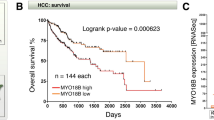
Abstract
Terminal differentiation failure is an important cause of rhabdomyosarcoma genesis, however, little is known about the epigenetic regulation of aberrant myogenic differentiation. Here, we show that GATA-4 recruits polycomb group proteins such as EZH2 to negatively regulate miR-29a in undifferentiated C2C12 myoblast cells, whereas recruitment of GRIP-1 to GATA-4 proteins displaces EZH2, resulting in the activation of miR-29a during myogenic differentiation of C2C12 cells. Moreover, in poorly differentiated rhabdomyosarcoma cells, EZH2 still binds to the miR-29a promoter with GATA-4 to mediate transcriptional repression of miR-29a. Interestingly, once re-differentiation of rhabdomyosarcoma cells toward skeletal muscle, EZH2 was dispelled from miR-29a promoter which is similar to that in myogenic differentiation of C2C12 cells. Eventually, this expression of miR-29a results in limited rhabdomyosarcoma cell proliferation and promotes myogenic differentiation. We thus establish that GATA-4 can function as a molecular switch in the up- and downregulation of miR-29a expression. We also demonstrate that GATA-4 acts as a tumor suppressor in rhabdomyosarcoma partly via miR-29a, which thus provides a potential therapeutic target for rhabdomyosarcoma.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The source data that support the findings of this study are available in figshare with the identifier https://doi.org/10.6084/m9.figshare.20173850.
References
Chen C, Dorado Garcia H, Scheer M, Henssen AG. Current and future treatment strategies for rhabdomyosarcoma. Front Oncol. 2019;9:1458.
Missiaglia E, Shepherd CJ, Aladowicz E, Olmos D, Selfe J, Pierron G, et al. MicroRNA and gene co-expression networks characterize biological and clinical behavior of rhabdomyosarcomas. Cancer Lett. 2017;385:251–60.
Hanna JA, Garcia MR, Lardennois A, Leavey PJ, Maglic D, Fagnan A, et al. PAX3-FOXO1 drives miR-486-5p and represses miR-221 contributing to pathogenesis of alveolar rhabdomyosarcoma. Oncogene 2018;37:1991–2007.
Pandey PR, Chatterjee B, Olanich ME, Khan J, Miettinen MM, Hewitt SM, et al. PAX3-FOXO1 is essential for tumour initiation and maintenance but not recurrence in a human myoblast model of rhabdomyosarcoma. J Pathol. 2017;241:626–37.
Megiorni F. Epigenetics in rhabdomyosarcoma: cues to new biomarkers and targeted therapies. EBioMedicine. 2020;52:102673.
Cieśla M, Dulak J, Józkowicz A. MicroRNAs and epigenetic mechanisms of rhabdomyosarcoma development. Int J Biochem Cell Biol. 2014;53:482–92.
Bersani F, Lingua MF, Morena D, Foglizzo V, Miretti S, Lanzetti L, et al. Deep sequencing reveals a novel miR-22 regulatory network with therapeutic potential in rhabdomyosarcoma. Cancer Res. 2016;76:6095–106.
Ballarino M, Morlando M, Fatica A, Bozzoni I. Non-coding RNAs in muscle differentiation and musculoskeletal disease. J Clin Investig. 2016;126:2021–30.
Sun M, Huang F, Yu D, Zhang Y, Xu H, Zhang L, et al. Autoregulatory loop between TGF-beta1/miR-411-5p/SPRY4 and MAPK pathway in rhabdomyosarcoma modulates proliferation and differentiation. Cell death Dis. 2015;6:e1859.
Vella S, Pomella S, Leoncini PP, Colletti M, Conti B, Marquez VE, et al. MicroRNA-101 is repressed by EZH2 and its restoration inhibits tumorigenic features in embryonal rhabdomyosarcoma. Clin Epigenetics. 2015;7:82.
Wang H, Garzon R, Sun H, Ladner KJ, Singh R, Dahlman J, et al. NF-kappaB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell. 2008;14:369–81.
Wei W, He HB, Zhang WY, Zhang HX, Bai JB, Liu HZ, et al. miR-29 targets Akt3 to reduce proliferation and facilitate differentiation of myoblasts in skeletal muscle development. Cell Death Dis. 2013;4:e668.
