Abstract
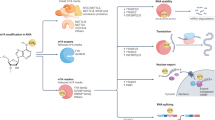
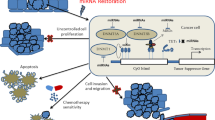
m6A modification, the most abundant and widespread RNA modification, is present and involved in the occurrence and development of various cancers. To date, most studies have mainly focused on the roles of a single m6A regulator (writer/eraser/reader) in various cancers, but cumulative evidence shows that aberrant m6A regulators and m6A levels exert dual effects (promoting and/or inhibiting roles) in cancer progression. Recently, studies have investigated the direct interactions between different m6A regulators (writer/eraser and reader) and mRNAs in a variety of cancers. In this review, we summarize the functions of m6A regulators and their roles in various types of cancers. We further propose the possible crosstalk mechanisms (Writer-m6A-Reader-mRNA axis and Eraser-m6A-Reader-mRNA axis) between different m6A regulators and mRNAs during cancer progression. We also discuss the clinical potential of m6A regulator‑targeting strategies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169:1187–200.
Frye M, Harada BT, Behm M, He C. RNA modifications modulate gene expression during development. Science. 2018;361:1346–9.
Huang H, Weng H, Chen J. m6A modification in coding and non-coding RNAs: roles and therapeutic implications in cancer. Cancer Cell. 2020;37:270–88.
Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from novikoff hepatoma cells. Proc Natl Acad Sci 1974;71:3971–5.
Adams JM, Cory S. Modified nucleosides and bizarre 5′-termini in mouse myeloma mRNA. Nature. 1975;255:28–33.
Beemon K, Keith J. Localization of N6-methyladenosine in the Rous sarcoma virus genome. J Mol Biol. 1977;113:165–79.
Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, et al. N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885–7.
Jiang X, Liu B, Nie Z, Duan L, Xiong Q, Jin Z, et al. The role of m6A modification in the biological functions and diseases. Signal Transduct Target Ther. 2021;6:74.
Uddin MB, Wang Z, Yang C. The m6A RNA methylation regulates oncogenic signaling pathways driving cell malignant transformation and carcinogenesis. Mol Cancer. 2021;20:61.
Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–6.
Blanco S, Bandiera R, Popis M, Hussain S, Lombard P, Aleksic J, et al. Stem cell function and stress response are controlled by protein synthesis. Nature. 2016;534:335–40.
Gkatza NA, Castro C, Harvey RF, Heiß M, Popis MC, Blanco S, et al. Cytosine-5 RNA methylation links protein synthesis to cell metabolism. Coller J, editor. PLoS Biol. 2019;17:e3000297.
Shi H, Wei J, He C. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol Cell. 2019;74:640–50.
Nombela P, Miguel-López B, Blanco S. The role of m6A, m5C and Ψ RNA modifications in cancer: Novel therapeutic opportunities. Mol Cancer. 2021;20:18.
Chen F, Chen Z, Guan T, Zhou Y, Ge L, Zhang H, et al. N 6 -methyladenosine regulates mRNA stability and translation efficiency of KRT7 to promote breast cancer lung metastasis. Cancer Res. 2021;81:2847–60.
Liu Z, Wang T, She Y, Wu K, Gu S, Li L, et al. N6-methyladenosine-modified circIGF2BP3 inhibits CD8+ T-cell responses to facilitate tumor immune evasion by promoting the deubiquitination of PD-L1 in non-small cell lung cancer. Mol Cancer. 2021;20:105.
Qing Y, Dong L, Gao L, Li C, Li Y, Han L, et al. R-2-hydroxyglutarate attenuates aerobic glycolysis in leukemia by targeting the FTO/m6A/PFKP/LDHB axis. Mol Cell. 2021;81:922–.e9.
Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, et al. m6A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017;31:591–606.e6.
Li H, Wu H, Wang Q, Ning S, Xu S, Pang D. Dual effects of N6-methyladenosine on cancer progression and immunotherapy. Mol Ther - Nucleic Acids. 2021;24:25–39.
Vu LP, Pickering BF, Cheng Y, Zaccara S, Nguyen D, Minuesa G, et al. The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med. 2017;23:1369–76.
Li F, Yi Y, Miao Y, Long W, Long T, Chen S, et al. N6-methyladenosine modulates nonsense-mediated mRNA decay in human glioblastoma. Cancer Res. 2019;79:5785–98.
