Abstract
Transcriptomic analyses of pancreatic ductal adenocarcinoma (PDAC) have identified two major epithelial subtypes with distinct biology and clinical behaviours. Here, we aimed to clarify the role of FGFR1 and FGFR4 in the definition of aggressive PDAC phenotypes. We found that the expression of FGFR4 is exclusively detected in epithelial cells, significantly elevated in the classical PDAC subtype, and associates with better outcomes. In highly aggressive basal-like/squamous PDAC, reduced FGFR4 expression aligns with hypermethylation of the gene and lower levels of histone marks associated with active transcription in its regulatory regions. Conversely, FGFR1 has more promiscuous expression in both normal and malignant pancreatic tissues and is strongly associated with the EMT phenotype but not with the basal-like cell lineage. Regardless of the genetic background, the increased proliferation of FGFR4-depleted PDAC cells correlates with hyperactivation of the mTORC1 pathway both in vitro and in vivo. Downregulation of FGFR4 in classical cell lines invariably leads to the enrichment of basal-like/squamous gene programs and is associated with either partial or full switch of phenotype. In sum, we show that endogenous levels of FGFR4 limit the malignant phenotype of PDAC cells. Finally, we propose FGFR4 as a valuable marker for the stratification of PDAC patients.
Similar content being viewed by others
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is a malignancy arising from the exocrine pancreatic epithelium and is the deadliest cancer worldwide [1, 2]. The abysmal prognosis of PDAC is contributed by late diagnosis, its complex biology, and lack of effective treatments. The molecular taxonomy of PDAC has been redefined by multiple studies that have used transcriptomic profiling to analyse bulk tissues, cell lines, and microdissected epithelia [3,4,5,6,7]. All classifications have highlighted the existence of two major subtypes based on characteristics of the neoplastic epithelium, namely the classical/progenitor and the basal-like/squamous subtypes. The classical/progenitor PDAC subtype is often regarded as the default pancreatic cancer subtype [5, 8] as it is characterised by the expression of transcription factors involved in specification and maintenance of pancreatic cell fate [3, 9, 10]. Conversely, the basal-like/squamous subtype is associated with loss of pancreatic endodermal identity, and expression of the master regulator of basal-like cells program ΔNp63 [3, 7, 11]. This subtype shows a significantly worse survival outcome and is enriched for inactivation of TP53 and chromatin regulators, including ARID1A and KDM6A [3, 12]. Preclinical data have showed that the two subtypes differ also in the response to chemotherapy [6, 10, 13], to agents targeting the cell cycle [14], and further display unique metabolic vulnerabilities [15]. While there is some clinical evidence for the predictive value of transcriptomic classifications [10, 16, 17], clinical investigations are ongoing to conclusively demonstrate the relevance of subtype-specific treatments. Previous works have elegantly demonstrated the causative involvement of pancreatic and endodermal transcription factors (e.g. GATA6, HNF1A, HNF4A) into the maintenance of subtype-specific gene programs [10, 15, 18, 19]. In particular, the emergence of the basal-like/squamous program is almost invariably associated with the loss of expression of transcription factors regulating pancreatic cell fates. Cell fate maintenance is also ensured by the signalling of growth factors through receptor tyrosine kinases (RTKs) [20, 21]. Following activation, RTKs transmit intracellular signals of varying qualities and quantities that alter the transcriptional landscape of a cell [20]. The fibroblast growth factors (FGFs) to Fibroblast growth factor receptors (FGFRs) axis is reportedly involved in the maturation of pancreatic cells from the endoderm [22,23,24,25]. Here, we explored the involvement of FGFRs in the definition of molecular subtypes of PDAC. We report the downregulation of FGFR4 in PDAC showing basal-like/squamous features. Integrating the analysis of transcriptomic, methylation, and chromatin accessibility datasets from patient-derived tissue specimens and cultures, we shed light on the mechanisms leading to downregulation of FGFR4 in basal-like/squamous tumours. Conversely, we found that elevated expression of FGFR1 is a functional marker of Epithelial to Mesenchymal Transition (EMT) rather than of the basal-like/squamous subtype. To elucidate whether FGFR4 is cause or consequence of more aggressive PDAC subtypes, we targeted the receptor using RNA interference approaches in both monolayer cell cultures and patient-derived organoids to show that its loss accelerates cell proliferation and in vivo growth regardless of the genetic and transcriptomic background of the models. Mechanistically, loss of FGFR4 was associated with increased fluxes through the mTORC1 pathway and accordingly increased protein synthesis. Downregulation of FGFR4 in classical/progenitor cell lines invariably led to the enrichment of basal-like/squamous gene programs which was associated with either partial of full switch of phenotype. Overall, our data provide direct evidence that the loss of FGFR4 promotes aggressive phenotypes of PDAC.
