Abstract
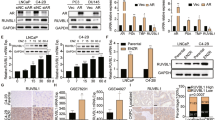
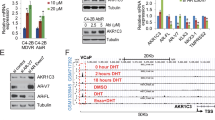
Prostate cancer (PCa) continues to threaten men’s health, and treatment targeting the androgen receptor (AR) pathway is the major therapy for PCa patients. Several second-generation androgen receptor inhibitors (SG-ARIs), including enzalutamide (ENZ), apalutamide (APA) and darolutamide (DARO), have been developed to better block the activity of AR. Unavoidably, emergence of resistance to these novel drugs still persists. Herein, we identified glutathione S-transferase Mu 2 (GSTM2) as an important determinant in the acquisition of resistance to SG-ARIs. Elevated GSTM2 was detected in enzalutamide-resistant (ENZ-R) PCa, and overexpression of GSTM2 in naïve enzalutamide-sensitive (ENZ-S) cells effectively transformed them to ENZ-R PCa. Aryl hydrocarbon receptor (AhR), the upstream transcription factor, was implicated in the overexpression of GSTM2 in ENZ-R cells. Mechanistically, GSTM2 antagonized the effect of ENZ by rescuing cells from oxidative stress-associated damage and activation of p38 MAPK pathway. Surprisingly, high GSTM2 levels also associated with cross-resistance to APA and DARO. Taking together, these results provide new insight to ameliorate resistance to SG-ARIs and improve treatment outcome.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7–33.
Teo MY, Rathkopf DE, Kantoff P. Treatment of advanced prostate cancer. Annu Rev Med. 2019;70:479–99.
Coutinho I, Day TK, Tilley WD, Selth LA. Androgen receptor signaling in castration-resistant prostate cancer: a lesson in persistence. Endocr Relat Cancer. 2016;23:T179–t197.
Higano C. Enzalutamide, apalutamide, or darolutamide: are apples or bananas best for patients?. Nat Rev Urol. 2019;16:335–6.
Scher HI, Fizazi K, Saad F, Taplin ME, Sternberg CN, Miller K, et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med. 2012;367:1187–97.
Smith MR, Saad F, Chowdhury S, Oudard S, Hadaschik BA, Graff JN, et al. Apalutamide treatment and metastasis-free survival in prostate cancer. N Engl J Med. 2018;378:1408–18.
Fizazi K, Shore N, Tammela TL, Ulys A, Vjaters E, Polyakov S, et al. Darolutamide in nonmetastatic, castration-resistant prostate cancer. N Engl J Med. 2019;380:1235–46.
Shiota M, Yokomizo A, Naito S. Oxidative stress and androgen receptor signaling in the development and progression of castration-resistant prostate cancer. Free Radic Biol Med. 2011;51:1320–8.
Kumar B, Koul S, Khandrika L, Meacham RB, Koul HK. Oxidative stress is inherent in prostate cancer cells and is required for aggressive phenotype. Cancer Res. 2008;68:1777–85.
Hayes JD, Dinkova-Kostova AT, Tew KD. Oxidative stress in cancer. Cancer Cell. 2020;38:167–97.
Sies H, Berndt C, Jones DP. Oxidative stress. Annu Rev Biochem. 2017;86:715–48.
Neha K, Haider MR, Pathak A, Yar MS. Medicinal prospects of antioxidants: a review. Eur J Med Chem. 2019;178:687–704.
Allocati N, Masulli M, Di C. Glutathione transferases: substrates, inihibitors and pro-drugs in cancer and neurodegenerative diseases. Oncogenesis. 2018;7:8.
Zhou SG, Wang P, Pi RB, Gao J, Fu JJ, Fang J, et al. Reduced expression of GSTM2 and increased oxidative stress in spontaneously hypertensive rat. Mol Cell Biochem. 2008;309:99–107.
Han I, Jeong SJ, Lee HJ, Koh W, Lee HJ, Lee EO, et al. Proteomic analysis of mesenchymal stem-like cells derived from ovarian teratoma: potential role of glutathione S-transferase M2 in ovarian teratoma. Proteomics. 2011;11:352–60.
Guo E, Wei H, Liao X, Wu L, Zeng X. Clinical significance and biological mechanisms of glutathione S-transferase mu gene family in colon adenocarcinoma. BMC Med Genet. 2020;21:130.