Liu J, Eckert MA, Harada BT, Liu SM, Lu Z, Yu K, et al. m6A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat Cell Biol. 2018;20:1074–83.
Jin D, Guo J, Wu Y, Yang L, Wang X, Du J, et al. m6A demethylase ALKBH5 inhibits tumor growth and metastasis by reducing YTHDFs-mediated YAP expression and inhibiting miR-107/LATS2–mediated YAP activity in NSCLC. Mol Cancer. 2020;19:40.
Zhong L, Liao D, Zhang M, Zeng C, Li X, Zhang R, et al. YTHDF2 suppresses cell proliferation and growth via destabilizing the EGFR mRNA in hepatocellular carcinoma. Cancer Lett. 2019;442:252–61.
Wang S, Chai P, Jia R, Jia R. Novel insights on m6A RNA methylation in tumorigenesis: a double-edged sword. Mol Cancer. 2018;17:101.
Dong S, Wu Y, Liu Y, Weng H, Huang HN. 6 ‐methyladenosine steers RNA metabolism and regulation in cancer. Cancer Commun. 2021;41:538–59.
Yang Y, Hsu PJ, Chen YS, Yang YG. Dynamic transcriptomic m6A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018;28:616–24.
Wang P, Doxtader KA, Nam Y. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol Cell. 2016;63:306–17.
Brown JA, Kinzig CG, DeGregorio SJ, Steitz JA. Methyltransferase-like protein 16 binds the 3′-terminal triple helix of MALAT1 long noncoding RNA. Proc Natl Acad Sci. 2016;113:14013–8.
van Tran N, Ernst FGM, Hawley BR, Zorbas C, Ulryck N, Hackert P, et al. The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res. 2019;47:7719–33.
Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–89.
Wen J, Lv R, Ma H, Shen H, He C, Wang J, et al. Zc3h13 regulates nuclear RNA m6A methylation and mouse embryonic stem cell self-renewal. Mol Cell. 2018;69:1028–.e6.
Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18–29.
Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, et al. Nuclear m 6 A reader YTHDC1 regulates mRNA splicing. Mol Cell. 2016;61:507–19.
Zhou B, Liu C, Xu L, Yuan Y, Zhao J, Zhao W, et al. N 6 ‐methyladenosine reader protein YT521‐B homology domain‐containing 2 suppresses liver steatosis by regulation of mRNA stability of lipogenic genes. Hepatology. 2021;73:91–103.
Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, et al. N6-methyladenosine modulates messenger RNA translation efficiency. Cell . 2015;161:1388–99.
Zaccara S, Jaffrey SR. A unified model for the function of YTHDF proteins in regulating m6A-Modified mRNA. Cell. 2020;181:1582–.e18.
Dixit D, Prager BC, Gimple RC, Poh HX, Wang Y, Wu Q, et al. The RNA m6A reader YTHDF2 maintains oncogene expression and is a targetable dependency in glioblastoma stem cells. Cancer Discov. 2021;11:480–99.
Ma S, Chen C, Ji X, Liu J, Zhou Q, Wang G, et al. The interplay between m6A RNA methylation and noncoding RNA in cancer. J Hematol Oncol. 2019;12:121.
Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, et al. Recognition of RNA N 6 -methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20:285–95.
Zhang L, Wan Y, Zhang Z, Jiang Y, Gu Z, Ma X, et al. IGF2BP1 overexpression stabilizes PEG10 mRNA in an m6A-dependent manner and promotes endometrial cancer progression. Theranostics. 2021;11:1100–14.
Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, et al. 5′ UTR m6A promotes cap-independent translation. Cell. 2015;163:999–1010.
Wu R, Li A, Sun B, Sun JG, Zhang J, Zhang T, et al. A novel m6A reader Prrc2a controls oligodendroglial specification and myelination. Cell Res. 2019;29:23–41.
Baquero-Perez B, Antanaviciute A, Yonchev ID, Carr IM, Wilson SA, Whitehouse A. The Tudor SND1 protein is an m6A RNA reader essential for replication of Kaposi’s sarcoma-associated herpesvirus. eLife. 2019;8:e47261.
Wang X, Ma R, Zhang X, Cui L, Ding Y, Shi W, et al. Crosstalk between N6-methyladenosine modification and circular RNAs: current understanding and future directions. Mol Cancer. 2021;20:121.
