Abstract
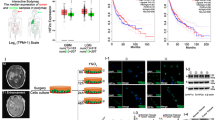
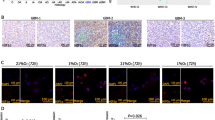
Circular RNAs (circRNAs) play important roles in the malignant progression of tumours. Herein, we identified an unreported circRNA (hsa-circ-0072688, also named circADAMTS6) that is specifically upregulated in the hypoxic microenvironment of glioblastoma and closely correlated with poor prognosis of gliblastoma patients. We found that circADAMTS6 promotes the malignant progression of glioblastoma by promoting cell proliferation and inhibiting apoptosis. Mechanistically, the hypoxic tumour microenvironment upregulates circADAMTS6 expression through transcription factor activator protein 1 (AP-1) and RNA-binding protein TAR DNA-binding protein 43 (TDP43). Moreover, circADAMTS6 accelerates glioblastoma progression by recruiting and stabilising annexin A2 (ANXA2) in a proteasomes-dependent manner. Furthermore, we found T-5224 (AP-1 inhibitor) treatment induces downregulation of circADAMTS6 and then inhibits tumour growth. In conclusion, our findings highlight the important role of the circADAMTS6/ANXA2 axis based on hypoxic microenvironment in glioblastoma progression, as well as its regulation in NF-κB pathway. Targeting circADAMTS6 is thus expected to become a novel therapeutic strategy for glioblastoma.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The datasets used and analysed during the current study are available from the corresponding author upon reasonable request.
References
Ostrom QT, Cioffi G, Waite K, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2014-2018. Neuro Oncol. 2021;23:iii1–iii105. https://doi.org/10.1093/neuonc/noab200
Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D, et al. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol 2021;23:1231–51. https://doi.org/10.1093/neuonc/noab106
Stupp R, Taillibert S, Kanner AA, Kesari S, Steinberg DM, Toms SA, et al. Maintenance therapy with tumor-treating fields plus temozolomide vs temozolomide alone for glioblastoma: a randomized clinical trial. JAMA. 2015;314:2535–43. https://doi.org/10.1001/jama.2015.16669
Stupp R, Hegi ME, Gorlia T, Erridge SC, Perry J, Hong YK, et al. Cilengitide combined with standard treatment for patients with newly diagnosed glioblastoma with methylated MGMT promoter (CENTRIC EORTC 26071-22072 study): a multicentre, randomised, open-label, phase 3 trial. Lancet Oncol. 2014;15:1100–8. https://doi.org/10.1016/S1470-2045(14)70379-1
Wick W, Gorlia T, Bendszus M, Taphoorn M, Sahm F, Harting I, et al. Lomustine and bevacizumab in progressive glioblastoma. N. Engl J Med. 2017;377:1954–63. https://doi.org/10.1056/NEJMoa1707358
Reardon DA, Rich JN, Friedman HS, Bigner DD. Recent advances in the treatment of malignant astrocytoma. J Clin Oncol. 2006;24:1253–65. https://doi.org/10.1200/JCO.2005.04.5302
Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675–91. https://doi.org/10.1038/s41576-019-0158-7
Gao X, Xia X, Li F, Zhang M, Zhou H, Wu X, et al. Circular RNA-encoded oncogenic E-cadherin variant promotes glioblastoma tumorigenicity through activation of EGFR-STAT3 signalling. Nat Cell Biol. 2021;23:278–91. https://doi.org/10.1038/s41556-021-00639-4
Liu Y, Li Z, Zhang M, Zhou H, Wu X, Zhong J, et al. Rolling-translated EGFR variants sustain EGFR signaling and promote glioblastoma tumorigenicity. Neuro Oncol. 2021;23:743–56. https://doi.org/10.1093/neuonc/noaa279
Lou J, Hao Y, Lin K, Lyu Y, Chen M, Wang H, et al. Circular RNA CDR1as disrupts the p53/MDM2 complex to inhibit gliomagenesis. Mol Cancer. 2020;19:138 https://doi.org/10.1186/s12943-020-01253-y