Andonova IE, Justenhoven C, Winter S, Hamann U, Baisch C, Rabstein S, et al. No evidence for glutathione S-transferases GSTA2, GSTM2, GSTO1, GSTO2, and GSTZ1 in breast cancer risk. Breast Cancer Res Treat. 2010;121:497–502.
Tang SC, Wu CH, Lai CH, Sung WW, Yang WJ, Tang LC, et al. Glutathione S-transferase mu2 suppresses cancer cell metastasis in non-small cell lung cancer. Mol Cancer Res. 2013;11:518–29.
Peng L, Zhuang L, Lin K, Yao Y, Zhang Y, Arumugam T, et al. Downregulation of GSTM2 enhances gemcitabine chemosensitivity of pancreatic cancer in vitro and in vivo. Pancreatology. 2021;21:115–23.
Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol. 2015;109:21.29.1–9.
Li C, Lanman NA, Kong Y, He D, Mao F, Farah E, et al. Inhibition of the erythropoietin-producing receptor EPHB4 antagonizes androgen receptor overexpression and reduces enzalutamide resistance. J Biol Chem. 2020;295:5470–83.
Ricci G, De Maria F, Antonini G, Turella P, Bullo A, Stella L, et al. 7-Nitro-2,1,3-benzoxadiazole derivatives, a new class of suicide inhibitors for glutathione S-transferases. Mechanism of action of potential anticancer drugs. J Biol Chem. 2005;280:26397–405.
Bhattacharjee P, Paul S, Banerjee M, Patra D, Banerjee P, Ghoshal N, et al. Functional compensation of glutathione S-transferase M1 (GSTM1) null by another GST superfamily member, GSTM2. Sci Rep. 2013;3:2704.
Sagar YK. Does glutathione transferase loading into exosomes from prostate cancer cells influence progression? 2016. https://eprints.qut.edu.au/98015/.
Li Y, Chan SC, Brand LJ, Hwang TH, Silverstein KA, Dehm SM. Androgen receptor splice variants mediate enzalutamide resistance in castration-resistant prostate cancer cell lines. Cancer Res. 2013;73:483–9.
Zhao L, Au JL, Wientjes MG. Comparison of methods for evaluating drug-drug interaction. Front Biosci. 2010;2:241–9.
Hayes JD, Dinkova-Kostova AT, McMahon M. Cross-talk between transcription factors AhR and Nrf2: lessons for cancer chemoprevention from dioxin. Toxicol Sci. 2009;111:199–201.
Rothhammer V, Quintana FJ. The aryl hydrocarbon receptor: an environmental sensor integrating immune responses in health and disease. Nat Rev Immunol. 2019;19:184–97.
Shiota M, Yokomizo A, Tada Y, Inokuchi J, Kashiwagi E, Masubuchi D, et al. Castration resistance of prostate cancer cells caused by castration-induced oxidative stress through Twist1 and androgen receptor overexpression. Oncogene. 2010;29:237–50.
Dolado I, Swat A, Ajenjo N, De Vita G, Cuadrado A, Nebreda AR. p38alpha MAP kinase as a sensor of reactive oxygen species in tumorigenesis. Cancer Cell. 2007;11:191–205.
Canovas B, Nebreda AR. Diversity and versatility of p38 kinase signalling in health and disease. Nat Rev Mol Cell Biol. 2021;22:346–66.
Zhao J, Ning S, Lou W, Yang JC, Armstrong CM, Lombard AP, et al. Cross-resistance among next-generation antiandrogen drugs through the AKR1C3/AR-V7 axis in advanced prostate cancer. Mol Cancer Ther. 2020;19:1708–18.
Clegg NJ, Wongvipat J, Joseph JD, Tran C, Ouk S, Dilhas A, et al. ARN-509: a novel antiandrogen for prostate cancer treatment. Cancer Res. 2012;72:1494–503.
Moilanen AM, Riikonen R, Oksala R, Ravanti L, Aho E, Wohlfahrt G, et al. Discovery of ODM-201, a new-generation androgen receptor inhibitor targeting resistance mechanisms to androgen signaling-directed prostate cancer therapies. Sci Rep. 2015;5:12007.
Cancer Genome Atlas Research Network. The molecular taxonomy of primary prostate cancer. Cell. 2015;163:1011–25. https://pubmed.ncbi.nlm.nih.gov/26544944/.
Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22.