Alarcón CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF. HNRNPA2B1 is a mediator of m6A-dependent nuclear RNA processing events. Cell . 2015;162:1299–308.
Roundtree IA, Luo GZ, Zhang Z, Wang X, Zhou T, Cui Y, et al. YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. eLife. 2017;6:e31311.
Lee Y, Choe J, Park OH, Kim YK. Molecular mechanisms driving mRNA degradation by m6A modification. Trends Genet. 2020;36:177–88.
Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, et al. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 2017;27:315–28.
Hsu PJ, Zhu Y, Ma H, Guo Y, Shi X, Liu Y, et al. Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017;27:1115–27.
Choe J, Lin S, Zhang W, Liu Q, Wang L, Ramirez-Moya J, et al. mRNA circularization by METTL3–eIF3h enhances translation and promotes oncogenesis. Nature. 2018;561:556–60.
Zhou Z, Lv J, Yu H, Han J, Yang X, Feng D, et al. Mechanism of RNA modification N6-methyladenosine in human cancer. Mol Cancer. 2020;19:104.
Li Z, Weng H, Su R, Weng X, Zuo Z, Li C, et al. FTO plays an oncogenic role in acute myeloid leukemia as a N6-Methyladenosine RNA demethylase. Cancer Cell. 2017;31:127–41.
Su R, Dong L, Li C, Nachtergaele S, Wunderlich M, Qing Y, et al. R-2HG exhibits anti-tumor activity by targeting FTO/m6A/MYC/CEBPA signaling. Cell . 2018;172:90–105.e23.
Shen C, Sheng Y, Zhu AC, Robinson S, Jiang X, Dong L, et al. RNA demethylase ALKBH5 selectively promotes tumorigenesis and cancer stem cell self-renewal in acute myeloid leukemia. Cell Stem Cell. 2020;27:64–80.e9.
Weng H, Huang H, Wu H, Qin X, Zhao BS, Dong L, et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m6A modification. Cell Stem Cell. 2018;22:191–205.e9.
Paris J, Morgan M, Campos J, Spencer GJ, Shmakova A, Ivanova I, et al. Targeting the RNA m6A reader YTHDF2 selectively compromises cancer stem cells in acute myeloid leukemia. Cell Stem Cell. 2019;25:137–.e6.
Visvanathan A, Patil V, Arora A, Hegde AS, Arivazhagan A, Santosh V, et al. Essential role of METTL3-mediated m 6 A modification in glioma stem-like cells maintenance and radioresistance. Oncogene. 2018;37:522–33.
Tassinari V, Cesarini V, Tomaselli S, Ianniello Z, Silvestris DA, Ginistrelli LC, et al. ADAR1 is a new target of METTL3 and plays a pro-oncogenic role in glioblastoma by an editing-independent mechanism. Genome Biol. 2021;22:51.
Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G, et al. m6A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 2017;18:2622–34.
Dixit D, Prager BC, Gimple RC, Poh HX, Wang Y, Wu Q, et al. The rna m6a reader ythdf2 maintains oncogene expression and is a targetable dependency in glioblastoma stem cells. Cancer Discov. 2021;11:480–99.
Song P, Yang F, Jin H, Wang X. The regulation of protein translation and its implications for cancer. Signal Transduct Target Ther. 2021;6:68.
Jin H, Ying X, Que B, Wang X, Chao Y, Zhang H, et al. N6-methyladenosine modification of ITGA6 mRNA promotes the development and progression of bladder cancer. EBioMedicine. 2019;47:195–207.
Yang F, Jin H, Que B, Chao Y, Zhang H, Ying X, et al. Dynamic m6A mRNA methylation reveals the role of METTL3-m6A-CDCP1 signaling axis in chemical carcinogenesis. Oncogene. 2019;38:4755–72.
Xie J, Ba J, Zhang M, Wan Y, Jin Z, Yao Y. The m6A methyltransferase METTL3 promotes the stemness and malignant progression of breast cancer by mediating m6A modification on SOX2. J BUON: Off J Balk Union Oncol. 2021;26:444–9.
Wan W, Ao X, Chen Q, Yu Y, Ao L, Xing W, et al. METTL3/IGF2BP3 axis inhibits tumor immune surveillance by upregulating N6-methyladenosine modification of PD-L1 mRNA in breast cancer. Mol Cancer. 2022;21:60.
Li E, Xia M, Du Y, Long K, Ji F, Pan F, et al. METTL3 promotes homologous recombination repair and modulates chemotherapeutic response in breast cancer by regulating the EGF/RAD51 axis. eLife. 2022;11:e75231.