Khan IN, Ullah N, Hussein D, Saini KS. Current and emerging biomarkers in tumors of the central nervous system: possible diagnostic, prognostic and therapeutic applications. Semin Cancer Biol. 2018;52:85–102. https://doi.org/10.1016/j.semcancer.2017.07.004
Chen LL. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol. 2020;21:475–90. https://doi.org/10.1038/s41580-020-0243-y
Zhang Y, Qian J, Gu C, Yang Y. Alternative splicing and cancer: a systematic review. Signal Transduct Target Ther. 2021;6:78 https://doi.org/10.1038/s41392-021-00486-7
Li S, Han L. Circular RNAs as promising biomarkers in cancer: detection, function, and beyond. Genome Med. 2019;11:15 https://doi.org/10.1186/s13073-019-0629-7
Shang Q, Yang Z, Jia R, Ge S. The novel roles of circRNAs in human cancer. Mol Cancer. 2019;18:6 https://doi.org/10.1186/s12943-018-0934-6
Goodall GJ, Wickramasinghe VO. RNA in cancer. Nat Rev Cancer. 2021;21:22–36. https://doi.org/10.1038/s41568-020-00306-0
Wei Y, Lu C, Zhou P, Zhao L, Lyu X, Yin J, et al. EIF4A3-induced circular RNA ASAP1 promotes tumorigenesis and temozolomide resistance of glioblastoma via NRAS/MEK1/ERK1-2 signaling. Neuro Oncol. 2021;23:611–24. https://doi.org/10.1093/neuonc/noaa214
Zhang S, Liao K, Miao Z, Wang Q, Miao Y, Guo Z, et al. CircFOXO3 promotes glioblastoma progression by acting as a competing endogenous RNA for NFAT5. Neuro Oncol. 2019;21:1284–96. https://doi.org/10.1093/neuonc/noz128
Chen D, Chou FJ, Chen Y, Tian H, Wang Y, You B, et al. Targeting the radiation-induced TR4 nuclear receptor-mediated QKI/circZEB1/miR-141-3p/ZEB1 signaling increases prostate cancer radiosensitivity. Cancer Lett. 2020;495:100–11. https://doi.org/10.1016/j.canlet.2020.07.040
Shen S, Yang Y, Shen P, Ma J, Fang B, Wang Q, et al. circPDE4B prevents articular cartilage degeneration and promotes repair by acting as a scaffold for RIC8A and MID1. Ann Rheum Dis. 2021;80:1209–19. https://doi.org/10.1136/annrheumdis-2021-219969
Chen J, Luo H, Liu Y, Zhang W, Li H, Luo T, et al. Oxygen-self-produced nanoplatform for relieving hypoxia and breaking resistance to sonodynamic treatment of pancreatic cancer. ACS Nano. 2017;11:12849–62. https://doi.org/10.1021/acsnano.7b08225.
Man J, Yu X, Huang H, Zhou W, Xiang C, Huang H, et al. Hypoxic induction of vasorin regulates notch1 turnover to maintain glioma stem-like cells. Cell Stem Cell. 2018;22:104–.e6. https://doi.org/10.1016/j.stem.2017.10.005
Hsieh CH, Lin YJ, Wu CP, Lee HT, Shyu WC, Wang CC. Livin contributes to tumor hypoxia-induced resistance to cytotoxic therapies in glioblastoma multiforme. Clin Cancer Res. 2015;21:460–70. https://doi.org/10.1158/1078-0432.CCR-14-0618
Hambardzumyan D, Bergers G. Glioblastoma: defining tumor niches. Trends Cancer. 2015;1:252–65. https://doi.org/10.1016/j.trecan.2015.10.009
Fornes O, Castro-Mondragon JA, Khan A, van der Lee R, Zhang X, Richmond PA, et al. JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2020;48:D87–D92. https://doi.org/10.1093/nar/gkz1001
Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13:34–42. https://doi.org/10.1080/15476286.2015.1128065
Qiu W, Guo X, Li B, Wang J, Qi Y, Chen Z, et al. Exosomal miR-1246 from glioma patient body fluids drives the differentiation and activation of myeloid-derived suppressor cells. Mol Ther. 2021;29:3449–64. https://doi.org/10.1016/j.ymthe.2021.06.023
Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17:272–83. https://doi.org/10.1038/nrg.2016.20
