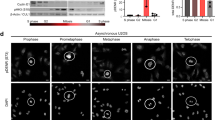
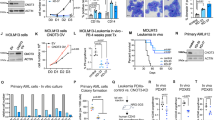
Abstract
Cyclin-dependent kinase 13 (CDK13) has been suggested to phosphorylate RNA polymerase II and is involved in transcriptional activation. However, whether CDK13 catalyzes other protein substrates and how CDK13 contributes to tumorigenesis remain largely unclear. We here identify key translation machinery components, 4E-BP1 and eIF4B, as novel CDK13 substrates. CDK13 directly phosphorylates 4E-BP1 at Thr46 and eIF4B at Ser422; genetically or pharmacologically inhibiting CDK13 disrupts mRNA translation. Polysome profiling analysis shows that MYC oncoprotein synthesis strictly depends on CDK13-regulated translation in colorectal cancer (CRC), and CDK13 is required for CRC cell proliferation. As mTORC1 is implicated in 4E-BP1 and eIF4B phosphorylation, inactivation of CDK13 in combination with the mTORC1 inhibitor rapamycin further dephosphorylates 4E-BP1 and eIF4B and blocks protein synthesis. As a result, dual inhibition of CDK13 and mTORC1 induces more profound tumor cell death. These findings clarify the pro-tumorigenic role of CDK13 by direct phosphorylation of translation initiation factors and enhancing protein synthesis. Therefore, therapeutic targeting of CDK13 alone or in combination with rapamycin may pave a new way for cancer treatment.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The accession number for the RNA-Seq data reported in this study have been submitted to the Gene Expression Omnibus under accession number GSE224287.
References
Malumbres M. Cyclin-dependent kinases. Genome Biol. 2014;15:122.
Chou J, Quigley DA, Robinson TM, Feng FY, Ashworth A. Transcription-Associated Cyclin-Dependent Kinases as Targets and Biomarkers for Cancer Therapy. Cancer Discov. 2020;10:351–70.
Hortobagyi GN, Stemmer SM, Burris HA, Yap YS, Sonke GS, Paluch-Shimon S, et al. Ribociclib as First-Line Therapy for HR-Positive, Advanced Breast Cancer. N Engl J Med. 2016;375:1738–48.
Finn RS, Martin M, Rugo HS, Jones S, Im SA, Gelmon K, et al. Palbociclib and Letrozole in Advanced Breast Cancer. N Engl J Med. 2016;375:1925–36.
Sledge GW Jr, Toi M, Neven P, Sohn J, Inoue K, Pivot X, et al. MONARCH 2: Abemaciclib in Combination With Fulvestrant in Women With HR+/HER2- Advanced Breast Cancer Who Had Progressed While Receiving Endocrine Therapy. J Clin Oncol. 2017;35:2875–84.
Kumar SK, LaPlant B, Chng WJ, Zonder J, Callander N, Fonseca R, et al. Dinaciclib, a novel CDK inhibitor, demonstrates encouraging single-agent activity in patients with relapsed multiple myeloma. Blood. 2015;125:443–8.
Marqués F, Moreau JL, Peaucellier G, Lozano JC, Schatt P, Picard A, et al. A new subfamily of high molecular mass CDC2-related kinases with PITAI/VRE motifs. Biochem Biophys Res Commun. 2000;279:832–7.
Greenleaf AL. Human CDK12 and CDK13, multi-tasking CTD kinases for the new millenium. Transcription. 2019;10:91–110.
Fan Z, Devlin JR, Hogg SJ, Doyle MA, Harrison PF, Todorovski I, et al. CDK13 cooperates with CDK12 to control global RNA polymerase II processivity. Sci Adv. 2020;6:eaaz5041.
Bartkowiak B, Liu P, Phatnani HP, Fuda NJ, Cooper JJ, Price DH, et al. CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev. 2010;24:2303–16.
