Abstract
TRAF6, a well-known adapter molecule, plays pivotal role in TLR/IL-1R associated signaling pathway. Although TRAF6 has been shown to have oncogenic activity in various malignant tumors, the details remain unclear. In this study, we demonstrated that TRAF6 facilitates Ras (G12V) and EGF-induced cellular transformation through EGFR. Silencing of TRAF6 expression significantly downregulated AP-1 activity, as well as MMP-2,9 expression after EGF stimulation. Furthermore, we found that TRAF6 plays an essential role in cutaneous squamous cell carcinoma (cSCC) malignant phenotypes, affecting cell growth and migration. CD147/Basigin, a transmembrane glycoprotein belonging to the immunoglobulin superfamily, is over-expressed in tumors and induces tumorigenesis. Our results showed that CD147 formed complex with EGFR and TRAF6. Knockdown of TRAF6 disrupted the CD147-EGFR complex, thereby inducing EGFR endocytosis. Therefore, TRAF6 might be a novel molecular target for cSCC prevention or therapy.
Similar content being viewed by others
Introduction
Cutaneous squamous cell carcinoma (cSCC) is second incidence to basal cell carcinoma (BCC) among malignant non-melanoma skin cancers1. Although the incidence of cSCC is much lower than that of BCC in the general population, the clinical characteristics of cSCC manifest more aggressively. cSCC refers to the cellular malignant transformation and abnormal growth of keratinocyte cells, which are the major cell type in the epidermis. Unlike BCC, cSCC also shows aggressive behavior, which showed the risk for metastasis relevent to advanced high-risk lesions with, and ~4.0–12.5% of patients have nodal metastasis2, 3. In addition, the increased incidence of recurrence and metastasis after surgical resection is associated with several features, including tumor diameters of >2 cm, deep invasion (>2 mm), localization to chronically damaged or diseased skin and poor histological differentiation4.
Most cSCC occurs on the head, neck, and extremities, where there is a large possibility of exposure to the sun, as ultraviolet (UV) exposure is the major cause and a direct contributor to the occurrence of cSCC. cSCC pathogenesis follows the classic tumor model, involving multiple steps from precancerous lesion, such as actinic keratosis (AK) to carcinoma in situ, final to invasive of cSCC. Subsequent steps include malignant transformation, abnormal cell growth, angiogenesis, invasion of the surrounding tissue and formation distant organ metastasis.
EGFR (epidermal growth factor receptor) is a transmembrane member of the ErbB receptor tyrosine kinase family, which is located on the cellular surface5. After associating with its ligands, such as EGF or transforming growth factor-a (TGF-a), EGFR dimerizes and triggers auto-phosphorylation of tyrosine kinases, thus leading to activation of various intracellular downstream pathways, including the PI3K/AKT, RAF/MEK/ERK, and STAT3 signaling pathways6. Dysregulation of EGFR is closely linked to tumorigenesis and has been implicated in a number of tumors7,8,9,10,11. EGFR is over-expressed on cSCC cells, particularly in advanced or metastatic tumor tissue12,13,14. Genetic analysis has indicated that EGFR has a very low frequency of mutation in cSCC15, 16, and RAS mutations are also very rarely observed in cSCC16, 17. Mutated RAS may activate molecules downstream of EGFR, and consequently, inhibition of EGFR is largely ineffective in tumor entities with RAS mutations. Because of dysregulated EGFR activation in the absence of EGFR or RAS mutations, targeting EGFR is a promising therapeutic strategy in cSCC18,19,20.
Tumor necrosis factor receptor-associated factor 6 (TRAF6), a member of the TRAF family, was first identified as an adaptor of the signals induced by the TNFR. The TRAF family comprises signal transducers of TLR/interleukin-1 (IL-1) family members, which triggers signaling transduction in innate immune responses21. In addition, TRAF6 has an E3 ubiquitin ligase activity mediatesd conjugation of lysine-63 (K63)–linked polyubiquitin chains to proteins22, 23. Recent studies have reported that TRAF6 promotes oncogenesis by inhibiting apoptosis and stimulating proliferation and invasion in cancer. TRAF6 alters the expression of Bcl2, Bax, and MMP9, thereby regulating cell apoptosis and invasive ability in gastric cancer24. TRAF6 upregulates HIF-1a expression and promote tumor angiogenesis in colon cancer25. Luo et al. have demonstrated that TRAF6 directly interacts with CD147, thereby promoting melanoma invasion and metastasis, whereas inhibition of TRAF6 expression or activity reverses the malignant phenotype of melanoma cells26, 27. In addition, TRAF6 is highly expressed in human pancreatic cancer28, colon cancer29, gliomas30, breast cancer31, and lung cancer32. However, the role of TRAF6 in cSCC remains unknown. In this study, we found that TRAF6 is required for EGF-induced cell transformation and plays critical roles in cSCC cell growth and metastasis through EGFR signaling pathways.
Results
TRAF6 mediates EGF-induced cell transformation and cell migration
TRAF6 has a critical function in the LPS/IL-1β-induced signaling pathway through the TAK1-Ikkα/β pathway, but whether TRAF6 is involved in the oncogenic stimuli-induced transduction pathway remains unknown. Our findings showed that TRAF6 promotes Ras (G12V)-induced cell transformation in NIH3T3 cells. The NIH3T3 cells had been transfected with TRAF6 alone, Ras (G12V) alone or TRAF6 with Ras (G12V). As shown in Fig. 1a, overexpression of Ras (G12V) alone could induce foci formation clearly, whereas TRAF6 failed to induce NIH3T3 cells foci formation. However, the combination of TRAF6 with Ras (G12V) induced more foci formation than Ras (G12V) alone (Fig. 1a), thus indicating that TRAF6 may play a role in cell transformation. To clarify the role of TRAF6 in cell transformation, we constructed stable TRAF6-silenced HaCaT cells by using two independent targeting sequences (Fig. 1b, left panel). As expected, EGF mediated anchorage-independent cell growth was significantly blocked in TRAF6 expresison inhibition HaCaT cells (Fig. 1b, right panel). In addition, wound healing and Transwell assays were performed to study the function of TRAF6 in cell migration. The number of migratory cells was dramatically decreased in TRAF6-knockdown HaCaT cells (Fig. 1c, d). In addition, overexpression of TRAF6-wt could promote cell proliferation and migration, but not for TRAF6-DN in HaCaT cells (sFig. 1), which suggest that the activity of TRAF6 regulates cell transformation.