Li T, Hu PS, Zuo Z, Lin JF, Li X, Wu QN, et al. METTL3 facilitates tumor progression via an m6A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol Cancer. 2019;18:112.
Zhang Y, Kang M, Zhang B, Meng F, Song J, Kaneko H, et al. M6A modification-mediated CBX8 induction regulates stemness and chemosensitivity of colon cancer via upregulation of LGR5. Mol Cancer. 2019;18:185.
Xiang S, Liang X, Yin S, Liu J, Xiang Z. N6-methyladenosine methyltransferase METTL3 promotes colorectal cancer cell proliferation through enhancing MYC expression. Am J Transl Res. 2020;12:1789–806.
Shen C, Xuan B, Yan T, Ma Y, Xu P, Tian X, et al. m6A-dependent glycolysis enhances colorectal cancer progression. Mol Cancer. 2020;19:72.
Xiong J, He J, Zhu J, Pan J, Liao W, Ye H, et al. Lactylation-driven METTL3-mediated RNA m6A modification promotes immunosuppression of tumor-infiltrating myeloid cells. Mol Cell. 2022;82:1660–.e10.
Liu X, He H, Zhang F, Hu X, Bi F, Li K, et al. m6A methylated EphA2 and VEGFA through IGF2BP2/3 regulation promotes vasculogenic mimicry in colorectal cancer via PI3K/AKT and ERK1/2 signaling. Cell Death Dis. 2022;13:483.
He H, Wu W, Sun Z, Chai L. MiR-4429 prevented gastric cancer progression through targeting METTL3 to inhibit m6A-caused stabilization of SEC62. Biochemical Biophysical Res Commun. 2019;517:581–7.
Wang Q, Chen C, Ding Q, Zhao Y, Wang Z, Chen J, et al. METTL3-mediated m 6 A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut. 2020;69:1193–205.
Huo FC, Zhu ZM, Zhu WT, Du QY, Liang J, Mou J. METTL3-mediated m6A methylation of SPHK2 promotes gastric cancer progression by targeting KLF2. Oncogene. 2021;40:2968–81.
Liu L, He J, Sun G, Huang N, Bian Z, Xu C, et al. The N6-methyladenosine modification enhances ferroptosis resistance through inhibiting SLC7A11 mRNA deadenylation in hepatoblastoma. Clin Transl Med. 2022;12:e778.
Lin X, Chai G, Wu Y, Li J, Chen F, Liu J, et al. RNA m 6 A methylation regulates the epithelial mesenchymal transition of cancer cells and translation of Snail. Nature. Communications. 2019;10:2065.
Fan Z, Gao Y, Zhang W, Yang G, Liu P, Xu L, et al. METTL3/IGF2BP1/CD47 contributes to the sublethal heat treatment induced mesenchymal transition in HCC. Biochemical Biophysical Res Commun. 2021;546:169–77.
Cai X, Chen Y, Man D, Yang B, Feng X, Zhang D, et al. RBM15 promotes hepatocellular carcinoma progression by regulating N6-methyladenosine modification of YES1 mRNA in an IGF2BP1-dependent manner. Cell Death Discovery 2021;7:315.
Jin D, Guo J, Wu Y, Du J, Yang L, Wang X, et al. m6A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1-miR-1914-3p-YAP axis to induce NSCLC drug resistance and metastasis. J Hematol Oncol. 2019;12:135.
Chen WW, Qi JW, Hang Y, Wu JX, Zhou XX, Chen JZ, et al. Simvastatin is beneficial to lung cancer progression by inducing METTL3-induced m6A modification on EZH2 mRNA. Eur Rev Med Pharmacol Sci. 2020;24:4263–70.
Yu X, Zhao H, Cao Z. The m6A methyltransferase METTL3 aggravates the progression of nasopharyngeal carcinoma through inducing EMT by m6A-modified Snail mRNA. Minerva Med. 2022;113:309–14.
Zhang R, Li SW, Liu L, Yang J, Huang G, Sang Y. TRIM11 facilitates chemoresistance in nasopharyngeal carcinoma by activating the β-catenin/ABCC9 axis via p62-selective autophagic degradation of Daple. Oncogenesis. 2020;9:45.