Results
FGFR4 is associated with the classical phenotype of PDAC
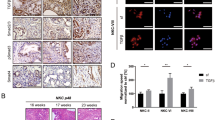
Receptor-Tyrosine Kinases (RTKs) initiated signaling is critical to cell fate determination [20, 21]. To investigate the functional relevance of RTKs in determining PDAC cell lineages and promoting its malignant phenotype, we started by exploring transcriptomic data of human PDAC specimens from the TCGA consortium [4] and found the selective enrichment of FGFR4 in the classical subtype (Fig. 1A, B). We reproduced this finding in transcriptomic data from two additional cohorts [3, 5] (Fig. 1B). While relatively enriched in basal-like PDAC (Fig. 1A), the expression of FGFR1 did not significantly discriminate basal-like from classical tumours in the 3 PDAC cohorts [3,4,5] investigated (Fig. 1B). Next, we explored available single-cell RNA-Seq (scRNA-Seq) data of human PDAC tissues to localise the expression of both FGFR1 and FGFR4. We integrated four PDAC scRNA-seq data sets [5, 26,27,28] using Harmony [29] and performed cell type annotation to find that FGFR4 is predominantly expressed by epithelial cells, while FGFR1 could be detected in epithelial, stromal (fibroblasts, endothelial cells), and immune cells (i.e. macrophages) (Fig. 1C). Next, we explored scRNA-seq data from normal pancreas [30,31,32] (Supplementary Fig. 1A) and found that the expression of FGFR4 is mostly restricted to the epithelial cells while FGFR1 is detectable in many cell types. In situ hybridisation (ISH) analyses of normal pancreatic tissues confirmed scRNA-seq data (Supplementary Fig. 1B). To better understand the association of epithelial FGFRs expression with PDAC molecular subtypes, we then focused on transcriptomic datasets derived from either tissue with high neoplastic cellularity (i.e. ICGC [3], average qPure score > 60%) or microdissected neoplastic epithelia (i.e. PanCuRx [5]). First, we divided samples of the two cohorts in four different groups based on FGFR1 and FGFR4 expression statuses (see methods and Supplementary Fig. 1C). To explore the association of FGFR1 and FGFR4 with the aggressive basal-like and squamous molecular subtypes as defined by Moffitt [7] and Bailey [3] (Fig. 1D, Supplementary Fig. 1D), we calculated the basal-like and squamous signature scores for each sample of the ICGC and the PanCuRx cohorts [3, 5] (see methods). Tumours with low levels of FGFR4 showed the highest basal-like/squamous scores regardless of the FGFR1 status (either high or low); indeed, the signature scores between the FGFR4lowFGFR1high and FGFR4lowFGFR1low were not significantly different (Fig. 1D and Supplementary Fig. 1D). Accordingly, all the FGFR4high tumours of the ICGC cohort classified as classical, while only 9.7% (13/134) of the FGFR4high tumours of the PanCuRx were defined as basal-like (Supplementary Fig. 1E). These results were substantially similar to those obtained when tumours of the two cohorts were separated based on the expression level of the known classical driver GATA6 (Supplementary Fig. 1F). When using the molecular classification proposed in the PanCuRx study [5], the group of FGFR4low tumours was highly enriched for basal subtypes (Supplementary Fig. 1G). Reduced FGFR4 expression in PDAC tissues from the ICGC [3] was associated with inferior overall survival (Fig. 1E). Conversely, the levels of FGFR1 had no