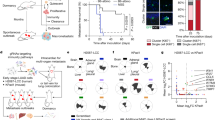
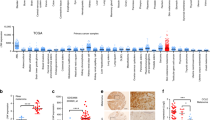
Abstract
Interferon-gamma (IFNG) has long been regarded as the flag-bearer for the anti-cancer immunosurveillance mechanisms. However, relatively recent studies have suggested a dual role of IFNG, albeit there is no direct experimental evidence for its potential pro-tumor functions. Here we provide in vivo evidence that treatment of mouse melanoma cell lines with Ifng enhances their tumorigenicity and metastasis in lung colonization allograft assays performed in immunocompetent syngeneic host mice, but not in immunocompromised host mice. We also show that this enhancement is dependent on downstream signaling via Stat1 but not Stat3, suggesting an oncogenic function of Stat1 in melanoma. The experimental results suggest that melanoma cell-specific Ifng signaling modulates the tumor microenvironment and its pro-tumorigenic effects are partially dependent on the γδ T cells, as Ifng-enhanced tumorigenesis was inhibited in the TCR-δ knockout mice. Overall, these results show that Ifng signaling may have tumor-promoting effects in melanoma by modulating the immune cell composition of the tumor microenvironment.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
RNA-Seq data have been deposited in the Gene Expression Omnibus (GEO) and can be accessed through the accession number GSE214873.
References
Gandini S, Sera F, Cattaruzza MS, Pasquini P, Picconi O, Boyle P, et al. Meta-analysis of risk factors for cutaneous melanoma: II. Sun exposure. Eur J Cancer. 2005;41:45–60.
Nikolaou V, Stratigos AJ. Emerging trends in the epidemiology of melanoma. Br J Dermatol. 2014;170:11–9.
Tran TT, Schulman J, Fisher DE. UV and pigmentation: molecular mechanisms and social controversies. Pigment Cell Melanoma Res. 2008;21:509–16.
Garibyan L, Fisher DE. How sunlight causes melanoma. Curr Oncol Rep. 2010;12:319–26.
Garland CF, Garland FC, Gorham ED. Epidemiologic evidence for different roles of ultraviolet A and B radiation in melanoma mortality rates. Ann Epidemiol. 2003;13:395–404.
Moan J, Porojnicu AC, Dahlback A. Ultraviolet radiation and malignant melanoma. Adv Exp Med Biol. 2008;624:104–16.
Austin MT, Xing Y, Hayes-Jordan AA, Lally KP, Cormier JN. Melanoma incidence rises for children and adolescents: an epidemiologic review of pediatric melanoma in the United States. J Pediatr Surg. 2013;48:2207–13.
Bennett DC. Ultraviolet wavebands and melanoma initiation. Pigment Cell Melanoma Res. 2008;21:520–4.
Maddodi N, Setaluri V. Role of UV in cutaneous melanoma. Photochem Photobiol. 2008;84:528–36.
Slade AD, Austin MT. Childhood melanoma: an increasingly important health problem in the USA. Curr Opin Pediatr. 2014;26:356–61.
Whiteman DC, Whiteman CA, Green AC. Childhood sun exposure as a risk factor for melanoma: a systematic review of epidemiologic studies. Cancer Causes Control. 2001;12:69–82.
Hocker T, Tsao H. Ultraviolet radiation and melanoma: a systematic review and analysis of reported sequence variants. Hum Mutat. 2007;28:578–88.
Matsumura Y, Ananthaswamy HN. Molecular mechanisms of photocarcinogenesis. Front Biosci. 2002;7:d765–783.
Norval M, McLoone P, Lesiak A, Narbutt J. The effect of chronic ultraviolet radiation on the human immune system. Photochem Photobiol. 2008;84:19–28.
Zaidi MR, Davis S, Noonan FP, Graff-Cherry C, Hawley TS, Walker RL, et al. Interferon-gamma links ultraviolet radiation to melanomagenesis in mice. Nature. 2011;469:548–53.
