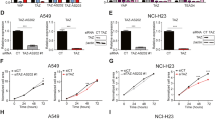
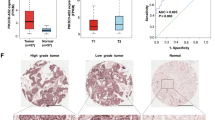
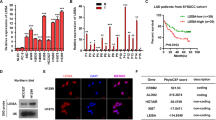
Abstract
Non-coding RNAs have an integral regulatory role in numerous functions related to lung cancer development. Here, we report identification of a novel lncRNA, termed TP53-inhibiting lncRNA (TILR), which was found to function as a constitutive negative regulator of p53 expression, including activation of downstream genes such as p21 and MDM2, and induction of apoptosis. A proteomic search for TILR-associated proteins revealed an association with PCBP2, while the mid-portion of TILR was found to be required for both PCBP2 and p53 mRNA binding. In addition, depletion of PCBP2 resulted in phenocopied effects of TILR silencing. TILR was also shown to suppress p53 expression in a post-transcriptional manner, as well as via a positive feedback loop involving p53 and Fanconi anemia pathway genes. Taken together, the present findings clearly demonstrate that TILR constitutively inhibits p53 expression in cooperation with PCBP2, thus maintaining p53 transcriptional activity at a level sufficiently low for avoidance of spurious apoptosis induction.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The microarray data generated in this study have been deposited in the NCBI Gene Expression Omnibus database under the accession code GSE180940 and GSE180944.
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49.
Takahashi T, Nau MM, Chiba I, Birrer MJ, Rosenberg RK, Vinocour M, et al. p53: a frequent target for genetic abnormalities in lung cancer. Science. 1989;246:491–4.
Cancer Genome Atlas Research N. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–50.
Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer. 2014;14:359–70.
Skoulidis F, Heymach JV. Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy. Nat Rev Cancer. 2019;19:495–509.
Kastenhuber ER, Lowe SW. Putting p53 in context. Cell. 2017;170:1062–78.
Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–6.
Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–32.
Zhang Y, Pitchiaya S, Cieslik M, Niknafs YS, Tien JC, Hosono Y, et al. Analysis of the androgen receptor-regulated lncRNA landscape identifies a role for ARLNC1 in prostate cancer progression. Nat Genet. 2018;50:814–24.
Cho SW, Xu J, Sun R, Mumbach MR, Carter AC, Chen YG, et al. Promoter of lncRNA Gene PVT1 is a tumor-suppressor DNA boundary element. Cell. 2018;173:1398–412.e22.
Long Y, Hwang T, Gooding AR, Goodrich KJ, Rinn JL, Cech TR. RNA is essential for PRC2 chromatin occupancy and function in human pluripotent stem cells. Nat Genet. 2020;52:931–8.
Kajino T, Shimamura T, Gong S, Yanagisawa K, Ida L, Nakatochi M, et al. Divergent lncRNA MYMLR regulates MYC by eliciting DNA looping and promoter-enhancer interaction. EMBO J. 2019;38:e98441.
Hsu F, Kent WJ, Clawson H, Kuhn RM, Diekhans M, Haussler D. The UCSC known genes. Bioinformatics. 2006;22:1036–46.
Takeuchi T, Tomida S, Yatabe Y, Kosaka T, Osada H, Yanagisawa K, et al. Expression profile-defined classification of lung adenocarcinoma shows close relationship with underlying major genetic changes and clinicopathologic behaviors. J Clin Oncol. 2006;24:1679–88.
Tomida S, Takeuchi T, Shimada Y, Arima C, Matsuo K, Mitsudomi T, et al. Relapse-related molecular signature in lung adenocarcinomas identifies patients with dismal prognosis. J Clin Oncol. 2009;27:2793–9.
Catherman AD, Li M, Tran JC, Durbin KR, Compton PD, Early BP, et al. Top-down proteomics of human membrane proteins from enriched mitochondrial fractions. Anal Chem. 2013;85:1880–8.
Chen Y, Li Y, Zhong J, Zhang J, Chen Z, Yang L, et al. Identification of missing proteins defined by chromosome-centric proteome project in the cytoplasmic detergent-insoluble proteins. J Proteome Res. 2015;14:3693–709.
Slack FJ, Chinnaiyan AM. The role of non-coding RNAs in oncology. Cell. 2019;179:1033–55.
Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2021;22:96–118.
Niraj J, Farkkila A, D’Andrea AD. The Fanconi anemia pathway in cancer. Annu Rev Cancer Biol. 2019;3:457–78.
Nalepa G, Clapp DW. Fanconi anaemia and cancer: an intricate relationship. Nat Rev Cancer. 2018;18:168–85.