a Ectopic TRAF6 promotes Ras (G12V)-induced foci formation. TRAF6, Ras (G12V) and TRAF6 together with Ras (G12V) were transfected into NIH3T3 cells as indicated, and a foci formation assay was performed as described in Materials and methods (upper panel). The histograms showed the result of three times repeats are calculated as means ± S.D. Significant differences were evaluated using a one-way ANOVA and the asterisk (*) indicates a significant difference (p < 0.05) (lower panel). The protein levels of transfected TRAF6 and Ras (G12V) were tested by western blotting with the indicated antibodies (lower panel). b Knockdown of TRAF6 attenuates EGF-induced cell transformation. Stable TRAF6-silenced HaCaT cells were generated as described in Materials and methods. Cells were seeded in 0.3% BME agar containing 20% FBS plus EGF (20 ng/ml) and maintained in a 37 °C, 5% CO2 incubator for 12 d, and colonies were counted using a microscope and the ImageJ computer software program. Representative photos are shown, and data from multiple experiments are expressed as the means ± S.D. The asterisk (*) indicates a significant difference between cells expressing mock or sh-TRAF6 (p < 0.05, Student’s t-test). c, d The effect of TRAF6 on cell migration. Cells that migrated across the membrane were stained with crystal violet and imaged at ×100 magnification. The multiple areas (n = 5) represent the means ± S.D. of each group. The asterisk (*) indicates a significant difference between cells expressing mock or sh-TRAF6 (p < 0.05, Student’s t-test), as shown in c. Wound healing assays were conducted to test the migration capability in vitro as described in Materials and methods. Images (at ×40 magnification) were taken at 24 h and 36 h, as shown in d
TRAF6 has critical roles in cSCC malignant phenotype
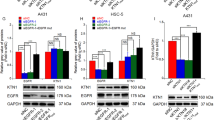
Cell transformation is a critical step in the process of cSCC, and our above data showed that TRAF6 affects Ras (G12V) and EGF-induced cell transformation, as well as cell migration in HaCaT cells, thus suggesting that TRAF6 might be involved in cSCC pathogenesis. To investigate the effect of TRAF6 on the cSCC malignant phenotype, we generated a stable TRAF6-knockdown A431 cell line (Fig. 2a). As shown in the right panel of Fig. 2b (upper panel), cell proliferation was significantly suppressed in TRAF6-silenced cells. Furthermore, knockdown of TRAF6-induced cell cycle G2/M arrest (Fig. 2b, lower panel) and raised p21 protein level (Fig. 2c, left panel), as well as transcriptional expression (Fig. 2c, right panel). As mentioned previously, ~4–10% of cSCC patients had distant metastasis, and therefore, we also tested the effect of TRAF6 on tumor cell migration. In agreement with our previous results, the migration of A431 cells was dramatically decreased after TRAF6 knockdown (Fig. 2d, e).
a Knockdown of TRAF6 attenuates the proliferation of A431 cells. TRAF6-knockdown cells were generated by using different sequences as described in Materials and methods. TRAF6 expression was assessed by western blotting as indicated (upper panel). Histograms indicated relative TRAF6 expression, as means ± S.D. Significant differences were evaluated using a one-way ANOVA and the asterisk (*) indicates a significant difference (p < 0.05) (lower panel). b The TRAF6-silenced cells were seeded into 96-well plates, and proliferation was assessed by using a CellTiter96 Aqueous One Solution detection kit. The data from multiple experiments are expressed as the means ± S.D. Significant differences were evaluated using one-way ANOVA, and the asterisk (*) indicates a significant difference (p < 0.05) (upper panel). The effect of TRAF6 knockdown on cell cycle distribution. sh-Mock, sh-TRAF6#1 cells were analyzed by FCM as described in Materials and methods (lower panel). c The protein level and transcriptional expression of p21 were measured by western-bloting and RT-real time PCR as described in Materials and methods. The data from triplicate experiments are expressed as the means ± S.D. Significant differences were evaluated with one-way ANOVA, and significant differences are indicated, as shown in (c). d, e Wound healing assays were conducted to test the migration capability in vitro as described in Materials and methods. Images (at ×40 magnification) were taken at 24 h and 48 h, as shown in (d). The effect of TRAF6 on cSCC cell migration. Cells that migrated across the membrane were stained with crystal violet and imaged at ×100 magnification. Data represent the means (n = 3) ± S.D. of each group. The asterisk (*) indicates a significant difference between cells expressing mock or sh-TRAF6 (p < 0.05, Student’s t-test), as shown in (e)
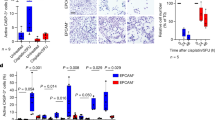
To validate the effect of TRAF6 on cSCC growth, we constructed xenograft models to test the role of TRAF6 in cell proliferation in vivo. In agreement with the in vitro data, silencing TRAF6 expression significantly blocked cSCC cell growth, thus resulting in smaller tumor sizes and delayed tumor formation (Fig. 3a, b). Histopathological findings of the tumors showed a decreased Ki-67 and EGFR expression in the knockdown of TRAF6 group, compared with control group (Fig. 3c). These results suggested that decreased levels of TRAF6 may lead to comparable attenuation of tumor growth via decreased cell proliferation.
a, b sh-Mock or sh-TRAF6 #1,2 cells (1 × 106) were injected into the right flanks of female athymic nude mice. The growth of tumors was monitored as described in Materials and methods. The results are shown as mean tumor volume ± S.D., and the asterisk (*) indicates a significant difference (p < 0.05 one-way ANOVA). c Tumor tissues were isolated from nude mice and immunohistochemistry (IHC) was conducted as described in Materials and methods to assess Ki-67 and EGFR expression
TRAF6 regulates EGF-induced signaling pathways through EGFR
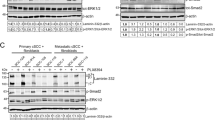
Our previous results indicated that TRAF6 promotes Ras (G12V) and EGF-induced cell transformation, as well as a malignant phenotype in A431 cells, thus suggesting that TRAF6 might, to some extent, exert role in EGF pathway. To investigate the details of the role of TRAF6 in the EGFR pathway, we used an EGFR Signaling Antibody Array and found that the expression of total EGFR and p-EGFR was significantly decreased in TRAF6-knockdown HaCaT cells (Fig. 4a). To confirm the effect of TRAF6 on EGF-induced pathways, we assessed the activation of key molecules in TRAF6-knockdown HaCaT cells EGF treatment. The results showed that silence TRAF6-expression attenuated the phosphorylation of EGFR, Stat3 and blocked c-Fos expression in presence of EGF (Fig. 4b). EGFR is known to trans-activate MMPs, such as MMP-2,933,34,35. Given TRAF6 regulation of EGFR, we next analyzed MMP-2,9 expression in TRAF6-silenced HaCaT cells after EGF treatment. As shown in Fig. 4c, knockdown of TRAF6 suppressed EGF-induced MMP-2,9 expression.