prognostic significance in different PDAC cohorts (n = 3, data not shown). To corroborate these findings, we used ISH to evaluate the expression of FGFR1 and FGFR4 in a cohort of 106 human pancreatic tissues from treatment naïve patients (Fig. 1F, Supplementary Fig. 2A, and Supplementary Table 1). A total of 97 tissues were suitable for evaluation of both FGFRs. We confirmed that FGFR4 is expressed by epithelial cells while FGFR1 is prominent in stromal elements (Fig. 1F and Supplementary Fig. 2A). Low expression of FGFR4 was observed in 23% (22/97) of cases (Fig. 1G) and was significantly enriched in high grade tumours (Supplementary Fig. 2B). Furthermore, FGFR4 was never detected in poorly differentiated areas while detectable with variable degree of expression in well-differentiated tumour glands (Supplementary Fig. 2C). Finally, low expression of FGFR4 identified patients showing inferior overall survival in our cohort (Fig. 1H). To identify suitable human models for genetic manipulation of the two FGFRs, we screened an initial array of cell lines (n = 6) and Patient-derived Organoids (n = 5). In accordance with our results, ISH analyses of 6 human PDAC cell lines revealed high FGFR4 in cells displaying an epithelial phenotype (Supplementary Fig. 2D–F), and higher levels of FGFR1 in mesenchymal-like cells showing expression of ZEB1 and/or Vimentin (Supplementary Fig. 2D–F). For PDOs, RNA-seq was used to classify cultures as either classical or basal-like using single-sample Gene Set Variation Analysis (ssgsea method) [33]. Of note, the organoid culture with the highest basal-like/squamous identity (PDA9-O) showed the lowest expression of FGFR4 (Supplementary Fig. 2G), thus representing a model for FGFR4low basal-like tumours.
A Receptor Tyrosine Kinases (RTKs) expression in basal-like and classical subtypes. The scatter plot shows the expressed RTK ranked by their mean log2 fold change in basal-like versus classical for samples of the TCGA cohort [4]. Highlighted: FGFR1, FGFR2, FGFR3, FGFR4. B Boxplot of FGFR1 (left) and FGFR4 (middle) Z-scores stratified by the Moffitt subtypes[7] in the TCGA [4], ICGC [3], and PanCuRx [5] datasets. ****p < 0.0001; and ns, not significant as determined by Wilcoxon test. C Violin plots of the normalised expression of FGFR1 (left panel) and FGFR4 (right panel) in each annotated cell cluster from the integration of four different scRNA-seq datasets of PDAC tissues [5, 26,27,28] (see methods). D GSVA score (using ssgsea method) for the basal-like signature [7] for each sample of the ICGC [3] and PanCuRx [5] cohort according to the expression of FGFR1 and FGFR4. **p < 0.01; ****p < 0.0001; and ns, not significant as determined by Wilcoxon test. E Kaplan–Meier plot comparing the overall survival of patients from the ICGC cohort [3] (n = 96) according to the expression of FGFR4. p, Log-rank (Mantel–Cox) test. F Representative ISH images showing expression of FGFR4 and FGFR1 in two different pancreatic cancer tissues. Scale Bar, 50 µm. Insets show magnification of selected areas, and red arrowheads indicate either epithelial or stromal cells. G Stacked bar plot showing the percentage of PDAC tissues (n = 97) with either low (blue) or high (orange) expression of FGFR4. H Kaplan–Meier plot comparing the overall survival of patients according to FGFR4 ISH status. p, Log-rank (Mantel–Cox) test.