Ma H, Zhang F, Zhong Q, Hou J. METTL3-mediated m6A modification of KIF3C-mRNA promotes prostate cancer progression and is negatively regulated by miR-320d. Aging. 2021;13:22332–44.
Zhao W, Cui Y, Liu L, Ma X, Qi X, Wang Y, et al. METTL3 facilitates oral squamous cell carcinoma tumorigenesis by enhancing c-Myc stability via YTHDF1-Mediated m6A Modification. Mol Ther - Nucleic Acids. 2020;20:1–12.
Wei J, Yin Y, Zhou J, Chen H, Peng J, Yang J, et al. METTL3 potentiates resistance to cisplatin through m 6 A modification of TFAP2C in seminoma. J Cell Mol Med. 2020;24:11366–80.
Wang X, Tian L, Li Y, Wang J, Yan B, Yang L, et al. RBM15 facilitates laryngeal squamous cell carcinoma progression by regulating TMBIM6 stability through IGF2BP3 dependent. J Exp Clin Cancer Res. 2021;40:80.
Zhou D, Tang W, Xu Y, Xu Y, Xu B, Fu S, et al. METTL3/YTHDF2 m6A axis accelerates colorectal carcinogenesis through epigenetically suppressing YPEL5. Molecular. Oncology. 2021;15:2172–84.
Chai RC, Chang YZ, Chang X, Pang B, An SY, Zhang KN, et al. YTHDF2 facilitates UBXN1 mRNA decay by recognizing METTL3-mediated m6A modification to activate NF-κB and promote the malignant progression of glioma. J Hematol Oncol. 2021;14:109.
Chen M, Wei L, Law CT, Tsang FHC, Shen J, Cheng CLH, et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67:2254–70.
Xu QC, Tien YC, Shi YH, Chen S, Zhu YQ, Huang XT, et al. METTL3 promotes intrahepatic cholangiocarcinoma progression by regulating IFIT2 expression in an m6A-YTHDF2-dependent manner. Oncogene. 2022;41:1622–33.
Cheng C, Wu Y, Xiao T, Xue J, Sun J, Xia H, et al. METTL3-mediated m6A modification of ZBTB4 mRNA is involved in the smoking-induced EMT in cancer of the lung. Mol Ther - Nucleic Acids. 2021;23:487–500.
Jin M, Li G, Liu W, Wu X, Zhu J, Zhao D, et al. Cigarette smoking induces aberrant N6-methyladenosine of DAPK2 to promote non-small cell lung cancer progression by activating NF-κB pathway. Cancer Lett. 2021;518:214–29.
Xu Y, Chen Y, Yao Y, Xie H, Lu G, Du C, et al. VIRMA contributes to non-small cell lung cancer progression via N6-methyladenosine-dependent DAPK3 post-transcriptional modification. Cancer Lett. 2021;522:142–54.
Wang W, Shao F, Yang X, Wang J, Zhu R, Yang Y, et al. METTL3 promotes tumour development by decreasing APC expression mediated by APC mRNA N6-methyladenosine-dependent YTHDF binding. Nature. Communications. 2021;12:3803.
Li J, Xie H, Ying Y, Chen H, Yan H, He L, et al. YTHDF2 mediates the mRNA degradation of the tumor suppressors to induce AKT phosphorylation in N6-methyladenosine-dependent way in prostate cancer. Mol Cancer. 2020;19:152.
Chen Y, Pan C, Wang X, Xu D, Ma Y, Hu J, et al. Silencing of METTL3 effectively hinders invasion and metastasis of prostate cancer cells. Theranostics 2021;11:7640–57.
Tian J, Zhu Y, Rao M, Cai Y, Lu Z, Zou D. et al. N 6 -methyladenosine mRNA methylation of PIK3CB regulates AKT signalling to promote PTEN-deficient pancreatic cancer progression. Gut. 2020;69:2180–92.
Chen X, Xu M, Xu X, Zeng K, Liu X, Pan B, et al. METTL14-mediated N6-methyladenosine modification of SOX4 mRNA inhibits tumor metastasis in colorectal cancer. Mol Cancer. 2020;19:106.
Chen S, Zhang L, Li M, Zhang Y, Sun M, Wang L, et al. Fusobacterium nucleatum reduces METTL3-mediated m6A modification and contributes to colorectal cancer metastasis. Nature. Communications. 2022;13:1248.