Chikenji A, Ando H, Nariyama M, Suga T, Iida R, Gomi K. MyoD is regulated by the miR-29a-Tet1 pathway in C2C12 myoblast cells. J Oral Sci. 2016;58:219–29.
Yao CX, Wei QX, Zhang YY, Wang WP, Xue LX, Yang F, et al. miR-200b targets GATA-4 during cell growth and differentiation. RNA Biol. 2013;10:465–80.
Mikhailov AT, Torrado M. Myocardial transcription factors in diastolic dysfunction: clues for model systems and disease. Heart Fail Rev. 2016;21:783–94.
Whitcomb J, Gharibeh L, Nemer M. From embryogenesis to adulthood: Critical role for GATA factors in heart development and function. IUBMB Life. 2020;72:53–67.
Wannenes F, Caprio M, Gatta L, Fabbri A, Bonini S, Moretti C. Androgen receptor expression during C2C12 skeletal muscle cell line differentiation. Mol Cell Endocrinol. 2008;292:11–19.
Chen SL, Dowhan DH, Hosking BM, Muscat GE. The steroid receptor coactivator, GRIP-1, is necessary for MEF-2C-dependent gene expression and skeletal muscle differentiation. Genes Dev. 2000;14:1209–28.
Chinenov Y, Gupte R, Dobrovolna J, Flammer JR, Liu B, Michelassi FE, et al. Role of transcriptional coregulator GRIP1 in the anti-inflammatory actions of glucocorticoids. Proc Natl Acad Sci USA. 2012;109:11776–81.
Di Croce L, Helin K. Transcriptional regulation by Polycomb group proteins. Nat Struct Mol Biol. 2013;20:1147–55.
He A, Shen X, Ma Q, Cao J, von Gise A, Zhou P, et al. PRC2 directly methylates GATA4 and represses its transcriptional activity. Genes Dev. 2012;26:37–42.
Cai R, Qimuge N, Ma M, Wang Y, Tang G, Zhang Q, et al. MicroRNA-664-5p promotes myoblast proliferation and inhibits myoblast differentiation by targeting serum response factor and Wnt1. J Biol Chem. 2018;293:19177–90.
Qiu H, Liu N, Luo L, Zhong J, Tang Z, Kang K, et al. MicroRNA-17-92 regulates myoblast proliferation and differentiation by targeting the ENH1/Id1 signaling axis. Cell Death Differ. 2016;23:1658–69.
Kouznetsova VL, Tchekanov A, Li X, Yan X, Tsigelny IF. Polycomb repressive 2 complex-Molecular mechanisms of function. Protein Sci. 2019;28:1387–99.
Healy E, Mucha M, Glancy E, Fitzpatrick DJ, Conway E, Neikes HK, et al. PRC2.1 and PRC2.2 synergize to coordinate H3K27 trimethylation. Mol Cell. 2019;76:437−52.
Cutter DiPiazza AR, Taneja N, Dhakshnamoorthy J, Wheeler D, Holla S, Grewal SIS. Spreading and epigenetic inheritance of heterochromatin require a critical density of histone H3 lysine 9 tri-methylation. Proc Natl Acad Sci USA. 2021;118:e2100699118.
Qu Y, Lu D, Jiang H, Chi X, Zhang H. EZH2 is required for mouse oocyte meiotic maturation by interacting with and stabilizing spindle assembly checkpoint protein BubRI. Nucleic Acids Res. 2016;44:7659–72.
Davicioni E, Finckenstein FG, Shahbazian V, Buckley JD, Triche TJ, Anderson MJ. Identification of a PAX-FKHR gene expression signature that defines molecular classes and determines the prognosis of alveolar rhabdomyosarcomas. Cancer Res. 2006;66:6936–46.
Hinson AR, Jones R, Crose LE, Belyea BC, Barr FG, Linardic CM. Human rhabdomyosarcoma cell lines for rhabdomyosarcoma research: utility and pitfalls. Front Oncol. 2013;3:183.
Bevilacqua A, Willis MS, Bultman SJ. SWI/SNF chromatin-remodeling complexes in cardiovascular development and disease. Cardiovasc Pathol. 2014;23:85–91.