FGFR1 is a functional marker of EMT in PDAC
The interrogation of the genomic data from the ICGC [3] and TCGA [4] cohorts in cBioportal (https://www.cbioportal.org) revealed no recurrent genetic alteration affecting FGFR1 in PDAC (Supplementary Fig. 3A, B). We then looked at the correlation between FGFR1 and the established markers/drivers of PDAC cell phenotypes. In all the interrogated transcriptomic datasets (n = 5) [3,4,5, 7, 34], FGFR1 expression positively correlated with the expression of EMT genes, in particular with the master regulator ZEB1, yet not with expression of squamous lineage genes (TP63, KRT5, KRT14) (Fig. 2A). To corroborate the suggested link with EMT, we found a significant positive correlation between the Hallmark EMT signature and FGFR1 in samples from the TCGA cohort [4] (Fig. 2B). Next, we investigated whether induction of EMT or downregulation of its master regulator ZEB1 would affect expression of FGFR1. Following 48 h of TGFβ1 treatment, the SMAD4-proficient cell line hM1 displayed significant elevation in the expression of several EMT markers/drivers as well as of FGFR1 both at mRNA and protein levels (Fig. 2C). The mRNA levels of CDH1 and FGFR4 were only moderately affected by the treatment. Furthermore, the transient downregulation of ZEB1 in the PDAC cell line showing the more prominent mesenchymal-like phenotype (i.e. PANC1) led to a significant reduction in the expression of FGFR1, while increasing expression of FGFR4 and the classical gene CDH1 (Fig. 2D). Then, we sought to assess the functional consequences of FGFR1 inhibition in PDAC. First, we treated 7 human PDAC cell lines with increasing concentrations of the FGFR1 inhibitor BGJ398 [35] and measured cell viability using an ATP-based assay (Fig. 2E). Four out of the seven cell lines tested were primary cell lines (PDA2, PDA9, PDA23, PDA6), which were classified as either basal-like or classical based on RNA-Seq data (Supplementary Fig. 3D and data not shown). BGJ398 demonstrated poor activity and modestly affected proliferation of PDA2, hT1, and of Hs766T only at the highest dose (1 µM) (Fig. 2E). Then, we performed genetic depletion of FGFR1 by RNA interference approaches. Stable silencing of FGFR1 in PANC1 was associated with reduced protein expression of the EMT driver ZEB1 but not of the marker Vimentin (Fig. 2F). In vitro proliferation of Hs766T, but not of PANC1, was significantly reduced following downregulation of FGFR1 (Fig. 2G). However, orthotopically transplanted PANC1 cells deficient for FGFR1 generated smaller tumours compared to parental cells transduced with the empty vector (Fig. 2H), thus suggesting cell extrinsic effects of FGFR1 downregulation in this cell line. In addition, we performed RNAi-based competition assay in the basal-like PDA9-O which displayed the highest protein levels of FGFR1 among the organoid cultures investigated. We observed the progressive drop-out of the vector targeting FGFR1 (Supplementary Fig. 3E), suggesting that FGFR1 is advantageous for the proliferation of this organoid culture. We next sought to explore whether loss of FGFR1 in a basal-like background could lead to changes in PDAC cell identity. Using RNAi, we targeted FGFR1 in the basal-like Hs766T (PDX1−CK5+, Supplementary Fig. 2E) and performed RNA-Seq analysis (Supplementary Fig. 3F and Supplementary Table 2). GSEA demonstrated that the loss of FGFR1 led to alteration of genes programs related to cell proliferation and Interferons’ response (Supplementary Fig. 3G, Supplementary Table 3). However, the loss of FGFR1 did not lead to significant changes in subtype expression signatures and accordingly FGFR1-deficient cells were classified as basal-like (Fig. 2I). Conversely, downregulation of FGFR1 led to the significant reduction of the EMT transcriptional phenotype (Fig. 2J). In sum, our analysis reveals that FGFR1 is a functional driver of EMT in pancreatic cancer.