a, b, c Silencing of TRAF6 attenuates p-EGFR and EGFR expression. HaCaT cells expressing sh-TRAF6#1,2 were starved for 36 h and then treated with EGF (100 ng/ml) for 30 min, and a protein array was performed to assess the effect of TRAF6 on the EGFR signaling pathway (a) or cells expressing sh-TRAF6 were starved for 36 h and then treated with EGF (100 ng/ml) for various times, as indicated. Immunoblotting was used to detect protein expression with the indicated antibodies. Anti-β-Actin or Gad were used to verify equal loading of protein (b, c). The histograms indicated relative MMP-2 and MMP-9 expression, as means ± S.D. Significant differences were evaluated using a one-way ANOVA and the asterisk (*) indicates a significant difference (p < 0.05) (lower panel). d TRAF6 regulates AP-1 activity. Flag-TRAF6-WT and Flag-TRAF6-DN cells were transfected with the AP-1-luciferase reporter gene (100 ng), as well as the Renilla luciferase gene (20 ng) for normalization. At 30 h after transfection, the firefly luciferase activity was determined in cell lysates and normalized to the Renilla luciferase activity. Significant differences were evaluated using Student’s t-test, and the respective asterisks indicate a significant difference (p < 0.05) (upper panel). TRAF6-silenced cells were co-transfected with a plasmid mixture containing the AP-1 luciferase reporter gene (0.8 µg) and the Renilla luciferase gene (0.2 µg) for normalization. At 20 h after transfection, cells were starved for 16 h and then treated with EGF (20 ng/mL) for various times, as indicated. Firefly luciferase activity was determined in cell lysates and normalized against Renilla luciferase activity. Significant differences were evaluated using Student’s t-test, and the respective asterisks indicate a significant difference (p < 0.05) (lower panel)
The dimeric AP-1 complex, which may include Jun, Fos, and other family members, is a critical downstream mediator of the EGF-induced transduction pathway. To explore whether TRAF6 might affect AP-1 activity, HEK293 cells were transiently transfected with the AP-1 reporter gene with different amount of TRAF6-WT or TRAF6-DN (ring domain mutation), as shown in Fig. 4d (upper panel), the activity of AP-1 had been dramatically raised with dose-dependent manner in cells expressing TRAF6-WT but not in those expressing TRAF6-DN. Next, to confirm the effect of TRAF6 on AP-1 activity, the AP-1 reporter gene together with Renilla luciferase gene were transfected into TRAF6 expression silence HaCaT cells and found that AP-1 activity was downregulated in TRAF6 low-expression HaCaT cells (Fig. 4d, lower panel).
TRAF6 regulates EGFR activation through CD147
To study the mechanism associated with TRAF6 regulation of EGFR, we tested the direct interaction between TRAF6 and EGFR, but the experiment failed (data not shown). Our previous results had demonstrated that TRAF6 directly interacts with CD147. CD147 is a cell surface protein belonging to the immunoglobulin super-family involved various tumor malignant phenotypes36,37,38. Silencing CD147 expression with siRNA or anti-CD147 antibody has been shown to significantly attenuate total EGFR expression or phosphorylation of EGFR39,40,41, thus indicating that CD147 is a key EGFR mediator. Therefore, we propose that CD147 acts as a “bridge” connecting TRAF6 and EGFR. To test this hypothesis, we performed endogenous immunoprecipitation with CD147 and found that TRAF6 and EGFR were detected in the CD147 complex (Fig. 5a), thus indicating that CD147 interacts with TRAF6 and EGFR. Interestingly, knockdown of TRAF6 impaired CD147 binding to EGFR (Fig. 5b). We also examined the effect of CD147 on phosphorylation of EGFR, and in agreement with the previous results, knockdown of CD147 dramatically blocked p-EGFR with EGF treatment (Fig. 5c).
a CD147 interacts with TRAF6 and EGFR. HaCaT cell and A431 cell extracts were used for immunoprecipitation with a CD147 antibody or control IgG. The immunoprecipitated complex was detected by western blotting with the indicated antibodies. b TRAF6 affects CD147 binding to EGFR. The extracts from TRAF6-silenced cells were immunoprecipitated with anti-CD147 antibody or control IgG, and then, WB was performed to detect EGFR expression. c CD147 regulates EGFR in the EGF-induced signaling pathway. HaCaT cells expressing sh-CD147 were starved for 36 h and then treated with EGF (100 ng/ml) for various time points. Immunoblotting was used to detect EGFR expression, and anti-Gad was used to verify equal loading of protein. d TRAF6 and CD147 regulate EGFR-protein stability. Cells expressing Sh-TRAF6 and sh-CD147 were constructed as described in “Materials and methods”. After serum starvation for 16 h, the cells were treated with CHX and then stimulated with EGF (100 ng/ml) for various time points, as indicated. Immunoblotting was used to detect EGFR expression, and anti-GAPDH was used to verify equal loading of protein. e TRAF6 and CD147 alter EGFR endocytosis. Cells expressing Sh-TRAF6 and sh-CD147 were seeded onto cover glasses. After serum starvation for 16 h, the cells were stimulated with EGF (100 ng/ml) for various time points, as indicated. The cells were then fixed and stained using an anti-EGFR fluorescence-conjugated antibody. The cover glasses were mounted and visualized by using confocal microscopy. The green color indicates endogenous EGFR staining in cells. Three independent experiments were performed and showed similar results
Under ligand stimulation, endocytosis of EGFR occurs and leads to a shift from diffuse membrane staining to punctate cytoplasmic staining (representing endocytic vesicles). To understand the role of TRAF6 and CD147 in EGFR trafficking, we utilized a fluorescence-conjugated primary antibody to track EGFR and examined the effect of TRAF6, CD147 on EGFR protein stability. We found that knockdown of TRAF6 and CD147 accelerated EGFR-protein degradation (Fig. 5d) and EGFR-containing endocytic vesicles were present in TRAF6- and CD147-knockdown HaCaT cells even without EGF treatment (Fig. 5e), because the EGFR distribution typically shifts from the cell membrane to the cytoplasm, with punctate fluorescence after EGF stimulation in control cells (Fig. 5e). Hence, our results suggested that TRAF6 and CD147 may be involved in EGFR endocytosis.