Abida W, Cyrta J, Heller G, Prandi D, Armenia J, Coleman I, et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc Natl Acad Sci USA. 2019;116:11428–36.
Hamdy FC, Donovan JL, Lane JA, Mason M, Metcalfe C, Holding P, et al. 10-year outcomes after monitoring, surgery, or radiotherapy for localized prostate cancer. N Engl J Med. 2016;375:1415–24.
Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–25.
Alumkal JJ, Sun D, Lu E, Beer TM, Thomas GV, Latour E, et al. Transcriptional profiling identifies an androgen receptor activity-low, stemness program associated with enzalutamide resistance. Proc Natl Acad Sci USA. 2020;117:12315–23.
He Y, Wei T, Ye Z, Orme JJ, Lin D, Sheng H, et al. A noncanonical AR addiction drives enzalutamide resistance in prostate cancer. Nat Commun. 2021;12:1521.
Zhao SG, Chen WS, Li H, Foye A, Zhang M, Sjöström M, et al. The DNA methylation landscape of advanced prostate cancer. Nat Genet. 2020;52:778–89.
Davies A, Nouruzi S, Ganguli D, Namekawa T, Thaper D, Linder S, et al. An androgen receptor switch underlies lineage infidelity in treatment-resistant prostate cancer. Nat Cell Biol. 2021;23:1023–34.
Zhang Z, Zhou C, Li X, Barnes SD, Deng S, Hoover E, et al. Loss of CHD1 promotes heterogeneous mechanisms of resistance to AR-targeted therapy via chromatin dysregulation. Cancer Cell. 2020;37:584–.e11.
Gibbons JA, Ouatas T, Krauwinkel W, Ohtsu Y, van der Walt JS, Beddo V, et al. Clinical pharmacokinetic studies of enzalutamide. Clin Pharmacokinet. 2015;54:1043–55.
de Vries R, Jacobs F, Mannens G, Snoeys J, Cuyckens F, Chien C, et al. Apalutamide absorption, metabolism, and excretion in healthy men, and enzyme reaction in human hepatocytes. Drug Metab Dispos. 2019;47:453–64.
Ji C, Guha M, Zhu X, Whritenour J, Hemkens M, Tse S, et al. Enzalutamide and apalutamide: in vitro chemical reactivity studies and activity in a mouse drug allergy model. Chem Res Toxicol. 2020;33:211–22.
Liu C, Lou W, Zhu Y, Yang JC, Nadiminty N, Gaikwad NW, et al. Intracrine androgens and AKR1C3 activation confer resistance to enzalutamide in prostate cancer. Cancer Res. 2015;75:1413–22.
Hayer A, Shao L, Chung M, Joubert LM, Yang HW, Tsai FC, et al. Engulfed cadherin fingers are polarized junctional structures between collectively migrating endothelial cells. Nat Cell Biol. 2016;18:1311–23.
Acknowledgements
The research is generously supported by NIH R01 CA157429 (XL), R01 CA196634 (XL), R01 CA264652 (XL), R01 CA256893 (XL). This research is also supported by the Biospecimen Procurement & Translational Pathology, Biostatistics and Bioinformatics, Redox Metabolism, and Flow Cytometry and Immune Monitoring Shared Resources of the University of Kentucky Markey Cancer Center (P30CA177558). We thanks Dr. Alumkal Joshi for the generous sharing of the RNA-seq raw data, and Eleanor Erikson for the critical reading and editing of the manuscript.
Author information
Authors and Affiliations
Contributions
XL: conceptualization; CL and XL: project administration; CL and XL: investigation; CL: validation and visualization; CL, JPL, DH, YZ, NAL, MK and XL: methodology; CL, JPL, DH, YZ and NAL: data curation; CL, JPL, DH, YZ and NAL: formal analysis; XR, JL, ZZ, YQZ, LL, CW and XL: supervision; FM, XR, EF, CMN, YK, LL, CW and XL: resources; JPL, DH, YZ, NAL and XL: software; XL: funding acquisition; CL, JPL and DH: writing—original draft; XL: writing—review and editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, C., Liu, J., He, D. et al. GSTM2 is a key molecular determinant of resistance to SG-ARIs. Oncogene 41, 4498–4511 (2022). https://doi.org/10.1038/s41388-022-02444-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-022-02444-1
This article is cited by
-
GSTM2 alleviates heart failure by inhibiting DNA damage in cardiomyocytes
Cell & Bioscience (2023)