A Heatmap showing correlation (Spearman’s correlation) between FGFR1 and the squamous lineage markers, and the EMT genes in five different transcriptomic datasets [3,4,5, 7, 34]. All annotated boxes, p < 0.001 B Scatter plot showing the positive correlation between FGFR1 mRNA expression and the Epithelial-to-Mesenchymal Transition (EMT) signature score from MsigDB for the TCGA cohort [4]. C Changes in the expression level of the indicated genes in hM1 cell line treated with 5 ng/mL of TGFβ1 alone or in combination with 500 nM of TGFβ1 inhibitor (A83-01) for 48 h. On the right, immunoblot analysis of ZEB1, FGFR1, and Vimentin in whole-cell lysates from hM1 treated with 500 nM TGFβ1 for 48 h. GAPDH was used as loading control. D Changes at 48 h in the expression level of the indicated genes in the Hs766T cell line transfected with either mock control or two different concentrations of siRNAs targeting ZEB1. On the right, immunoblot analysis of ZEB1 in whole-cell lysates of PANC1 transfected with non-targeting control (NTC) or 25 pmol of siRNA against ZEB1. GAPDH was used as loading control. In (C and D), results shown as mean ± SD of three replicates. **p < 0.01; ***p < 0.001 as determined by Student’s t test. E Cell viability of PDAC monolayer cultures treated with BGJ398 (n = 7 cell cultures) as indicated. Data are displayed as heatmap of the percentage of inhibition at each dose of the drugs and presented as mean of three independent experiments. F Immunoblot analysis of FGFR1, ZEB1, and Vimentin in whole-cell lysates of PANC1 cells stably expressing the control vector (NTC) or the shRNA targeting FGFR1 (shFGFR1). GAPDH was used as loading control. G Relative growth (as percentage of cell proliferation) of PANC1 and Hs766T cells stably transduced with either the control vector (NTC) or the vector targeting FGFR1. Data presented are means ± SD of three biological replicates. H Scatter dot plot showing differences in tumour volumes between tumour-bearing mice transplanted with PANC1/NTC (n = 5 mice) or PANC1/shFGFR1 (n = 10 mice). Tumour volumes were measured by ultrasound 5 weeks after transplantation. **p < 0.01 as determined by Student’s t test. I Boxplots of GSVA score (based on ssgsea method) for the Classical and the Basal-like signatures calculated for the Hs766T cell line transfected with non-targeting control or siRNA against FGFR1. J GSEA plot evaluating the EMT signature upon FGFR1 knockdown in Hs766T cell line.
FGFR4 is epigenetically downregulated in basal-like/squamous PDAC
The reduced expression of FGFR4 in aggressive subtypes of PDAC might be due to either genetic or epigenetic mechanisms. First, we explored the genomic data from the ICGC [3] and TCGA [4] cohorts and found no recurrent genetic alterations affecting FGFR4 (Supplementary Fig. 3A, C). Therefore, we interrogated the available methylation data from the ICGC cohort [3] and found that the downregulation of FGFR4 in the squamous subtype was associated with hypermethylation of the gene (Fig. 3A). Next, we reanalysed data from Diaferia et al. [36] that used ChIP-Seq to profile marks indicative of either active (e.g. H3K27ac) or inactive (e.g. H3K9me3) chromatin in PDAC cell lines representative of classical and basal-like tumours. We found that the genomic region proximal to the FGFR4 transcription start site showed differential levels of H3K27ac between basal-like and classical cell lines, with changes that were concordant with higher level of transcripts in the classical cell line (Fig. 3B and Supplementary Fig. 4A). A broader peak of the active enhancer chromatin mark H3K4me1 was observed for the classical cell line in the intron 1 of FGFR4 gene (Fig. 3B), which has been reported to contain an enhancer region [37]. In keeping with that, FGFR4 expression positively correlated with the expression of endodermal transcription factors and epithelial genes (CDH1 and ERBB3) (Fig. 3C) in 5 different transcriptomic datasets of PDAC [3,4,5, 7, 34]. In particular, FGFR4 showed significant positive correlation with the transcription factor HNF1A in all datasets [3,4,5, 7, 34] (Fig. 3C). Accordingly, putative target genes of HNF1A were significantly enriched in FGFR4 high tumours of the ICGC cohort (Fig. 3D). To prove direct regulation by the transcription factor, we reanalysed available data from pancreas-specific knockout of Hnf1a [19]. In Hnf1a-deficient versus proficient pancreatic cells, expression of Fgfr4 was significantly reduced and this was concordant with genomic regions proximal to the transcription start site of Fgfr4 showing reduced occupancy by HNF1A and reduced levels of H3K27ac (Fig. 3E). Overall, our analysis strongly suggests that FGFR4 is a marker of the classical subtype whose expression is epigenetically reduced in basal-like PDAC cells.