Discussion
Epidermal cell transformation plays a critical stage in cSCC development that involves oncogenic key molecules. In this study, we found that TRAF6 plays a key role in cell transformation. Although over-expression of TRAF6 alone did not effect on foci formation in NIH3T3 cells, more foci were observed with a combination of TRAF6 and Ras (G12V), thus indicating that TRAF6 enhances the cell transformation ability driven by oncogenes (Fig. 1a). SliencingTRAF6 expresison significantly suppressed the EGF meidated anchorage-independent cell growth of HaCaT cells (Fig. 1b), thus indicating that TRAF6 is involved in the EGF associated signaling pathway. Regarding the TLR/IL-1β pathway, TRAF6 is also an essential molecule for activation of other signaling pathways. After TGFβ stimulation, TRAF6 induces Lys63-linked ubiquitination of TβR1, which in turn promotes cleavage of TβR1 by TACE. After cleavage, the intracellular domain (ICD) is released from TβR1, translocates into the nucleus and subsequently forms a complex with p300 that regulates expression of genes, such as Snail, MMP2, and cyclinD142,43,44. In the IGF-induced pathway, TRAF6 acts as a direct E3 ligase, ubiquitinating AKT, and thereby inducing AKT cellular membrane recruitment and phosphorylation leading to activation45.
In this study, we provided the first evidence that TRAF6 affects the EGF-induced pathway. Through a protein array, we found that p-EGFR and total EGFR were altered in TRAF6-silenced cells (Fig. 4a). Furthermore, p-Stat3, p-EGFR, and c-Fos were dramatically downregulated by TRAF6 knockdown (Fig. 4b). EGFR is a key molecule orchestrating cell growth, survival, cell cycle progression, and migration and is over-expressed or dysregulated in many cancers46,47,48,49. EGFR exerts these functions through multiple signaling cascades. After ligand binding, tyrosine residues at the intracellular domain of EGFR are auto-phosphorylated, thus creating docking sites for adapter proteins, such as Grb2 or GAB1 (4). For example, after binding to EGFR, Grb2 recruits SoS1 (a Ras guanine nucleotide exchange factor) cell membrane localization, thereby activating the Ras/Raf/ERK signaling cascade50, 51. AP-1 is a downstream convergence point of different signaling cascades and is required for EGF-induced cell transformation and tumor development. In addition, EGFR has been reported to regulate invasion or migration by regulating MMP expression, in a manner dependent on AP-1 activity52,53,54. MMPs have a critical role in ECM remodeling, embryonic development and wound healing in physiological conditions and facilitate tumor cell migration and metastasis through ECM degradation, modulation of cell adhesion and promotion of angiogenesis. We found that AP-1 activity was decreased in both TRAF6-DN and TRAF6-knockdown cells (Fig. 4c), and knockdown of TRAF6 significantly attenuated MMP-2,9 expression upon EGF stimulation (Fig. 4d).
The cSCC is a common skin cancer, and cSCC incidence has increased over the past few decades55. Genetic alterations and signaling pathway dysregulation have been found to be associated with cSCC initiation and progression, including upregulation of EGFR, Stat3, and CD14756,57,58. Our results demonstrated that knockdown of TRAF6 decreases A431 cell proliferation and migration (Fig. 2a, d, e), induces cell cycle G2/M arrest (Fig. 2b) and increases p21 transcriptional expression (Fig. 2c). Previous studies have shown that blocking the EGFR/Stat3 axis leads to upregulation of p21 expression59, and p21 binds to cyclin-dependent kinases (CDKs) and inhibits CDKs activity, thus leading to growth arrest at G2 phase of the cell cycle60, 61. CD147/BSG, a transmembrane glycoprotein belonging to the immunoglobulin superfamily, has various physiological functions, such as fertilization, spermatogenesis, T-cell development, formation of the blood brain barrier and retinal development62. CD147 expression had been found to be elevated in different tumors, such as liver, pancreatic, lung, and colon cancers63,64,65. Moreover, CD147 is overexpressed in both head and neck cancer and oral squamous cell carcinomas and is associated with head and neck cancer metastasis and patient survival66,67,68,69. Interestingly, blocking CD147 expression with anti-CD147 mAb or si-CD147 inhibits EGFR expression and leads to cSCC growth suppression in vitro and in vivo70. Targeting both EGFR and CD147 efficiently decreases HNSCC cell proliferation and migration71. Our results showed that CD147 directly associates with EGFR, as well as with TRAF6 in HaCaT and A431 cells (Fig. 5a). Notably, knockdown of TRAF6 impairs the interaction between CD147 and EGFR (Fig. 5b) and attenuates EGF-induced EGFR activation (Fig. 5c), in agreement with previous results. In addition, silencing TRAF6 and CD147 expression accelerated EGFR degradation (Fig. 5d) and induced the formation of EGFR-containing endocytic vesicles (Fig. 5e). EGF-induced activation of EGFR and subsequent tyrosine phosphorylation leads to endocytosis of the EGF/EGFR complex into late endosomes, which is followed by lysosomal fusion and degradation of the receptor, thus decreasing EGFR expression on the cell membrane and ultimately downregulating EGFR signaling72, 73.
Overall, our study revealed that in addition to the role of TRAF6 in the TLR/IL-1β pathway, TRAF6 is required for Ras (G12V)- and EGF-induced cellular transformation through EGFR. Silencing TRAF6 expression significantly downregulates AP-1 activity, as well as MMP-2,9 expression after EGF stimulation. Furthermore, we confirmed that TRAF6 promotes a malignant cSCC phenotype, thus affecting cell growth and migration. In addition, CD147 associates with EGFR and TRAF6, and knockdown of TRAF6 disrupts the CD147-EGFR complex. Therefore, TRAF6 might be a novel molecular target for cSCC prevention or therapy.
Materials and methods
Reagents and antibodies
The cell culture reagents, such as medium (Dulbecco’s modified Eagle’s medium, DMEM) and fetal bovine serum (FBS) were obtained from Life Technologies, Inc. (Rockville, MD). The TRAF6 antibody (Santa Cruz, CA, USA) was used by 1:500, anti-GAPDH (Proteintech, USA) was prepared at 1:3000; the antibody of c-Myc, CD147 (Santa Cruz, CA, USA) had been prepared by 1:500 dilution; The Flag antibody (Sigma, Germany) was diluted at 1:5000 and the no-EGFR, p-EGFR, no-STAT3, p-STAT3 (Cell Signaling Technology, Danvers, MA) were used at 1:1000 dilution. The EGFR Signaling Antibody Array Kit (#12622) was purchased from Cell Signaling Technology (Danvers) Co., Ltd.