Greifenberg AK, Hönig D, Pilarova K, Düster R, Bartholomeeusen K, Bösken CA, et al. Structural and Functional Analysis of the Cdk13/Cyclin K Complex. Cell Rep. 2016;14:320–31.
Liang K, Gao X, Gilmore JM, Florens L, Washburn MP, Smith E, et al. Characterization of human cyclin-dependent kinase 12 (CDK12) and CDK13 complexes in C-terminal domain phosphorylation, gene transcription, and RNA processing. Mol Cell Biol. 2015;35:928–38.
Kim HE, Kim DG, Lee KJ, Son JG, Song MY, Park YM, et al. Frequent amplification of CENPF, GMNN and CDK13 genes in hepatocellular carcinomas. PloS ONE. 2012;7:e43223.
Wang J, Zhang Y, Lu L, Lu Y, Tang Q, Pu J. Insight into the molecular mechanism of LINC00152/miR-215/CDK13 axis in hepatocellular carcinoma progression. J Cell Biochem. 2019;120:18816–25.
Ramírez-Moya J, Miliotis C, Baker AR, Gregory RI, Slack FJ, Santisteban P. An ADAR1-dependent RNA editing event in the cyclin-dependent kinase CDK13 promotes thyroid cancer hallmarks. Mol Cancer. 2021;20:115.
Qi JC, Yang Z, Lin T, Ma L, Wang YX, Zhang Y, et al. CDK13 upregulation-induced formation of the positive feedback loop among circCDK13, miR-212-5p/miR-449a and E2F5 contributes to prostate carcinogenesis. J Exp Clin Cancer Res: CR. 2021;40:2.
Lee LJ, Papadopoli D, Jewer M, Del Rincon S, Topisirovic I, Lawrence MG, et al. Cancer Plasticity: The Role of mRNA Translation. Trends Cancer. 2021;7:134–45.
Truitt ML, Ruggero D. New frontiers in translational control of the cancer genome. Nat Rev Cancer. 2016;16:288–304.
Merrick WC. eIF4F: a retrospective. J Biol Chem. 2015;290:24091–9.
Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–45.
Parsyan A, Svitkin Y, Shahbazian D, Gkogkas C, Lasko P, Merrick WC, et al. mRNA helicases: the tacticians of translational control. Nat Rev Mol Cell Biol. 2011;12:235–45.
Böhm R, Imseng S, Jakob RP, Hall MN, Maier T, Hiller S. The dynamic mechanism of 4E-BP1 recognition and phosphorylation by mTORC1. Mol Cell. 2021;81:2403–16.e2405.
Raught B, Peiretti F, Gingras AC, Livingstone M, Shahbazian D, Mayeur GL, et al. Phosphorylation of eucaryotic translation initiation factor 4B Ser422 is modulated by S6 kinases. EMBO J. 2004;23:1761–9.
Chen H, Liu H, Qing G. Targeting oncogenic Myc as a strategy for cancer treatment. Signal Transduct Targeted Ther. 2018;3:5.
Quereda V, Bayle S, Vena F, Frydman SM, Monastyrskyi A, Roush WR, et al. Therapeutic Targeting of CDK12/CDK13 in Triple-Negative Breast Cancer. Cancer Cell. 2019;36:545–58.e547.
Even Y, Durieux S, Escande ML, Lozano JC, Peaucellier G, Weil D, et al. CDC2L5, a Cdk-like kinase with RS domain, interacts with the ASF/SF2-associated protein p32 and affects splicing in vivo. J Cell Biochem. 2006;99:890–904.
Chen HH, Wong YH, Geneviere AM, Fann MJ. CDK13/CDC2L5 interacts with L-type cyclins and regulates alternative splicing. Biochem Biophys Res Commun. 2007;354:735–40.
Żuryń A, Krajewski A, Klimaszewska-Wiśniewska A, Grzanka A, Grzanka D. Expression of cyclin B1, D1 and K in non‑small cell lung cancer H1299 cells following treatment with sulforaphane. Oncol Rep. 2019;41:1313–23.