A Hypermethylation of FGFR4 in squamous/basal-like tumours (blue) from the ICGC cohort [3] is concordant with the downregulation of the gene. Indicated is the CpG probe showing the highest correlation. B Representative snapshot of the genomic region of FGFR4 in PANC1 (high-grade, basal-like) and HPAF-II (low-grade, classical) from Diaferia et al. [36] showing histone modifications (H3K27ac, H3K4me1, H3K4me3, H3K9me3) and RNA-seq data. C Heatmap showing correlation (Spearman’s correlation) between FGFR4 and the expressed endodermal transcription factors and epithelial genes in five different transcriptomic datasets [3,4,5, 7, 34]. All annotated boxes, p < 0.001. D GSEA plot evaluating the enrichment of the geneset containing putative HNF1A target genes when comparing FGFR4 high versus FGFR4 low tumours of the ICGC cohort [3]. E Representative snapshot of the genomic region of Fgfr4 in mouse pancreatic cells proficient (control) or deficient (Hnf1a_KO) for HNF1A from Klasniz et al. [19] showing histone modifications (H3K27ac, H3K27me3), HNF1A occupancy, and RNA-seq data. F Changes in the expression levels of the indicated genes in the HPAF-II cell line transfected with either mock control or two different concentrations of siRNA against HNF1A. Results are shown as mean ± SD of three replicates. ***p < 0.001 as determined by Student’s t test. On the right, immunoblot analysis of HNF1A in whole-cell lysates of HPAF-II transfected with either non-targeting control (NTC) or 25 pmol of siRNA targeting HNF1A. β-Actin was used as loading control. G Enrichment of selected pathways upon HNF1A knockdown. The GSEA analysis was performed using gene sets from REACTOME, GO, Hallmark, and REACTOME databases in MsigDB library. Displayed gene sets that passed false-discovery rate < 0.05. See also Supplementary Table 5. H Expression of classical and basal-like genes in HNF1A-deficient HPAF-II cells (compared to parental cells) from RNA-Seq data. Data are presented as mean ± SD. ****, p < 0.0001 by Student t test. I GSEA plot evaluating the basal-like signature upon depletion of HNF1A in HPAF-II cell lines.
HNF1A blocks EMT in a classical PDAC cell line
To confirm the link between HNF1A and FGFR4 in the human PDAC setting, we manipulated the expression of HNF1A by RNAi in HPAF-II, which has been reportedly used as a model of classical PDAC [36, 38]. The successful downregulation of HNF1A was associated with the reduced expression of FGFR4 and increased expression of both ZEB1 and FGFR1 (Fig. 3F). Downregulation of HNF1A was associated with substantial changes in the transcriptome of the HPAF-II cell line (Supplementary Fig. 4B and Supplementary Table 4) and gene-set enrichment analysis (GSEA) [39] showed the enrichment of several pathways and terms related to extracellular matrix organization/deposition and the EMT (Fig. 3G, and Supplementary Table 5). Of the marker/drivers of PDAC epithelial subtypes, only HNF4A was significantly downregulated upon HNF1A knockdown (Fig. 3H). In keeping with a recent work [18], HFN1A downregulation was not sufficient to drive a full phenotype switch in HPAF-II cell line. Yet, HNF1A silencing significantly increased the expression of genes defining the basal-like signature [7] as assessed by GSEA (Fig. 3I).