Cell culture and transfections
The NIH3T3, Immortalized human keratinocyte cells (HaCaT) and human epidermoid carcinoma cells (A431) were purchased from the American Type Culture Collection (ATCC) and maintained in DMEM medium (10% FBS, 1% penicillin–streptomycin). The cytogenetically testing and authentication was performed by STR-PCR. For transfection experiments, cells were co-transfected with different plasmids using TurboFect (Thermo Scientific, MA, USA) following the manufacturer’s protocol.
Lentivirus infection
The stable knockdown TRAF6 expression cells in HaCaT or A431 were generated as described previous74. Briefly, pLKO1-TRAF6 plasmids together with pspAX2 and pMD2G were transfected into 293T cells, after 48 and 72 h transfection, the supernatant containing lentiviruses was collected, and then, along with 10 μg/ml polybrene, was perfomred to infect A431 and HaCaT cells. After infection 16 h, the infected cells were continued to culture in medium with 1.0 μg/ml puromycin until the control cells (uninfected) died (usually 2–3 d).
Immunoprecipitation
The extracts from cells were lysed with NP40 buffer containing protease inhibitor cocktail. For immunoprecipitation, extracts were pretreatment with 20 μl of agarose A/G-Sepharose beads (Beyotime Institute of Biotechnology) and incubated for 1 h at 4 °C with gentle shaking. Supernatants were collected and removed to a new tube, and then, 1.5 μg of antibodies and 40 μl of agarose A/G-Sepharose beads were added to the supernatants, and the mixture was rotated overnight at 4 °C. The beads were then washed three times in NP40 buffer, and the proteins analyzed by western blotting.
Immunoblotting and antibody array
The cells were collected and lysed by modified RIPA buffer containing 50 mM Tris-Cl pH 8.0, 150 mM NaCl, 0.5% NP-40, and protease-inhibitor cocktail. The protein of concentration had been tested with a BCA Kit, and appropriate amounts of protein were prepared for SDS-PAGE and then transferred to a PVDF membrane (Millipore). The membranes were blocked for 1 h with 5% non-fat dry milk, and then incubated with primary antibodies for overnight at 4 °C. The membranes washed with phosphate-buffered saline (PBS) buffer with 0.1% Tween 20 (PBS-T), reacted with horseradish peroxidase-conjugated secondary antibodies for 1 h, and visualized using an enhanced chemiluminescence substrate. The HaCaT cells were grown to 90% confluence and treated with serum-free medium for overnight. After treated with EGF at 100 ng/ml, cell extracts were prepared and analyzed by PathScan EGFR Signaling Antibody Array Kit (#12622). The antibody array was performed according to the kit protocols recommended by the manufacturer and detected with an imaging system (Bio-Rad, USA).
H-Ras(G12V) mediates cell transformation
The protocol of H-Ras(G12V) mediated cellular transformation followed previous described75. In brief, the H-RasG12V (100 ng) and TRAF6 (2.0 μg) were transiently transfected into NIH3T3 cells at six wells. After transfection 36 h, cells were splited into 10 cm dishes and cultured in DMEM(5% calf serum) for 2 week. The medium had been changed every 3 d. The Foci were fixed and stained with 0.5% crystal violet. Cell colony was calculated with a microscope (×40) and each assay was performed in triplicate.
Luciferase reporter gene assays
Cells were transfected with AP-1-Luc and SV-40-Renilla-Luc (Promega, Madison, WI). After 20 h transfection, cells were continued to culture with serum-free medium for 16 h and then treated with EGF (100 ng/ml) for indicated time points. Cell lysates were analyzed by firefly and Renilla luciferase activities with a dual-luciferase assay kit (Promega) following protocol.
Reverse transcription-real time PCR
The RNA was prepared from TRAF6 knocking-down HaCaT cells by the Trizol reagent. A total of 3 µg RNA was used as a template for the reverse transcription reaction (Invitrogen). The p21 primers used were as follows: (forward) 5′-ACTACCACTCACCCGCAGAC-3′ and (reverse) 5′-CCAGGTCCGTGCAGAAGT-3′.
Anchorage-independent cell growth
The EGF-induced cell transformation assay was performed as mentioned previous76. Cells mixed with Basal Medium Eagle (BME) medium containing 10% FBS and 0.33% agar, in presence of EGF or not, and then, seeded into 6-well plates (8 × 103/ml/well) with coated with 3 ml of solidified BME medium(10% FBS and 0.5% agar). Colonies were scored with a microscope and ImageJ software.
MTS for cell growth assay
The 2 × 103/well cells were cultured in 96-well plates in 100 µl DMEM; living cells were counted from 0 to 96 h by analyzed with 20 μl of sterile MTS dye each day. Colorimetric analysis was performed at 490 nm was measured using a microplate reader. Each cell line was established in five replicates, and each analysis was repeated three times.
In vivo tumor growth
Xenograft tumor models were established as mentioned previously74, 77, and the animal protocol was approved by the Ethics Committee of Xiangya Hospital (Central South University, China). The A431 cells with knockdown of TRAF6 expression by sh-Mock or sh-TRAF6#1 were collected and washed by PBS, and then resuspended by serum-free medium, the 4 or 6-weeks-old male nude mice (Shanghai SLAC Laboratory Animal Co. Ltd., Shanghai, China) had been subcutaneously injected with tumor cells (5 × 106/0.15 ml) on the right flanks. Tumor diameters and weights were recorded twice a week, and tumor volumes (mm3) were calculated by length × width × height × 0.5326. Tumor growth curves were drawn for each group (mean ± S.D.).
Cell transwell assay
To evaluate the ability of migration in HaCaT and A431 cell lines, an 8-mm-pore size chamber (BD Biosciences, Franklin Lakes, NJ, USA) was used. Briefly, a number of 5 × 104 cells in serum-free medium were put into the upper chamber, and then 500 µl of DMEM plus 20% FBS was added into the lower chambers. After a 16 h of incubation at the temperature of 37 °C, cells in the chamber were fixed with formaldehyde for 15 min and stained with 0.1% crystal violet. Cells passed through the transwell membrane were finally counted with a microscope (×40). Five fields were selected randomly at least. Each assay was performed in triplicate.