Lu XL, Zhan R, Zhao GM, Qian ZH, Gong CC, Li YQ. Expression of CDK13 Was Associated with Prognosis and Expression of HIF-1α and beclin1 in Breast Cancer Patients. J Investig Surg. 2022;35:442–7.
Duffy MJ, O’Grady S, Tang M, Crown J. MYC as a target for cancer treatment. Cancer Treatment Rev. 2021;94:102154.
Choi SH, Martinez TF, Kim S, Donaldson C, Shokhirev MN, Saghatelian A, et al. CDK12 phosphorylates 4E-BP1 to enable mTORC1-dependent translation and mitotic genome stability. Genes Dev. 2019;33:418–35.
Schmidt S, Denk S, Wiegering A. Targeting Protein Synthesis in Colorectal Cancer. Cancers. 2020;12:1298.
Benjamin D, Colombi M, Moroni C, Hall MN. Rapamycin passes the torch: a new generation of mTOR inhibitors. Nat Rev Drug Discov. 2011;10:868–80.
Choo AY, Yoon SO, Kim SG, Roux PP, Blenis J. Rapamycin differentially inhibits S6Ks and 4E-BP1 to mediate cell-type-specific repression of mRNA translation. Proceed Natl Acad Sci USA. 2008;105:17414–9.
Noh WC, Mondesire WH, Peng J, Jian W, Zhang H, Dong J, et al. Determinants of rapamycin sensitivity in breast cancer cells. Clin Cancer Res. 2004;10:1013–23.
Zhang Y, Zheng XF. mTOR-independent 4E-BP1 phosphorylation is associated with cancer resistance to mTOR kinase inhibitors. Cell Cycle. 2012;11:594–603.
Wang X, Beugnet A, Murakami M, Yamanaka S, Proud CG. Distinct signaling events downstream of mTOR cooperate to mediate the effects of amino acids and insulin on initiation factor 4E-binding proteins. Mol Cell Biol. 2005;25:2558–72.
Ito H, Ichiyanagi O, Naito S, Bilim VN, Tomita Y, Kato T, et al. GSK-3 directly regulates phospho-4EBP1 in renal cell carcinoma cell-line: an intrinsic subcellular mechanism for resistance to mTORC1 inhibition. BMC Cancer. 2016;16:393.
Mi W, Ye Q, Liu S, She QB. AKT inhibition overcomes rapamycin resistance by enhancing the repressive function of PRAS40 on mTORC1/4E-BP1 axis. Oncotarget. 2015;6:13962–77.
Humphrey SJ, Karayel O, James DE, Mann M. High-throughput and high-sensitivity phosphoproteomics with the EasyPhos platform. Nat Protoc. 2018;13:1897–916.
Acknowledgements
We thank all members of the Liu lab for helpful suggestions, and the Core Facility of Medical Research Institute at Wuhan University for providing immunofluorescence and histology platforms. This study was supported by grants from National Key Research and Development Program of China (2022YFA1103200), National Natural Science Foundation of China (81970152, 82025003, 82161138024) and Hubei Provincial Natural Science Fund for Creative Research Groups (2021CFA003).
Author information
Authors and Affiliations
Contributions
HL conceived and designed the study. HL and GQ supervised the study. HL and CW wrote the paper. CW and TX performed most of the experiments. KL and MQ performed phosphorylation mass spectrometry analysis and provided HCT116 CDK13 AS cells. YG and RH assisted with cell-based assays. DW provided technical assistance for mouse experiments and valuable help in RNA-Seq and proteomics data analysis.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wu, C., Xie, T., Guo, Y. et al. CDK13 phosphorylates the translation machinery and promotes tumorigenic protein synthesis. Oncogene 42, 1321–1330 (2023). https://doi.org/10.1038/s41388-023-02653-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-023-02653-2