Abstract
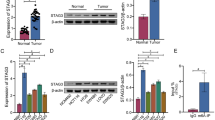
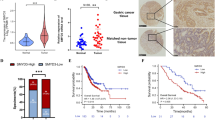
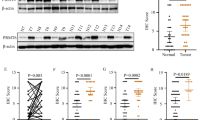
Perturbations in transforming growth factor-β (TGF-β) signaling can lead to a plethora of diseases, including cancer. Mutations and posttranslational modifications (PTMs) of the partner of SMAD complexes contribute to the dysregulation of TGF-β signaling. Here, we reported a PTM of SMAD4, R361 methylation, that was critical for SMAD complexes formation and TGF-β signaling activation. Through mass spectrometric, co-immunoprecipitation (Co-IP) and immunofluorescent (IF) assays, we found that oncogene protein arginine methyltransferase 5 (PRMT5) interacted with SMAD4 under TGF-β1 treatment. Mechanically, PRMT5 triggered SMAD4 methylation at R361 and induced SMAD complexes formation and nuclear import. Furthermore, we emphasized that PRMT5 interacting and methylating SMAD4 was required for TGF-β1-induced epithelial-mesenchymal transition (EMT) and colorectal cancer (CRC) metastasis, and SMAD4 R361 mutation diminished PRMT5 and TGF-β1-induced metastasis. In addition, highly expressed PRMT5 or high level of SMAD4 R361 methylation indicated worse outcomes in clinical specimens analysis. Collectively, our study highlights the critical interaction of PRMT5 and SMAD4 and the roles of SMAD4 R361 methylation for controlling TGF-β signaling during metastasis. We provided a new insight for SMAD4 activation. And this study indicated that blocking PRMT5-SMAD4 signaling might be an effective targeting strategy in SMAD4 wild-type CRC.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Source data of mass spectrometric analyses are provided with this paper. Other data that support the findings are available from the corresponding author upon reasonable request.
References
Batlle E, Massagué J. Transforming growth factor-β signaling in immunity and cancer. Immunity. 2019;50:924–40.
Massagué J. TGFβ in cancer. Cell. 2008;134:215–30.
Zhao M, Mishra L, Deng CX. The role of TGF-β/SMAD4 signaling in cancer. Int J Biol Sci. 2018;14:111–23.
Colak S, ten Dijke P. Targeting TGF-β signaling in cancer. Trends Cancer. 2017;3:56–71.
Xu J, Lamouille S, Derynck R. TGF-Β-induced epithelial to mesenchymal transition. Cell Res. 2009;19:156–72.
Miyaki M, Iijima T, Konishi M, Sakai K, Ishii A, Yasuno M, et al. Higher frequency of Smad4 gene mutation in human colorectal cancer with distant metastasis. Oncogene. 1999;18:3098–103.
Chen HB, Rud JG, Lin K, Xu L. Nuclear targeting of transforming growth factor-β-activated Smad complexes. J Biol Chem. 2005;280:21329–36. https://doi.org/10.1074/jbc.M500362200.
Van Hattem WA, Brosens LAA, De Leng WWJ, Morsink FH, Lens S, Carvalho R, et al. Large genomic deletions of SMAD4, BMPR1A and PTEN in juvenile polyposis. Gut. 2008;57:623–7.
Langeveld D, Van Hattem WA, De Leng WWJ, Morsink FH, Ten Kate FJW, Giardiello FM, et al. SMAD4 immunohistochemistry reflects genetic status in juvenile polyposis syndrome. Clin Cancer Res. 2010;16:4126–34.
Fleming NI, Jorissen RN, Mouradov D, Christie M, Sakthianandeswaren A, Palmieri M, et al. SMAD2, SMAD3 and SMAD4 mutations in colorectal cancer. Cancer Res. 2013;73:725–35.
Huang C, Hu F, Song D, Sun X, Liu A, Wu Q, et al. EZH2-triggered methylation of SMAD3 promotes its activation and tumor metastasis. J Clin Investig. 2022;132:e152394.
Wrighton KH, Lin X, Feng XH. Phospho-control of TGF-β superfamily signaling. Cell Res. 2009;19:8–20.
Dupont S, Zacchigna L, Cordenonsi M, Soligo S, Adorno M, Rugge M, et al. Germ-layer specification and control of cell growth by ectodermin, a Smad4 ubiquitin ligase. Cell. 2005;121:87–99.
Morén A, Imamura T, Miyazono K, Heldin CH, Moustakas A. Degradation of the tumor suppressor Smad4 by WW and HECT domain ubiquitin ligases. J Biol Chem. 2005;280:22115–23.