Wound-healing assay
Cells were cultured with complete medium in a 6-well plate at a density of 1 × 105 cells/well, and a straight line were scratched on the cell monolayer was by a 200-μl pipette tip. Then cells were washed by PBS three times to remove debris. Finally, cells were cultured in complete medium and photographed at 24 h and 48 h.
Cell cycle analysis
Collecting cells first, and washing with PBS, then fixing in ice-cold 70% ethanol in PBS and centrifuged at 4 °C overnight. Next, cells were centrifuged, resuspended with cold PBS and incubated in RNase at 37 °C for 30 min. Then cells were added with propidium iodide and incubated for 30 min at 37 °C. The cell cycle distribution was analyzed by flow cytometry (FCM).
Immunohistochemistry
Tumor samples from nude mice were cut and mounted on glass slides. Antigen retrieval was operated at 95 °C for 5 min in the microwave oven and cooling to room temperature, deparaffinized sections were treated with 3% hydrogen peroxide for 10 min. The slices were then washed in PBS for three times, 5 min each, and blocked in goat serum for 1 h. This was followed by the addition of anti-Ki-67 or EGFR antibody (1:200, Santa Cruz Biotechnology) in a humidified chamber overnight at 4 °C. After being washed by PBS, the slices were applied with a biotin-conjugated secondary antibody for 20 min and then peroxidase-conjugated streptavidin for an additional 30 min. After that, 3,3′-diaminobenzidine tetrahydrochloride was used to visualize the reaction and then the slices were counterstained with hematoxylin. Immunohistochemistry was performed at least three times.
Immunofluorescence analysis
Cells seeded in 18-mm cover glasses were washed with ice-cold 1× PBS. After EGF stimulation, 4% paraformaldehyde fixed cells for 15 min at room temperature. Cells were washed by 1× PBS for three times and then permeabilized with 0.5% Triton X-100 for 15 min at room temperature. Cells were blocked by 1% BSA for 15 min and incubated with anti-EGFR (Cell Signaling Technology, #5616) in 1% BSA overnight at 4 °C. Afterwards, the cover glasses were washed by 1× PBS for three times. Cells were counterstained with DAPI (0.5 μg/ml) for 30 min at room temperature. After being washed, the cover glass was mounted through fluorescent mounting medium. Thereafter, fluorescence microscopy images of cells were captured using a Leica SP8 Confocal laser scanning microscope.
Statistical analysis methods
The statistical results were represented with mean ± S.D. and analyzed by Student’s t-test or one-way ANOVA to examine the statistical differences. The p-value <0.05 was believed to be statistically significant.
References
Lansbury L. et al. Interventions for non-metastatic squamous cell carcinoma of the skin.Cochrane Database Syst. Rev. 14, Cd007869 (2010).
Silberstein, E., Sofrin, E., Bogdanov-Berezovsky, A., Nash, M. & Segal, N. Lymph node metastasis in cutaneous head and neck squamous cell carcinoma. Dermatol. Surg. 41, 1126–1129 (2015).
Shukla, M., Kumar, V. & Pandey, M. Distant cutaneous metastasis from oropharyngeal squamous cell carcinoma. Ear Nose Throat J. 91, E19–E21 (2012).
Thompson, A. K., Kelley, B. F., Prokop, L. J., Murad, M. H. & Baum, C. L. Risk factors for cutaneous squamous cell carcinoma recurrence, metastasis, and disease-specific death: a systematic review and meta-analysis. JAMA Dermatol. 152, 419–428 (2016).
Kovacs, E., Zorn, J. A., Huang, Y., Barros, T. & Kuriyan, J. A structural perspective on the regulation of the epidermal growth factor receptor. Annu. Rev. Biochem. 84, 739–764 (2015).
Ciardiello, F. & Tortora, G. EGFR antagonists in cancer treatment. N. Engl. J. Med. 358, 1160–1174 (2008).
Bian, Y., Yu, Y., Wang, S. & Li, L. Up-regulation of fatty acid synthase induced by EGFR/ERK activation promotes tumor growth in pancreatic cancer. Biochem. Biophys. Res. Commun. 463, 612–617 (2015).
Vene, R. et al. Celecoxib increases EGF signaling in colon tumor associated fibroblasts, modulating EGFR expression and degradation. Oncotarget 6, 12310–12325 (2015).
Suda, K. & Mitsudomi, T. Role of EGFR mutations in lung cancers: prognosis and tumor chemosensitivity. Arch. Toxicol. 89, 1227–1240 (2015).
Milagre, C. S. et al. Adaptive upregulation of EGFR limits attenuation of tumor growth by neutralizing IL6 antibodies, with implications for combined therapy in ovarian cancer. Cancer Res. 75, 1255–1264 (2015).
Scarpino, S., Pulcini, F., Di Napoli, A., Giubettini, M. & Ruco, L. EGFR mutation testing in pulmonary adenocarcinoma: evaluation of tumor cell number and tumor percent in paraffin sections of 120 small biopsies. Lung Cancer 87, 8–13 (2015).
Canueto, J. et al. EGFR expression is associated with poor outcome in cutaneous squamous cell carcinoma. Br. J. Dermatol. 176, 1279–1287 (2017).
Sweeny, L. et al. EGFR expression in advanced head and neck cutaneous squamous cell carcinoma. Head Neck 34, 681–686 (2012).
Bumpous, J. Metastatic cutaneous squamous cell carcinoma to the parotid and cervical lymph nodes: treatment and outcomes. Curr. Opin. Otolaryngol. Head Neck Surg. 17, 122–125 (2009).
Li, Y. Y. et al. Genomic analysis of metastatic cutaneous squamous cell carcinoma. Clin. Cancer Res. 21, 1447–1456 (2015).
Mauerer, A., Herschberger, E., Dietmaier, W., Landthaler, M. & Hafner, C. Low incidence of EGFR and HRAS mutations in cutaneous squamous cell carcinomas of a German cohort. Exp. Dermatol. 20, 848–850 (2011).
Ridd, K. & Bastian, B. C. Somatic mutation of epidermal growth factor receptor in a small subset of cutaneous squamous cell carcinoma. J. Invest. Dermatol. 130, 901–903 (2010).
Cobo, M. et al Afatinib in patients with squamous cell carcinoma of the lung: current context and the option of oral treatment. Med. Clin. 146, 25–29 (2016).Suppl 1.
Yin, V. T., Pfeiffer, M. L. & Esmaeli, B. Targeted therapy for orbital and periocular basal cell carcinoma and squamous cell carcinoma. Ophthal. Plast. Reconstr. Surg. 29, 87–92 (2013).