Ikeda H, Old LJ, Schreiber RD. The roles of IFN gamma in protection against tumor development and cancer immunoediting. Cytokine Growth Factor Rev. 2002;13:95–109.
Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science. 2011;331:1565–70.
Brown TJ, Lioubin MN, Marquardt H. Purification and characterization of cytostatic lymphokines produced by activated human T lymphocytes. Synergistic antiproliferative activity of transforming growth factor beta 1, interferon-gamma, and oncostatin M for human melanoma cells. J Immunol. 1987;139:2977–83.
Dummer R, Hassel JC, Fellenberg F, Eichmuller S, Maier T, Slos P, et al. Adenovirus-mediated intralesional interferon-gamma gene transfer induces tumor regressions in cutaneous lymphomas. Blood. 2004;104:1631–8.
Tamura K, Makino S, Araki Y, Imamura T, Seita M. Recombinant interferon beta and gamma in the treatment of adult T-cell leukemia. Cancer. 1987;59:1059–62.
Wall L, Burke F, Smyth JF, Balkwill F. The anti-proliferative activity of interferon-gamma on ovarian cancer: in vitro and in vivo. Gynecol Oncol. 2003;88:S149–151.
Kortylewski M, Komyod W, Kauffmann ME, Bosserhoff A, Heinrich PC, Behrmann I. Interferon-gamma-mediated growth regulation of melanoma cells: involvement of STAT1-dependent and STAT1-independent signals. J Investig Dermatol. 2004;122:414–22.
Zaidi MR, Merlino G. The two faces of interferon-gamma in cancer. Clin Cancer Res. 2011;17:6118–24.
Zaidi MR. The Interferon-Gamma Paradox in Cancer. J Interferon Cytokine Res. 2019;39:30–8.
Mo X, Kazmi HR, Preston-Alp S, Zhou B, Zaidi MR. Interferon-gamma Induces Melanogenesis Via Post-Translational Regulation of Tyrosinase. Pigment Cell Melanoma Res. 2022;35:342–55.
Qing Y, Stark GR. Alternative activation of STAT1 and STAT3 in response to interferon-gamma. J Biol Chem. 2004;279:41679–85.
Ribot JC, deBarros A, Pang DJ, Neves JF, Peperzak V, Roberts SJ, et al. CD27 is a thymic determinant of the balance between interferon-gamma- and interleukin 17-producing gammadelta T cell subsets. Nat Immunol. 2009;10:427–36.
Itohara S, Mombaerts P, Lafaille J, Iacomini J, Nelson A, Clarke AR, et al. T cell receptor delta gene mutant mice: independent generation of alpha beta T cells and programmed rearrangements of gamma delta TCR genes. Cell. 1993;72:337–48.
Mombaerts P, Clarke AR, Rudnicki MA, Iacomini J, Itohara S, Lafaille JJ, et al. Mutations in T-cell antigen receptor genes alpha and beta block thymocyte development at different stages. Nature. 1992;360:225–31.
Ilkovitch D, Lopez DM. Immune modulation by melanoma-derived factors. Exp Dermatol. 2008;17:977–85.
Grasso CS, Tsoi J, Onyshchenko M, Abril-Rodriguez G, Ross-Macdonald P, Wind-Rotolo M, et al. Conserved Interferon-gamma Signaling Drives Clinical Response to Immune Checkpoint Blockade Therapy in Melanoma. Cancer Cell. 2020;38:500–15 e503.
Natarajan VT, Ganju P, Singh A, Vijayan V, Kirty K, Yadav S, et al. IFN-gamma signaling maintains skin pigmentation homeostasis through regulation of melanosome maturation. Proc Natl Acad Sci U S A. 2014;111:2301–6.
Son J, Kim M, Jou I, Park KC, Kang HY. IFN-gamma inhibits basal and alpha-MSH-induced melanogenesis. Pigment Cell Melanoma Res. 2014;27:201–8.