Gibbings D, Mostowy S, Jay F, Schwab Y, Cossart P, Voinnet O. Selective autophagy degrades DICER and AGO2 and regulates miRNA activity. Nat Cell Biol. 2012;14:1314–21. https://doi.org/10.1038/ncb2611
Sheu-Gruttadauria J, MacRae IJ. Phase transitions in the assembly and function of human miRISC. Cell. 2018;173:946–.e16. https://doi.org/10.1016/j.cell.2018.02.051
Sablok G, Zhao H, Sun X. Plant circular RNAs (circRNAs): transcriptional regulation beyond miRNAs in plants. Mol Plant. 2016;9:192–4. https://doi.org/10.1016/j.molp.2015.12.021
Huang S, Li X, Zheng H, Si X, Li B, Wei G, et al. Loss of super-enhancer-regulated circRNA Nfix induces cardiac regeneration after myocardial infarction in adult mice. Circulation. 2019;139:2857–76. https://doi.org/10.1161/CIRCULATIONAHA.118.038361
Wen G, Zhou T, Gu W. The potential of using blood circular RNA as liquid biopsy biomarker for human diseases. Protein Cell. 2021;12:911–46. https://doi.org/10.1007/s13238-020-00799-3
Niu Y, Lin Z, Wan A, Sun L, Yan S, Liang H, et al. Loss-of-function genetic screening identifies aldolase A as an essential driver for liver cancer cell growth under hypoxia. Hepatology. 2021;74:1461–79. https://doi.org/10.1002/hep.31846
Ni J, Wang X, Stojanovic A, Zhang Q, Wincher M, Bühler L, et al. Single-cell RNA sequencing of tumor-infiltrating NK cells reveals that inhibition of transcription factor HIF-1α Unleashes NK Cell Activity. Immunity. 2020;52:1075–.e8. https://doi.org/10.1016/j.immuni.2020.05.001
Li L, Yang L, Fan Z, Xue W, Shen Z, Yuan Y, et al. Hypoxia-induced GBE1 expression promotes tumor progression through metabolic reprogramming in lung adenocarcinoma. Signal Transduct Target Ther. 2020;5:54 https://doi.org/10.1038/s41392-020-0152-8
Wilson GK, Tennant DA, McKeating JA. Hypoxia inducible factors in liver disease and hepatocellular carcinoma: current understanding and future directions. J Hepatol. 2014;61:1397–406. https://doi.org/10.1016/j.jhep.2014.08.025
Zhang P, Zhang XO, Jiang T, Cai L, Huang X, Liu Q, et al. Comprehensive identification of alternative back-splicing in human tissue transcriptomes. Nucleic Acids Res. 2020;48:1779–89. https://doi.org/10.1093/nar/gkaa005
Qing G, Lu Q, Xiong Y, Zhang L, Wang H, Li X, et al. New opportunities and challenges of smart polymers in post-translational modification proteomics. Adv Mater. 2017;29. https://doi.org/10.1002/adma.201604670.
Liu J, Qian C, Cao X. Post-translational modification control of innate immunity. Immunity. 2016;45:15–30. https://doi.org/10.1016/j.immuni.2016.06.020
Vucic D, Dixit VM, Wertz IE. Ubiquitylation in apoptosis: a post-translational modification at the edge of life and death. Nat Rev Mol Cell Biol. 2011;12:439–52. https://doi.org/10.1038/nrm3143
Wei WS, Chen X, Guo LY, Li XD, Deng MH, Yuan GJ, et al. TRIM65 supports bladder urothelial carcinoma cell aggressiveness by promoting ANXA2 ubiquitination and degradation. Cancer Lett. 2018;435:10–22. https://doi.org/10.1016/j.canlet.2018.07.036
Deng S, Jing B, Xing T, Hou L, Yang Z. Overexpression of annexin A2 is associated with abnormal ubiquitination in breast cancer. Genomics Proteom Bioinform. 2012;10:153–7. https://doi.org/10.1016/j.gpb.2011.12.001
Tu Y, Xie P, Du X, Fan L, Bao Z, Sun G, et al. S100A11 functions as novel oncogene in glioblastoma via S100A11/ANXA2/NF-κB positive feedback loop. J Cell Mol Med. 2019;23:6907–18. https://doi.org/10.1111/jcmm.14574
Wang YS, Li H, Li Y, Zhu H, Jin YH. Identification of natural compounds targeting Annexin A2 with an anti-cancer effect. Protein Cell. 2018;9:568–79. https://doi.org/10.1007/s13238-018-0513-z
