Biochem. 2019;66:808–14.
Xie W, Qiao X, Shang L, Dou J, Yang X, Qiao S, et al. Knockdown of ZNF233 suppresses hepatocellular carcinoma cell proliferation and tumorigenesis. Gene 2018;679:179–85.
Balogh A, Reiniger L, Hetey S, Kiraly P, Toth E, Karaszi K, et al. Decreased expression of ZNF554 in gliomas is associated with the activation of tumor pathways and shorter patient survival. Int J Mol Sci. 2020;21:5762.
Crowe AR, Yue W. Semi-quantitative determination of protein expression using immunohistochemistry staining and analysis: an integrated protocol. Bio Protoc. 2019;9:e3465.
Rio DC, Ares M Jr, Hannon GJ, Nilsen TW. Purification of RNA using TRIzol (TRI reagent). Cold Spring Harb Protoc 2010;2010:pdb prot5439.
Tang Z, Takahashi Y. Analysis of protein-protein interaction by Co-IP in human cells. Methods Mol Biol. 2018;1794:289–96.
Kumar P, Nagarajan A, Uchil PD. Analysis of cell viability by the MTT assay. Cold Spring Harb Protoc. 2018. https://doi.org/10.1101/pdb.prot095505.
Martinotti S, Ranzato E. Scratch wound healing assay. Methods Mol Biol. 2020;2109:225–9.
Chen B, Lai J, Dai D, Chen R, Liao N, Gao G, et al. PARPBP is a prognostic marker and confers anthracycline resistance to breast cancer. Ther Adv Med Oncol. 2020;12:1758835920974212.
Murugan AK. Special issue: PI3K/Akt signaling in human cancer. Semin Cancer Biol. 2019;59:1–2.
Huang J, Chen P, Liu K, Liu J, Zhou B, Wu R, et al. CDK1/2/5 inhibition overcomes IFNG-mediated adaptive immune resistance in pancreatic cancer. Gut 2021;70:890–9.
Qiao L, Zheng J, Tian Y, Zhang Q, Wang X, Chen JJ, et al. Regulator of chromatin condensation 1 abrogates the G1 cell cycle checkpoint via Cdk1 in human papillomavirus E7-expressing epithelium and cervical cancer cells. Cell Death Dis. 2018;9:583.
van Heek NT, Meeker AK, Kern SE, Yeo CJ, Lillemoe KD, Cameron JL, et al. Telomere shortening is nearly universal in pancreatic intraepithelial neoplasia. Am J Pathol. 2002;161:1541–7.
Chen X, Liu C, Zhang Z, Wang M, Guo S, Li T, et al. ZNF655 promotes the progression of glioma through transcriptional regulation of AURKA. Front Oncol. 2022;12:770013.
Teng Z, Yao J, Zhu L, Zhao L, Chen G. ZNF655 is involved in development and progression of non-small-cell lung cancer. Life Sci. 2021;280:119727.
Yang C, Zheng J, Liu X, Xue Y, He Q, Dong Y, et al. Role of ANKHD1/LINC00346/ZNF655 feedback loop in regulating the glioma angiogenesis via Staufen1-mediated mRNA decay. Mol Ther Nucleic Acids. 2020;20:866–78.
Zinczuk J, Zareba K, Guzinska-Ustymowicz K, Kedra B, Kemona A, Pryczynicz A. p16, p21, and p53 proteins play an important role in development of pancreatic intraepithelial neoplastic. Ir J Med Sci. 2018;187:629–37.
Guo J, Xie K, Zheng S. Molecular biomarkers of pancreatic intraepithelial neoplasia and their implications in early diagnosis and therapeutic intervention of pancreatic cancer. Int J Biol Sci. 2016;12:292–301.
Peng B, Fleming JB, Breslin T, Grau AM, Fojioka S, Abbruzzese JL, et al. Suppression of tumorigenesis and induction of p15(ink4b) by Smad4/DPC4 in human pancreatic cancer cells. Clin Cancer Res. 2002;8:3628–38.
Santamaria D, Barriere C, Cerqueira A, Hunt S, Tardy C, Newton K, et al. Cdk1 is sufficient to drive the mammalian cell cycle. Nature 2007;448:811–5.
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646–74.
He J, Gao R, Yang J, Li F, Fu Y, Cui J, et al. NCAPD2 promotes breast cancer progression through E2F1 transcriptional regulation of CDK1. Cancer Sci. 2022. https://doi.org/10.1111/cas.15347.
Author information
Authors and Affiliations
Contributions
HF designed the subject of this project. ZS and CL performed in vitro cell experiments. QW, XZha, and YD completed the tissue collection and related experiments. SL and XL conducted the xenograft experiment in mice. Data collection and statistical analysis were performed by XZhe and JZ. ZSha finished writing the manuscript and HF reviewed it.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics
The experimental procedures performed on mice were approved by the Ethics. Committee of Beijing Chaoyang Hospital and the Animal Protection Association.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Shao, Z., Li, C., Wu, Q. et al. ZNF655 accelerates progression of pancreatic cancer by promoting the binding of E2F1 and CDK1. Oncogenesis 11, 44 (2022). https://doi.org/10.1038/s41389-022-00418-2
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41389-022-00418-2
This article is cited by
-
E2F1 rs3213150 polymorphism influences cytarabine sensitivity and prognosis in patients with acute myeloid leukemia
Annals of Hematology (2023)
-
Catalpol inhibits hepatic stellate cell activation by reducing the formation and changing the contents of hepatocyte-derived extracellular vesicles
Journal of Cell Communication and Signaling (2023)