Lewis, C. M. et al. A phase II study of gefitinib for aggressive cutaneous squamous cell carcinoma of the head and neck. Clin. Cancer Res. 18, 1435–1446 (2012).
Inoue, J., Gohda, J. & Akiyama, T. Characteristics and biological functions of TRAF6. Adv. Exp. Med. Biol. 597, 72–79 (2007).
Deng, L. et al. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 103, 351–361 (2000).
Sun, L., Deng, L., Ea, C. K., Xia, Z. P. & Chen, Z. J. The TRAF6 ubiquitin ligase and TAK1 kinase mediate IKK activation by BCL10 and MALT1 in T lymphocytes. Mol. Cell 14, 289–301 (2004).
Han, F., Zhang, L., Qiu, W. & Yi, X. TRAF6 promotes the invasion and metastasis and predicts a poor prognosis in gastric cancer. Pathol. Res. Pract. 212, 31–37 (2016).
Sun, H. et al. TRAF6 upregulates expression of HIF-1alpha and promotes tumor angiogenesis. Cancer Res. 73, 4950–4959 (2013).
Luo, Z. et al. TRAF6 regulates melanoma invasion and metastasis through ubiquitination of Basigin. Oncotarget 7, 7179–7192 (2016).
Zhang, J. et al. Epigallocatechin-3-gallate(EGCG) suppresses melanoma cell growth and metastasis by targeting TRAF6 activity. Oncotarget 7, 79557–79571 (2016).
Rong, Y. et al. TRAF6 is over-expressed in pancreatic cancer and promotes the tumorigenicity of pancreatic cancer cells. Med. Oncol. 31, 260 (2014).
Sun, H. et al. TRAF6 is upregulated in colon cancer and promotes proliferation of colon cancer cells. Int. J. Biochem. Cell Biol. 53, 195–201 (2014).
Liu, J. et al. miR-146b-5p functions as a tumor suppressor by targeting TRAF6 and predicts the prognosis of human gliomas. Oncotarget 6, 29129–29142 (2015).
Bilir, C. et al. Increased serum tumor necrosis factor receptor-associated factor-6 expression in patients with non-metastatic triple-negative breast cancer. Oncol. Lett. 9, 2819–2824 (2015).
He, Z., Huang, C., Lin, G. & Ye, Y. siRNA-induced TRAF6 knockdown promotes the apoptosis and inhibits the invasion of human lung cancer SPC-A1 cells. Oncol. Rep. 35, 1933–1940 (2016).
Paye, A. et al. EGFR activation and signaling in cancer cells are enhanced by the membrane-bound metalloprotease MT4-MMP. Cancer Res. 74, 6758–6770 (2014).
Qin, Y. et al. MMP-2/9-oriented combinations enhance antitumor efficacy of EGFR/HER2-targeting fusion proteins and gemcitabine. Oncol. Rep. 32, 121–130 (2014).
Zuo, J. H. et al. Activation of EGFR promotes squamous carcinoma SCC10A cell migration and invasion via inducing EMT-like phenotype change and MMP-9-mediated degradation of E-cadherin. J. Cell Biochem. 112, 2508–2517 (2011).
Su, J., Chen, X. & Kanekura, T. A CD147-targeting siRNA inhibits the proliferation, invasiveness, and VEGF production of human malignant melanoma cells by down-regulating glycolysis. Cancer Lett. 273, 140–147 (2009).
Zeng, W. et al. CD147 promotes melanoma progression through hypoxia-induced MMP2 activation. Curr. Mol. Med. 14, 163–173 (2014).
Luo, Z. et al. CD147 interacts with NDUFS6 in regulating mitochondrial complex I activity and the mitochondrial apoptotic pathway in human malignant melanoma cells. Curr. Mol. Med. 14, 1252–1264 (2014).
Xu, B. Q. et al. Gemcitabine enhances cell invasion via activating HAb18G/CD147-EGFR-pSTAT3 signaling. Oncotarget 7, 62177–62193 (2016).
Hatanaka, M. et al. CD147-targeted siRNA in A375 malignant melanoma cells induces the phosphorylation of EGFR and downregulates cdc25C and MEK phosphorylation. Oncol. Lett. 11, 2424–2428 (2016).
Grass, G. D., Tolliver, L. B., Bratoeva, M. & Toole, B. P. CD147, CD44, and the epidermal growth factor receptor (EGFR) signaling pathway cooperate to regulate breast epithelial cell invasiveness. J. Biol. Chem. 288, 26089–26104 (2013).
Thakur, N., Sorrentino, A., Heldin, C. H. & Landstrom, M. TGF-beta uses the E3-ligase TRAF6 to turn on the kinase TAK1 to kill prostate cancer cells. Future Oncol. 5, 1–3 (2009).
Yamashita, M. et al. TRAF6 mediates Smad-independent activation of JNK and p38 by TGF-beta. Mol. Cell 31, 918–924 (2008).
Sorrentino, A. et al. The type I TGF-beta receptor engages TRAF6 to activate TAK1 in a receptor kinase-independent manner. Nat. Cell Biol. 10, 1199–1207 (2008).
Yang, W. L. et al. The E3 ligase TRAF6 regulates Akt ubiquitination and activation. Science 325, 1134–1138 (2009).
Chen, G., Kronenberger, P., Teugels, E., Umelo, I. A. & De Greve, J. Effect of siRNAs targeting the EGFR T790M mutation in a non-small cell lung cancer cell line resistant to EGFR tyrosine kinase inhibitors and combination with various agents. Biochem. Biophys. Res. Commun. 431, 623–629 (2013).
Dan, L., Jian, D., Na, L. & Xiaozhong, W. Crosstalk between EGFR and integrin affects invasion and proliferation of gastric cancer cell line, SGC7901. Oncol. Targets Ther. 5, 271–277 (2012).
Yang, J. J. et al. Clinical modes of EGFR tyrosine kinase inhibitor failure and subsequent management in advanced non-small cell lung cancer. Lung. Cancer 79, 33–39 (2013).
Brugger, W. & Thomas, M. EGFR-TKI resistant non-small cell lung cancer (NSCLC): new developments and implications for future treatment. Lung Cancer 77, 2–8 (2012).
Toki, M. I. et al. EGFR-GRB2 protein colocalization is a prognostic factor unrelated to overall EGFR expression or EGFR mutation in lung adenocarcinoma. J. Thorac. Oncol. 11, 1901–1911 (2016).