Kpetemey M, Dasgupta S, Rajendiran S, Das S, Gibbs LD, Shetty P, et al. MIEN1, a novel interactor of Annexin A2, promotes tumor cell migration by enhancing AnxA2 cell surface expression. Mol Cancer. 2015;14:156 https://doi.org/10.1186/s12943-015-0428-8
Sarkar S, Swiercz R, Kantara C, Hajjar KA, Singh P. Annexin A2 mediates up-regulation of NF-κB, β-catenin, and stem cell in response to progastrin in mice and HEK-293 cells. Gastroenterology. 2011;140:583–.e4. https://doi.org/10.1053/j.gastro.2010.08.054
Wang Y, Chen K, Cai Y, Cai Y, Yuan X, Wang L, et al. Annexin A2 could enhance multidrug resistance by regulating NF-κB signaling pathway in pediatric neuroblastoma. J Exp Clin Cancer Res. 2017;36:111 https://doi.org/10.1186/s13046-017-0581-6
Liu R, Tan J, Shen X, Jiang K, Wang C, Zhu G, et al. Therapeutic targeting of FOS in mutant TERT cancers through removing TERT suppression of apoptosis via regulating survivin and TRAIL-R2. Proc Natl Acad Sci USA. 2021;118:e2022779118 https://doi.org/10.1073/pnas.2022779118
Kamide D, Yamashita T, Araki K, Tomifuji M, Tanaka Y, Tanaka S, et al. Selective activator protein-1 inhibitor T-5224 prevents lymph node metastasis in an oral cancer model. Cancer Sci. 2016;107:666–73. https://doi.org/10.1111/cas.12914
Acknowledgements
We are grateful to Dr. Frederick F Lang and Dr. Krishna P.L. Bhat for providing GSC cell lines used in our study; and CGGA, TCGA, GEO, Granvendeel and Rembrandt for public sequence data. This work was supported by grants from the National Natural Science Foundation of China (Nos. 82273195; 81874083; 82072776; 82273286; 82072775; 82203419;), Natural Science Foundation of Shandong Province of China (Nos. ZR2019BH057;ZR2020QH174; ZR2021LSW025), the Jinan Science and Technology Bureau of Shandong Province (2021GXRC029), Key Clinical Research Project of Clinical Research Center of Shandong University (2020SDUCRCA011) and Taishan Pandeng Scholar Program of Shandong Province (No. tspd20210322).
Author information
Authors and Affiliations
Contributions
GL, HX and SZ conceived and devised the study; BL, RZ, ZP, SZ, WQ, QG, YQ, ZG, YF, HX, ML and JZ performed the experiments; HW, XJ, SW and QW performed bioinformatics and statistical analysis; JQ and LD analysed the data. XG and PZ supervised the research. SZ, BL and RZ wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhao, S., Li, B., Zhao, R. et al. Hypoxia-induced circADAMTS6 in a TDP43-dependent manner accelerates glioblastoma progression via ANXA2/ NF-κB pathway. Oncogene 42, 138–153 (2023). https://doi.org/10.1038/s41388-022-02542-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-022-02542-0
This article is cited by
-
The integrate profiling of single-cell and spatial transcriptome RNA-seq reveals tumor heterogeneity, therapeutic targets, and prognostic subtypes in ccRCC
Cancer Gene Therapy (2024)
-
Mechanisms of immune checkpoint inhibitors: insights into the regulation of circular RNAS involved in cancer hallmarks
Cell Death & Disease (2024)
-
CircRNF10 triggers a positive feedback loop to facilitate progression of glioblastoma via redeploying the ferroptosis defense in GSCs
Journal of Experimental & Clinical Cancer Research (2023)
-
New insight into circRNAs: characterization, strategies, and biomedical applications
Experimental Hematology & Oncology (2023)
-
Hypoxia-induced circRTN4IP1 promotes progression and glycolysis of hepatocellular carcinoma cells
Functional & Integrative Genomics (2023)