Lee PSW, Chang C, Liu D, Derynck R. Sumoylation of Smad4, the common Smad mediator of transforming growth factor-β family signaling. J Biol Chem. 2003;278:27853–63.
Lin X, Liang M, Liang YY, Brunicardi FC, Melchior F, Feng XH. Activation of transforming growth factor-β signaling by SUMO-1 modification of tumor suppressor Smad4/DPC4. J Biol Chem. 2003;278:18714–9.
Carlson SM, Gozani O. Emerging technologies to map the protein methylome. J Mol Biol. 2014;426:3350–62. https://doi.org/10.1016/j.jmb.2014.04.024.
Albert M, Helin K. Histone methyltransferases in cancer. Semin Cell Dev Biol. 2010;21:209–20. https://doi.org/10.1016/j.semcdb.2009.10.007.
Biggar KK, Li SSC. Non-histone protein methylation as a regulator of cellular signalling and function. Nat Rev Mol Cell Biol. 2015;16:5–17.
Wu Q, Schapira M, Arrowsmith CH, Barsyte-Lovejoy D. Protein arginine methylation: from enigmatic functions to therapeutic targeting. Nat Rev Drug Discov. 2021;20:509–30. https://doi.org/10.1038/s41573-021-00159-8.
Hamamoto R, Saloura V, Nakamura Y. Critical roles of non-histone protein lysine methylation in human tumorigenesis. Nat Rev Cancer. 2015;15:110–24. https://doi.org/10.1038/nrc3884.
Jarrold J, Davies CC. PRMTs and arginine methylation: cancer’s best-kept secret? Trends Mol Med. 2019;25:993–1009. https://doi.org/10.1016/j.molmed.2019.05.007.
Blanc RS, Richard S. Arginine methylation: the coming of age. Mol Cell. 2017;65:8–24.
Guccione E, Richard S. The regulation, functions and clinical relevance of arginine methylation. Nat Rev Mol Cell Biol. 2019;20:642–57. https://doi.org/10.1038/s41580-019-0155-x.
Rodríguez-Paredes M, Lyko F. The importance of non-histone protein methylation in cancer therapy. Nat Rev Mol Cell Biol. 2019;20:569–70. https://doi.org/10.1038/s41580-019-0147-x.
Stopa N, Krebs JE, Shechter D. The PRMT5 arginine methyltransferase: many roles in development, cancer and beyond. Cell Mol Life Sci. 2015;72:2041–59. https://doi.org/10.1007/s00018-015-1847-9.
Wang Y, Hu W, Yuan Y. Protein arginine methyltransferase 5 (PRMT5) as an anticancer target and its inhibitor discovery. J Med Chem. 2018;61:9429–41.
Kim H, Ronai ZA. PRMT5 function and targeting in cancer. Cell Stress. 2020;4:199–215.
Shi Y, Hata A, Lo RS, Massagué J, Pavletich NP. A structural basis for mutational inactivation of the tumour suppressor Smad4. Nature. 1997;388:87–93.
Inman GJ, Nicolás FJ, Hill CS. Nucleocytoplasmic shuttling of Smads 2, 3, and 4 permits sensing of TGF-β receptor activity. Mol Cell. 2002;10:283–94.
Xu P, Lin X, Feng XH. Posttranslational regulation of smads. Cold Spring Harb Perspect Biol. 2016;8:1–28.
Wang F, Xia X, Yang C, Shen J, Mai J, Kim HC, et al. SMAD4 gene mutation renders pancreatic cancer resistance to radiotherapy through promotion of autophagy. Clin Cancer Res. 2018;24:3176–85.
Oyanagi H, Shimada Y, Nagahashi M, Ichikawa H, Tajima Y, Abe K, et al. SMAD4 alteration associates with invasive-front pathological markers and poor prognosis in colorectal cancer. Histopathology. 2019;74:873–82.
Wesche J, Kühn S, Kessler BM, Salton M, Wolf A. Protein arginine methylation: a prominent modification and its demethylation. Cell Mol Life Sci. 2017;74:3305–15.
Hao Y, Baker D, Ten Dijke P. TGF-β-mediated epithelial-mesenchymal transition and cancer metastasis. Int J Mol Sci. 2019;20:2767.
Ilyas M, Efstathiou JA, Straub J, Kim HC, Bodmer WF. Transforming growth factor β stimulation of colorectal cancer cell lines: Type II receptor bypass and changes in adhesion molecule expression. Proc Natl Acad Sci USA. 1999;96:3087–91.