Schwartz YB, Pirrotta V. Ruled by ubiquitylation: a new order for polycomb recruitment. Cell Rep. 2014;8:321–5.
Kim KH, Roberts CW. Targeting EZH2 in cancer. Nat Med. 2016;22:128–34.
Achour C, Aguilo F. Long non-coding RNA and Polycomb: an intricate partnership in cancer biology. Front Biosci . 2018;23:2106–32.
Dai X, Chen C, Yang Q, Xue J, Chen X, Sun B, et al. Exosomal circRNA_100284 from arsenite-transformed cells, via microRNA-217 regulation of EZH2, is involved in the malignant transformation of human hepatic cells by accelerating the cell cycle and promoting cell proliferation. Cell Death Dis. 2018;9:454.
Akiyama Y, Watkins N, Suzuki H, Jair KW, van Engeland M, Esteller M, et al. GATA-4 and GATA-5 transcription factor genes and potential downstream antitumor target genes are epigenetically silenced in colorectal and gastric cancer. Mol Cell Biol. 2003;23:8429–39.
Charron F, Tsimiklis G, Arcand M, Robitaille L, Liang Q, Molkentin JD, et al. Tissue-specific GATA factors are transcriptional effectors of the small GTPase RhoA. Genes Dev. 2001;15:2702–19.
Ma CX, Song YL, Xiao L, Xue LX, Li WJ, Laforest B, et al. EGF is required for cardiac differentiation of P19CL6 cells through interaction with GATA-4 in a time- and dose-dependent manner. Cell Mol Life Sci. 2015;72:2005–22.
Irrcher I, Ljubicic V, Kirwan AF, Hood DA. AMP-activated protein kinase-regulated activation of the PGC-1alpha promoter in skeletal muscle cells. PLoS ONE. 2008;3:e3614.
Vu LP, Luciani L, Nimer SD. Histone-modifying enzymes: their role in the pathogenesis of acute leukemia and their therapeutic potential. Int J Hematol. 2013;97:198–209.
Xiong CJ, Li PF, Song YL, Xue LX, Jia ZQ, Yao CX, et al. Insulin induces C2C12 cell proliferation and apoptosis through regulation of cyclin D1 and BAD expression. J Cell Biochem. 2013;114:2708–17.
Yao CX, Xiong CJ, Wang WP, Yang F, Zhang SF, Wang TQ, et al. Transcription factor GATA-6 recruits PPARalpha to cooperatively activate Glut4 gene expression. J Mol Biol. 2012;415:143–58.
Zhang S, Yin Z, Dai FF, Wang H, Zhou MJ, Yang MH, et al. miR-29a attenuates cardiac hypertrophy through inhibition of PPARdelta expression. J Cell Physiol. 2019;234:13252–62.
Yao CX, Shi JC, Ma CX, Xiong CJ, Song YL, Zhang SF, et al. EGF protects cells against Dox-induced growth arrest through activating cyclin D1 expression. J Cell Biochem. 2015;116:1755–65.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (No. 82072975, 81771631, 81672091, and 91749107), the High-Level Talents of Henan Province, particularly the support for the Central Plains Thousand Talents Program, which are the leading talents of Central Plains Basic Research (ZYQR201810120), 2022 Henan Province Science and Technology R&D Program Joint Fund (Cultivation of Superior Disciplines), and the key project of discipline construction of Zhengzhou University in 2020 (XKZDQY202002).
Author information
Authors and Affiliations
Contributions
MXZ, LXX and WGZ conceived, designed, and directed the study. YLS performed most ChIP, GST pull-down, and coimmunoprecipitation experiments. MHY performed real-time PCR and transfection experiments. SZ finishes cell proliferation assay and Xenograft tumor experiment. HW performed immunofluorescence and immunohistochemistry assays. KLK, CXY, FFD, MJZ, JBL, ZRW, and ZNY analyzed data.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Song, YL., Yang, MH., Zhang, S. et al. A GRIP-1–EZH2 switch binding to GATA-4 is linked to the genesis of rhabdomyosarcoma through miR-29a. Oncogene 41, 5223–5237 (2022). https://doi.org/10.1038/s41388-022-02521-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-022-02521-5