Fan Z, Yang G, Zhang W, Liu Q, Liu G, Liu P, et al. Hypoxia blocks ferroptosis of hepatocellular carcinoma via suppression of METTL14 triggered YTHDF2‐dependent silencing of SLC7A11. J Cell Mol Med. 2021;25:10197–212.
He J, Zhou M, Yin J, Wan J, Chu J, Jia J, et al. METTL3 restrains papillary thyroid cancer progression via m6A/c-Rel/IL-8-mediated neutrophil infiltration. Mol Ther. 2021;29:1821–37.
Wang S, Gan M, Chen C, Zhang Y, Kong J, Zhang H, et al. Methyl CpG binding protein 2 promotes colorectal cancer metastasis by regulating N 6 ‐methyladenosine methylation through methyltransferase‐like 14. Cancer Sci. 2021;112:3243–54.
Lin Z, Niu Y, Wan A, Chen D, Liang H, Chen X, et al. RNA m 6 A methylation regulates sorafenib resistance in liver cancer through FOXO 3‐mediated autophagy. EMBO J. 2020;39:e103181.
Yin H, Zhang X, Yang P, Zhang X, Peng Y, Li D, et al. RNA m6A methylation orchestrates cancer growth and metastasis via macrophage reprogramming. Nat Commun. 2021;12:1394.
Zhu Y, Peng X, Zhou Q, Tan L, Zhang C, Lin S, et al. METTL3-mediated m6A modification of STEAP2 mRNA inhibits papillary thyroid cancer progress by blocking the Hedgehog signaling pathway and epithelial-to-mesenchymal transition. Cell Death Dis. 2022;13:358.
Zhang L, Luo X, Qiao S. METTL14-mediated N6-methyladenosine modification of Pten mRNA inhibits tumour progression in clear-cell renal cell carcinoma. Br J Cancer. 2022;127:30–42.
Yang Z, Yang S, Cui YH, Wei J, Shah P, Park G, et al. METTL14 facilitates global genome repair and suppresses skin tumorigenesis. Proc Natl Acad Sci 2021;118:e2025948118.
Zhang D, Ning J, Okon I, Zheng X, Satyanarayana G, Song P, et al. Suppression of m6A mRNA modification by DNA hypermethylated ALKBH5 aggravates the oncological behavior of KRAS mutation/LKB1 loss lung cancer. Cell Death Dis. 2021;12:518.
Xiao P, Liu YK, Han W, Hu Y, Zhang BY, Liu WL. Exosomal delivery of FTO confers gefitinib resistance to recipient cells through ABCC10 regulation in an m6A-dependent manner. Mol Cancer Res. 2021;19:726–38.
Qiu X, Yang S, Wang S, Wu J, Zheng B, Wang K, et al. M 6 A demethylase ALKBH5 regulates PD-L1 expression and tumor immunoenvironment in intrahepatic cholangiocarcinoma. Cancer Res. 2021;81:4778–93.
Yang S, Wei J, Cui YH, Park G, Shah P, Deng Y, et al. m6A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nature. Communications. 2019;10:2782.
Xu A, Zhang J, Zuo L, Yan H, Chen L, Zhao F, et al. FTO promotes multiple myeloma progression by posttranscriptional activation of HSF1 in an m6A-YTHDF2-dependent manner. Mol Ther. 2022;30:1104–18.
Wang F, Liao Y, Zhang M, Zhu Y, Wang W, Cai H, et al. N6-methyladenosine demethyltransferase FTO-mediated autophagy in malignant development of oral squamous cell carcinoma. Oncogene. 2021;40:3885–98.
Tan Z, Shi S, Xu J, Liu X, Lei Y, Zhang B, et al. RNA N6-methyladenosine demethylase FTO promotes pancreatic cancer progression by inducing the autocrine activity of PDGFC in an m6A-YTHDF2-dependent manner. Oncogene. 2022;41:2860–72.
Wu G, Suo C, Yang Y, Shen S, Sun L, Li S, et al. MYC promotes cancer progression by modulating m 6 A modifications to suppress target gene translation. EMBO Rep. 2021;22:e51519.
Tsuchiya K, Yoshimura K, Iwashita Y, Inoue Y, Ohta T, Watanabe H, et al. m6A demethylase ALKBH5 promotes tumor cell proliferation by destabilizing IGF2BPs target genes and worsens the prognosis of patients with non-small-cell lung cancer. Cancer Gene Therapy. 2022. https://doi.org/10.1038/s41417-022-00451-8.