Balkhy HH, Heinzel FP. Endotoxin fails to induce IFN-gamma in endotoxin-tolerant mice: deficiencies in both IL-12 heterodimer production and IL-12 responsiveness. J Immunol. 1999;162:3633–8.
Nguyen KB, Biron CA. Synergism for cytokine-mediated disease during concurrent endotoxin and viral challenges: roles for NK and T cell IFN-gamma production. J Immunol. 1999;162:5238–46.
Gordon-Alonso M, Hirsch T, Wildmann C, van der Bruggen P. Galectin-3 captures interferon-gamma in the tumor matrix reducing chemokine gradient production and T-cell tumor infiltration. Nat Commun. 2017;8:793.
Murtas D, Maric D, De Giorgi V, Reinboth J, Worschech A, Fetsch P, et al. IRF-1 responsiveness to IFN-gamma predicts different cancer immune phenotypes. Br J Cancer. 2013;109:76–82.
Zhang Y, Liu Z. STAT1 in cancer: friend or foe? Discov Med. 2017;24:19–29.
Hsu KS, Zhao X, Cheng X, Guan D, Mahabeleshwar GH, Liu Y, et al. Dual regulation of Stat1 and Stat3 by the tumor suppressor protein PML contributes to interferon alpha-mediated inhibition of angiogenesis. J Biol Chem. 2017;292:10048–60.
Kachroo P, Lee MH, Zhang L, Baratelli F, Lee G, Srivastava MK, et al. IL-27 inhibits epithelial-mesenchymal transition and angiogenic factor production in a STAT1-dominant pathway in human non-small cell lung cancer. J Exp Clin Cancer Res. 2013;32:97.
Zhang Y, Molavi O, Su M, Lai R. The clinical and biological significance of STAT1 in esophageal squamous cell carcinoma. BMC Cancer. 2014;14:791.
Brucet M, Marques L, Sebastian C, Lloberas J, Celada A. Regulation of murine Tap1 and Lmp2 genes in macrophages by interferon gamma is mediated by STAT1 and IRF-1. Genes Immun. 2004;5:26–35.
Leibowitz MS, Andrade Filho PA, Ferrone S, Ferris RL. Deficiency of activated STAT1 in head and neck cancer cells mediates TAP1-dependent escape from cytotoxic T lymphocytes. Cancer Immunol Immunother. 2011;60:525–35.
Rodriguez T, Mendez R, Del Campo A, Jimenez P, Aptsiauri N, Garrido F, et al. Distinct mechanisms of loss of IFN-gamma mediated HLA class I inducibility in two melanoma cell lines. BMC Cancer. 2007;7:34.
Chan SR, Vermi W, Luo J, Lucini L, Rickert C, Fowler AM, et al. STAT1-deficient mice spontaneously develop estrogen receptor alpha-positive luminal mammary carcinomas. Breast Cancer Res. 2012;14:R16.
Lesinski GB, Anghelina M, Zimmerer J, Bakalakos T, Badgwell B, Parihar R, et al. The antitumor effects of IFN-alpha are abrogated in a STAT1-deficient mouse. J Clin Investig. 2003;112:170–80.
Gentles AJ, Newman AM, Liu CL, Bratman SV, Feng W, Kim D, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med. 2015;21:938–45.
Bialasiewicz AA, Ma JX, Richard G. Alpha/beta- and gamma/delta TCR(+) lymphocyte infiltration in necrotising choroidal melanomas. Br J Ophthalmol. 1999;83:1069–73.
Raspollini MR, Castiglione F, Rossi Degl’innocenti D, Amunni G, Villanucci A, Garbini F, et al. Tumour-infiltrating gamma/delta T-lymphocytes are correlated with a brief disease-free interval in advanced ovarian serous carcinoma. Ann Oncol. 2005;16:590–6.