Ceccaldi R, Parmar K, Mouly E, Delord M, Kim JM, Regairaz M, et al. Bone marrow failure in Fanconi anemia is triggered by an exacerbated p53/p21 DNA damage response that impairs hematopoietic stem and progenitor cells. Cell Stem Cell. 2012;11:36–49.
Ceccaldi R, Briot D, Larghero J, Vasquez N, Dubois d’Enghien C, Chamousset D, et al. Spontaneous abrogation of the G(2)DNA damage checkpoint has clinical benefits but promotes leukemogenesis in Fanconi anemia patients. J Clin Invest. 2011;121:184–94.
Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303.
Shieh SY, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–34.
Bode AM, Dong Z. Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer. 2004;4:793–805.
Tang Y, Zhao W, Chen Y, Zhao Y, Gu W. Acetylation is indispensable for p53 activation. Cell. 2008;133:612–26.
Mazan-Mamczarz K, Galban S, Lopez de Silanes I, Martindale JL, Atasoy U, Keene JD, et al. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc Natl Acad Sci USA. 2003;100:8354–9.
Wedeken L, Singh P, Klempnauer KH. Tumor suppressor protein Pdcd4 inhibits translation of p53 mRNA. J Biol Chem. 2011;286:42855–62.
Jaber S, Toufektchan E, Lejour V, Bardot B, Toledo F. p53 downregulates the Fanconi anaemia DNA repair pathway. Nat Commun. 2016;7:11091.
Adriaens C, Standaert L, Barra J, Latil M, Verfaillie A, Kalev P, et al. p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat Med. 2016;22:861–8.
Zhou Y, Zhong Y, Wang Y, Zhang X, Batista DL, Gejman R, et al. Activation of p53 by MEG3 non-coding RNA. J Biol Chem. 2007;282:24731–42.
Liu CY, Zhang YH, Li RB, Zhou LY, An T, Zhang RC, et al. LncRNA CAIF inhibits autophagy and attenuates myocardial infarction by blocking p53-mediated myocardin transcription. Nat Commun. 2018;9:29.
Choi HS, Hwang CK, Song KY, Law PY, Wei LN, Loh HH. Poly(C)-binding proteins as transcriptional regulators of gene expression. Biochem Biophys Res Commun. 2009;380:431–6.
Ebi H, Tomida S, Takeuchi T, Arima C, Sato T, Mitsudomi T, et al. Relationship of deregulated signaling converging onto mTOR with prognosis and classification of lung adenocarcinoma shown by two independent in silico analyses. Cancer Res. 2009;69:4027–35.
Arima C, Kajino T, Tamada Y, Imoto S, Shimada Y, Nakatochi M, et al. Lung adenocarcinoma subtypes definable by lung development-related miRNA expression profiles in association with clinicopathologic features. Carcinogenesis. 2014;35:2224–31.
Tanaka H, Yanagisawa K, Shinjo K, Taguchi A, Maeno K, Tomida S, et al. Lineage-specific dependency of lung adenocarcinomas on the lung development regulator TTF-1. Cancer Res. 2007;67:6007–11.
Stein CS, Jadiya P, Zhang X, McLendon JM, Abouassaly GM, Witmer NH, et al. Mitoregulin: A lncRNA-encoded microprotein that supports mitochondrial supercomplexes and respiratory efficiency. Cell Rep. 2018;23:3710–20.e8.
Lin Y, Nakatochi M, Hosono Y, Ito H, Kamatani Y, Inoko A, et al. Genome-wide association meta-analysis identifies GP2 gene risk variants for pancreatic cancer. Nat Commun. 2020;11:3175.
Feng Y, Absher D, Eberhart DE, Brown V, Malter HE, Warren ST. FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol Cell. 1997;1:109–18.
Acknowledgements
We thank Naoe Hotta and Keiko Kano for assistance with data collection, as well as Sebastian Griesing and Atsuko Niimi for the helpful discussion. This work was supported in part by a Grant-in-Aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan, Grants-in-Aid for Scientific Research (A) and (C) from the Japan Society for the Promotion of Science, Takeda Science Foundation, and a grant from the Princess Takamatsu Cancer Research Fund.
Author information
Authors and Affiliations
Contributions
MI, TK, and TT conceived the project. MI, TK, KY, YH, HI, and YS performed the experiments. MI, TK, KY, MS, AT, and TT analyzed the data. TK and MN performed the informatics analysis. MI, TK, and TT wrote the paper, with input from all authors. TT supervised the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Iwai, M., Kajino, T., Nakatochi, M. et al. Long non-coding RNA TILR constitutively represses TP53 and apoptosis in lung cancer. Oncogene 42, 364–373 (2023). https://doi.org/10.1038/s41388-022-02546-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-022-02546-w