Lv D, Ding S, Zhong L, Tu J, Li H, Yao H, et al. M6A demethylase FTO-mediated downregulation of DACT1 mRNA stability promotes Wnt signaling to facilitate osteosarcoma progression. Oncogene. 2022;41:1727–41.
Yuan Y, Yan G, He M, Lei H, Li L, Wang Y, et al. ALKBH5 suppresses tumor progression via an m6A-dependent epigenetic silencing of pre-miR-181b-1/YAP signaling axis in osteosarcoma. Cell Death Dis. 2021;12:60.
Yang X, Shao F, Guo D, Wang W, Wang J, Zhu R, et al. WNT/β-catenin-suppressed FTO expression increases m6A of c-Myc mRNA to promote tumor cell glycolysis and tumorigenesis. Cell Death Dis. 2021;12:462.
Ruan DY, Li T, Wang YN, Meng Q, Li Y, Yu K, et al. FTO downregulation mediated by hypoxia facilitates colorectal cancer metastasis. Oncogene . 2021;40:5168–81.
Han S, Zhu L, Zhu Y, Meng Y, Li J, Song P, et al. Targeting ATF4-dependent pro-survival autophagy to synergize glutaminolysis inhibition. Theranostics . 2021;11:8464–79.
Yang Z, Cai Z, Yang C, Luo Z, Bao X. ALKBH5 regulates STAT3 activity to affect the proliferation and tumorigenicity of osteosarcoma via an m6A-YTHDF2-dependent manner. eBioMedicine. 2022;80:104019.
Cheng M, Sheng L, Gao Q, Xiong Q, Zhang H, Wu M, et al. The m 6 A methyltransferase METTL3 promotes bladder cancer progression via AFF4/NF-κB/MYC signaling network. Oncogene. 2019;38:3667–80.
Yang Z, Jiang X, Li D, Jiang X. HBXIP promotes gastric cancer via METTL3-mediated MYC mRNA m6A modification. Aging. 2020;12:24967–82.
Wu H, Li F, Zhu R. miR-338-5p inhibits cell growth and migration via inhibition of the METTL3/m6A/c-Myc pathway in lung cancer. Acta Biochimica et Biophysica Sin. 2021;53:304–16.
Yuan Y, Du Y, Wang L, Liu X. The M6A methyltransferase METTL3 promotes the development and progression of prostate carcinoma via mediating MYC methylation. J Cancer. 2020;11:3588–95.
Iaiza A, Tito C, Ianniello Z, Ganci F, Laquintana V, Gallo E, et al. METTL3-dependent MALAT1 delocalization drives c-Myc induction in thymic epithelial tumors. Clin Epigenetics. 2021;13:173.
Yang Z, Jiang X, Zhang Z, Zhao Z, Xing W, Liu Y, et al. HDAC3-dependent transcriptional repression of FOXA2 regulates FTO/m6A/MYC signaling to contribute to the development of gastric cancer. Cancer Gene Ther. 2021;28:141–55.
Tang X, Liu S, Chen D, Zhao Z, Zhou J. The role of the fat mass and obesity‑associated protein in the proliferation of pancreatic cancer cells. Oncol Lett. 2018;17:2473–8.
Gutschner T, Hämmerle M, Pazaitis N, Bley N, Fiskin E, Uckelmann H, et al. Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) is an important protumorigenic factor in hepatocellular carcinoma. Hepatology. 2014;59:1900–11.
Ye M, Dong S, Hou H, Zhang T, Shen M. Oncogenic role of long noncoding RNAMALAT1 in thyroid cancer progression through regulation of the miR-204/IGF2BP2/m6A-MYC signaling. Mol Ther - Nucleic Acids. 2021;23:1–12.
Xie F, Huang C, Liu F, Zhang H, Xiao X, Sun J, et al. CircPTPRA blocks the recognition of RNA N6-methyladenosine through interacting with IGF2BP1 to suppress bladder cancer progression. Mol Cancer. 2021;20:68.
Zhu P, He F, Hou Y, Tu G, Li Q, Jin T, et al. A novel hypoxic long noncoding RNA KB-1980E6.3 maintains breast cancer stem cell stemness via interacting with IGF2BP1 to facilitate c-Myc mRNA stability. Oncogene. 2021;40:1609–27.