Morén A, Raja E, Heldin CH, Moustakas A. Negative regulation of TGFβ signaling by the kinase LKB1 and the scaffolding protein LIP1. J Biol Chem. 2011;286:341–53.
Seong HA, Jung H, Ha H. Murine protein serine/threonine kinase 38 stimulates TGF-β signaling in a kinase-dependent manner via direct phosphorylation of smad proteins. J Biol Chem. 2010;285:30959–70.
He W, Dorn DC, Erdjument-Bromage H, Tempst P, Moore MAS, Massagué J. Hematopoiesis controlled by distinct TIF1γ and Smad4 branches of the TGFβ pathway. Cell. 2006;125:929–41.
Xi Q, Wang Z, Zaromytidou AI, Zhang XHF, Chow-Tsang LF, Liu JX, et al. A poised chromatin platform for TGF-β access to master regulators. Cell. 2011;147:1511–24.
Wang G, Long J, Gao Y, Zhang W, Han F, Xu C, et al. SETDB1-mediated methylation of Akt promotes its K63-linked ubiquitination and activation leading to tumorigenesis. Nat Cell Biol. 2019;21:214–25. https://doi.org/10.1038/s41556-018-0266-1.
Song D, Lan J, Chen Y, Liu A, Wu Q, Zhao C, et al. NSD2 promotes tumor angiogenesis through methylating and activating STAT3 protein. Oncogene. 2021;40:2952–67.
Gao J, Liu R, Feng D, Huang W, Huo M, Zhang J, et al. Snail/PRMT5/NuRD complex contributes to DNA hypermethylation in cervical cancer by TET1 inhibition. Cell Death Differ. 2021;28:2818–36. https://doi.org/10.1038/s41418-021-00786-z.
Wang N, Yan H, Wu D, Zhao Z, Chen X, Long Q, et al. PRMT5/Wnt4 axis promotes lymph-node metastasis and proliferation of laryngeal carcinoma. Cell Death Dis. 2020;11:864.
Zhu F, Guo H, Bates PD, Zhang S, Zhang H, Nomie KJ, et al. PRMT5 is upregulated by B-cell receptor signaling and forms a positive-feedback loop with PI3K/AKT in lymphoma cells. Leukemia. 2019;33:2898–911.
Rasco D, Tolcher A, Siu LL, Heinhuis K, Postel-Vinay S, Barbash O, et al. Abstract CT038: A phase I, open-label, dose-escalation study to investigate the safety, pharmacokinetics, pharmacodynamics, and clinical activity of GSK3326595 in subjects with solid tumors and non-Hodgkin’s lymphoma. Cancer Res. 2017. 77 (13_Supplement): CT038.
AbuHammad S, Cullinane C, Martin C, Bacolas Z, Ward T, Chen H, et al. Regulation of PRMT5-MDM4 axis is critical in the response to CDK4/6 inhibitors in melanoma. Proc Natl Acad Sci USA. 2019;116:17990–18000.
Luo Y, Gao Y, Liu W, Yang Y, Jiang J, Wang Y, et al. Myelocytomatosis-protein arginine N-methyltransferase 5 axis defines the tumorigenesis and immune response in hepatocellular carcinoma. Hepatology. 2021;74:1932–51.
Acknowledgements
This study was supported by the National Key Research and Development Program of China (No. 2022YFA1105303 GW) and National Natural Science Foundation of China (NSFC) (Nos. 81922053 GW, 81974432 GW, 81874186 JH, No. 82273254 JH, No. 82260521 XS) and Hubei Provincial Finance Department (SCZ202203 GW). We would like to acknowledge the support from the Molecular Medical Center of Tongji Hospital, Huazhong University of Science and Technology. We appreciate the assistance from members of Guihua Wang’s laboratory and Junbo Hu’s laboratory. We thank the Protein Chemistry and Proteomics Facility, Tsinghua University for the contribution of mass spectrometric analyses.
Author information
Authors and Affiliations
Contributions
GW and JH designed the research. AL performed the majority of the experiments. QW, YC, CH and XL helped analyze the data. DS and XWS contributed to the bioinformatics analysis. ML and ZW collected clinical samples. CQ and CY performed most of the phenotype experiments. KW, LL and KL assisted in animal experiments. CZ and HD did the mass spectrometry detection and analysis. XLS, SL, XLL and FX provided advice for our study. All authors approved this study.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, A., Yu, C., Qiu, C. et al. PRMT5 methylating SMAD4 activates TGF-β signaling and promotes colorectal cancer metastasis. Oncogene 42, 1572–1584 (2023). https://doi.org/10.1038/s41388-023-02674-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-023-02674-x