Yamazaki, T. et al. Role of Grb2 in EGF-stimulated EGFR internalization. J. Cell Sci. 115, 1791–1802 (2002).
Zhang, H. W., Wang, X., Zong, Z. H., Huo, X. & Zhang, Q. AP-1 inhibits expression of MMP-2/9 and its effects on rat smooth muscle cells. J. Surg. Res. 157, e31–e37 (2009).
Kajanne, R. et al. EGF-R regulates MMP function in fibroblasts through MAPK and AP-1 pathways. J. Cell Physiol. 212, 489–497 (2007).
Bergman, M. R. et al. A functional activating protein 1 (AP-1) site regulates matrix metalloproteinase 2 (MMP-2) transcription by cardiac cells through interactions with JunB-Fra1 and JunB-FosB heterodimers. Biochem. J. 369, 485–496 (2003).
Rogers, H. W. et al. Incidence estimate of nonmelanoma skin cancer in the United States, 2006. Arch. Dermatol. 146, 283–287 (2010).
Kolev, V. et al. EGFR signalling as a negative regulator of Notch1 gene transcription and function in proliferating keratinocytes and cancer. Nat. Cell Biol. 10, 902–911 (2008).
Toll, A. et al. MYC gene numerical aberrations in actinic keratosis and cutaneous squamous cell carcinoma. Br. J. Dermatol. 161, 1112–1118 (2009).
Boukamp, P. Non-melanoma skin cancer: what drives tumor development and progression? Carcinogenesis 26, 1657–1667 (2005).
Wang, S. T., Ho, H. J., Lin, J. T., Shieh, J. J. & Wu, C. Y. Simvastatin-induced cell cycle arrest through inhibition of STAT3/SKP2 axis and activation of AMPK to promote p27 and p21 accumulation in hepatocellular carcinoma cells. Cell Death Dis. 8, e2626 (2017).
Malumbres, M. & Barbacid, M. Cell cycle, CDKs and cancer: a changing paradigm. Nat. Rev. Cancer 9, 153–166 (2009).
Abbas, T. & Dutta, A. p21 in cancer: intricate networks and multiple activities. Nat. Rev. Cancer 9, 400–414 (2009).
Muramatsu, T. & Miyauchi, T. Basigin (CD147): a multifunctional transmembrane protein involved in reproduction, neural function, inflammation and tumor invasion. Histol. Histopathol. 18, 981–987 (2003).
Chen, Y., Gou, X., Ke, X., Cui, H. & Chen, Z. Human tumor cells induce angiogenesis through positive feedback between CD147 and insulin-like growth factor-I. PLoS ONE 7, e40965 (2012).
Zhu, C. et al. Inhibition of CD147 gene expression via RNA interference reduces tumor cell invasion, tumorigenicity and increases chemosensitivity to cisplatin in laryngeal carcinoma Hep2 cells. Oncol. Rep. 25, 425–432 (2011).
Su, J. et al. CD147 silencing inhibits tumor growth by suppressing glucose transport in melanoma. Oncotarget 7, 64778–64784 (2016).
Ishibashi, Y. et al. CD147 and matrix metalloproteinase-2 protein expression as significant prognostic factors in esophageal squamous cell carcinoma. Cancer 101, 1994–2000 (2004).
Yu, Y. H. et al. CD147 and Ki-67 overexpression confers poor prognosis in squamous cell carcinoma of oral tongue: a tissue microarray study. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 119, 553–565 (2015).
Huang, X. Q. et al. Co-expression of CD147 and GLUT-1 indicates radiation resistance and poor prognosis in cervical squamous cell carcinoma. Int J. Clin. Exp. Pathol. 7, 1651–1666 (2014).
Huang, Z. et al. Overexpression of CD147 contributes to the chemoresistance of head and neck squamous cell carcinoma cells. J. Oral Pathol. Med. 42, 541–546 (2013).
Frederick, J. W., Sweeny, L., Hartman, Y., Zhou, T. & Rosenthal, E. L. Epidermal growth factor receptor inhibition by anti-CD147 therapy in cutaneous squamous cell carcinoma. Head Neck 38, 247–252 (2016).
Suzuki, S. & Ishikawa, K. Combined inhibition of EMMPRIN and epidermal growth factor receptor prevents the growth and migration of head and neck squamous cell carcinoma cells. Int. J. Oncol. 44, 912–917 (2014).
Scita, G. & Di Fiore, P. P. The endocytic matrix. Nature 463, 464–473 (2010).
Goh, L. K., Huang, F., Kim, W., Gygi, S. & Sorkin, A. Multiple mechanisms collectively regulate clathrin-mediated endocytosis of the epidermal growth factor receptor. J. Cell Biol. 189, 871–883 (2010).
Li, W. et al. TRAF4 is a critical molecule for Akt activation in lung cancer. Cancer Res. 73, 6938–6950 (2013).
Xiao, T. et al. Phosphorylation of NFAT3 by CDK3 induces cell transformation and promotes tumor growth in skin cancer. Oncogene 36, 2835–2845 (2017).
Zhu, F. et al. Phosphorylation of H2AX at Ser139 and a new phosphorylation site Ser16 by RSK2 decreases H2AX ubiquitination and inhibits cell transformation. Cancer Res. 71, 393–403 (2011).
Xu, T., Xiao, D. & Zhang, X. ECRG4 inhibits growth and invasiveness of squamous cell carcinoma of the head and neck in vitro and in vivo. Oncol. Lett. 5, 1921–1926 (2013).
Acknowledgements
This work was financially supported by the Major Projects of International Cooperation and Exchanges NSFC Grand No. 81620108024 and Grant No. 81572679, 81472852, 81472882, 81430075, and 81772917 from the National Natural Science Foundation; and Grant No. 2015JJ2161 from the Natural Science Foundation of Hunan Province. The Strategy-Oriented Special Project of Central South University in China (ZLXD2017003)
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhang, X., Wu, L., Xiao, T. et al. TRAF6 regulates EGF-induced cell transformation and cSCC malignant phenotype through CD147/EGFR. Oncogenesis 7, 17 (2018). https://doi.org/10.1038/s41389-018-0030-1
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41389-018-0030-1
This article is cited by
-
CD147 mediates epidermal malignant transformation through the RSK2/AP-1 pathway
Journal of Experimental & Clinical Cancer Research (2022)
-
A novel chalcone derivative suppresses melanoma cell growth through targeting Fyn/Stat3 pathway
Cancer Cell International (2020)
-
The relationship between TRAF6 and tumors
Cancer Cell International (2020)