Xu H, Wang H, Zhao W, Fu S, Li Y, Ni W, et al. SUMO1 modification of methyltransferase-like 3 promotes tumor progression via regulating Snail mRNA homeostasis in hepatocellular carcinoma. Theranostics. 2020;10:5671–86.
Zhang B, Wu Q, Li B, Wang D, Wang L, Zhou YL. m6A regulator-mediated methylation modification patterns and tumor microenvironment infiltration characterization in gastric cancer. Mol Cancer. 2020;19:53.
Qiu X, Yang S, Wang S, Wu J, Zheng B, Wang K, et al. M 6 A demethylase ALKBH5 regulates PD-L1 expression and tumor immunoenvironment in intrahepatic cholangiocarcinoma. Cancer Res. 2021;81:4778–93.
Ma J, Yang F, Zhou C, Liu F, Yuan J, Wang F, et al. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N 6 ‐methyladenosine‐dependent primary MicroRNA processing. Hepatology. 2017;65:529–43.
Yang X, Zhang S, He C, Xue P, Zhang L, He Z, et al. METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Mol Cancer. 2020;19:46.
Guimarães-Teixeira C, Barros-Silva D, Lobo J, Soares-Fernandes D, Constâncio V, Leite-Silva P, et al. Deregulation of N6-Methyladenosine RNA modification and its erasers FTO/ALKBH5 among the main renal cell tumor subtypes. J Personalized Med. 2021;11:996.
Singh B, Kinne HE, Milligan RD, Washburn LJ, Olsen M, Lucci A. Important role of FTO in the survival of rare panresistant triple-negative inflammatory breast cancer cells facing a severe metabolic challenge. Tan M, editor. PLOS One. 2016;11:e0159072.
Huang Y, Su R, Sheng Y, Dong L, Dong Z, Xu H, et al. Small-molecule targeting of oncogenic FTO demethylase in acute myeloid leukemia. Cancer Cell. 2019;35:677–e10.
Yan F, Al-Kali A, Zhang Z, Liu J, Pang J, Zhao N, et al. A dynamic N 6-methyladenosine methylome regulates intrinsic and acquired resistance to tyrosine kinase inhibitors. Cell Res. 2018;28:1062–76.
Fukumoto T, Zhu H, Nacarelli T, Karakashev S, Fatkhutdinov N, Wu S, et al. N6-methylation of adenosine of FZD10 mRNA contributes to PARP inhibitor resistance. Cancer Res. 2019;79:2812–20.
Yue B, Song C, Yang L, Cui R, Cheng X, Zhang Z, et al. METTL3-mediated N6-methyladenosine modification is critical for epithelial-mesenchymal transition and metastasis of gastric cancer. Mol Cancer. 2019;18:142.
Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, Peer E, Kol N, Ben-Haim MS, et al. The dynamic N1 -methyladenosine methylome in eukaryotic messenger RNA. Nature. 2016;530:441–6.
Li X, Xiong X, Wang K, Wang L, Shu X, Ma S, et al. Transcriptome-wide mapping reveals reversible and dynamic N1-methyladenosine methylome. Nat Chem Biol. 2016;12:311–6.
Kasowitz SD, Ma J, Anderson SJ, Leu NA, Xu Y, Gregory BD, et al. Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development. Yan W, editor. PLoS Genetics. 2018;14:e1007412.
Liu J, Yue Y, Liu J, Cui X, Cao J, Luo G, et al. VIRMA mediates preferential m6A mRNA methylation in 3′UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018;4:10.
Huang Y, Yan J, Li Q, Li J, Gong S, Zhou H, et al. Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic Acids Res. 2015;43:373–84.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81902532 to YY and 82072773 to QM), the fellowship of China Postdoctoral Science Foundation (2022M712255 to XDN), the National Key Research and Development Program of China, Stem Cell and Translational Research (2017YFA0106500 to YW), the Distinguished Young Scientists Program of Sichuan Province (2019JDJQ0029 to YW), and the 1·3·5 project for disciplines of excellence, West China Hospital, Sichuan University (ZYYC20019 to YW).
Author information
Authors and Affiliations
Contributions
Manuscript conception and design: XDN, YY, QM, and YW. Literature search and manuscript drafting: XDN and YY. Manuscript editing, figure design, and review: all authors. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Niu, X., Yang, Y., Ren, Y. et al. Crosstalk between m6A regulators and mRNA during cancer progression. Oncogene 41, 4407–4419 (2022). https://doi.org/10.1038/s41388-022-02441-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-022-02441-4