Cordova A, Toia F, La Mendola C, Orlando V, Meraviglia S, Rinaldi G, et al. Characterization of human gammadelta T lymphocytes infiltrating primary malignant melanomas. PLoS ONE. 2012;7:e49878.
Peng G, Wang HY, Peng W, Kiniwa Y, Seo KH, Wang RF. Tumor-infiltrating gammadelta T cells suppress T and dendritic cell function via mechanisms controlled by a unique toll-like receptor signaling pathway. Immunity. 2007;27:334–48.
Ma C, Zhang Q, Ye J, Wang F, Zhang Y, Wevers E, et al. Tumor-infiltrating gammadelta T lymphocytes predict clinical outcome in human breast cancer. J Immunol. 2012;189:5029–36.
Singh S, Kumar S, Srivastava RK, Nandi A, Thacker G, Murali H et al. Loss of ELF5-FBXW7 stabilizes IFNGR1 to promote the growth and metastasis of triple-negative breast cancer through interferon-gamma signalling. Nat Cell Biol. 2020;22:591–602.
Togashi Y, Nishikawa H. Regulatory T Cells: Molecular and Cellular Basis for Immunoregulation. Curr Top Microbiol Immunol. 2017;410:3–27.
Jin C, Lagoudas GK, Zhao C, Bullman S, Bhutkar A, Hu B, et al. Commensal Microbiota Promote Lung Cancer Development via gammadelta T Cells. Cell. 2019;176:998–1013 e1016.
Zhao Y, Niu C, Cui J. Gamma-delta (gammadelta) T cells: friend or foe in cancer development? J Transl Med. 2018;16:3.
Byeseda SE, Burns AR, Dieffenbaugher S, Rumbaut RE, Smith CW, Li Z. ICAM-1 is necessary for epithelial recruitment of gammadelta T cells and efficient corneal wound healing. Am J Pathol. 2009;175:571–9.
Kabelitz D, Wesch D. Features and functions of gamma delta T lymphocytes: focus on chemokines and their receptors. Crit Rev Immunol. 2003;23:339–70.
Callahan MK, Postow MA, Wolchok JD. Immunomodulatory therapy for melanoma: ipilimumab and beyond. Clin Dermatol. 2013;31:191–9.
Drake CG, Lipson EJ, Brahmer JR. Breathing new life into immunotherapy: review of melanoma, lung and kidney cancer. Nat Rev Clin Oncol. 2014;11:24–37.
Noonan FP, Recio JA, Takayama H, Duray P, Anver MR, Rush WL, et al. Neonatal sunburn and melanoma in mice. Nature. 2001;413:271–2.
Noonan FP, Zaidi MR, Wolnicka-Glubisz A, Anver MR, Bahn J, Wielgus A, et al. Melanoma induction by ultraviolet A but not ultraviolet B radiation requires melanin pigment. Nat Commun. 2012;3:884.
Meeth K, Wang JX, Micevic G, Damsky W, Bosenberg MW. The YUMM lines: a series of congenic mouse melanoma cell lines with defined genetic alterations. Pigment Cell Melanoma Res. 2016;29:590–7.
Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–308.
Funding
USA National Institutes of Health, R01CA193711 (MRZ); R01CA236391 (DJK), R01AI068907 (DJK), P30CA006927 (FCCC Comprehensive Cancer Center Core Grant).
Author information
Authors and Affiliations
Contributions
BZ and JB: Conceptualization, methodology, investigation, data acquisition, and paper writing. HRK, XM, and SPA: methodology and data acquisition. KQC: Histopathological analyses and quantification. KNC: Bioinformatic analysis of RNA-Seq data. DJK: Investigation, acquisition of the grant, data interpretation, review and editing of paper. MRZ: Conceptualization, acquisition of grant, data interpretation, project supervision, and paper writing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhou, B., Basu, J., Kazmi, H.R. et al. Interferon-gamma signaling promotes melanoma progression and metastasis. Oncogene 42, 351–363 (2023). https://doi.org/10.1038/s41388-022-02561-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-022-02561-x