Loss of FGFR4 enhances the malignant behaviour of PDAC cells
To evaluate the functional relevance of FGFR4 in dictating PDAC phenotypes, we performed genetic and pharmacological perturbation experiments (Fig. 4 and Supplementary Fig. 4). Cell viability of established cell lines and primary PDAC cells (n = 7) was not significantly inhibited by the continuous treatment with the FGFR4 inhibitor BLU9331 [40] (FGFR4i) (Fig. 4A). Of note, low doses of FGFR4i slightly increased the proliferation of some PDAC cell lines. Also considering the dose-response analysis of PDAC cells with the FGFR1 inhibitor BGJ398 (Fig. 2E), our results show that pharmacological inhibition of FGFRs is not a viable strategy to reduce PDAC cells proliferation. Next, we targeted FGFR4 in the classical cell line HPAF-II using three different shRNAs (Fig. 4B). FGFR4 was successfully downregulated by all shRNAs with the #884 and the #419 showing the highest and lowest efficiency, respectively (Fig. 4B). The majority of results were obtained using the two most efficient shRNA, namely #884 and #885. Loss of FGFR4 in HPAF-II cell line was associated with downregulation of the epithelial markers E-Cadherin and ERBB3 (Fig. 4B) and increased in vitro proliferation (Fig. 4C). Interestingly, downregulation of FGFR4 expression altered the proliferative response to three different FGF ligands (Fig. 4D). In particular, continuous treatment with FGF2 and FGF19 significantly reduced the proliferation of control HPAF-II cell line but did not affect the proliferation of cells displaying downregulation of FGFR4 (Fig. 4D). Furthermore, we orthotopically transplanted HPAF-II transduced with non-targeting and targeting vectors into immunocompromised mice and monitored tumour growth for 4 weeks. At the experimental endpoint, loss of FGFR4 in HPAF-II cell line generated larger tumours with increased number of mitotic figures (Fig. 4E, and data not shown), and significantly increased the metastatic burden at the lungs and the liver (Fig. 4F). In agreement with that, the group of FGFR4low tumours of the PanCuRx cohort [5] was significantly enriched for metastases (Supplementary Fig. 4C). The in vivo pro-tumorigenic effect driven by the loss of FGFR4 was further validated in organoid-based xenografts with the classical PDO hT3 (Supplementary Fig. 2G). Immunodeficient mice we orthotopically transplanted with equal number of cells from either parental PDOs (n = 7) or cultures stably transduced with the shRNA targeting FGFR4 (n = 7) (Supplementary Fig. 4D). We monitored tumour growth with high-contrast ultrasound imaging starting at day 15 post transplantation. In agreement with the results obtained with established cell lines, PDOs expressing high levels of FGFR4 displayed reduced proliferation in vivo (Fig. 4G) and less efficient engraftment rate at earlier time points as only 1 out of 7 transplanted mice showed a detectable mass at day 15 as opposed to the FGFR4 deficient PDO (5/7 mice with detectable masses). Finally, we downregulated FGFR4 in the organoid model of FGFR4low basal-like tumours (PDA9-O) prior to in vivo transplantation in immunocompromised hosts. At the endpoint, the tumour masses were significantly larger in animals transplanted with FGFR4 deficient cells (Fig. 4H). ISH analyses of transplanted tissues confirmed the downregulation of FGFR4 as opposed to control mice (Fig. 4I and Supplementary Fig. 4E). To further generalise our results, we transiently downregulated FGFR4 in four PDAC cell lines (including HPAF-II) and observed increased proliferation regardless of the molecular subtype of the cells (Supplementary Fig. 4F). In agreement with that, GSEA comparing the transcriptomes of FGFR4low versus FGFR4high tumours in the ICGC cohort revealed the significant enrichment of terms related to “cell proliferation” and “cell cycle” in FGFR4low PDAC (Supplementary Fig. 4G). Overall, these data suggest that endogenous levels of FGFR4 limit the malignant phenotype of PDAC regardless of the genetic background of the cells.
A Cell viability of PDAC monolayer cultures treated with BLU3391 (n = 7 cell cultures) as indicated. Data are displayed as heatmap of the percentage of inhibition at each dose of the drugs and presented as mean of three independent experiments. B Immunoblot analysis of FGFR4, ErbB3, and E-Cadherin (loading control, β-Actin) in HPAF-II stably expressing the control vector (NTC) or the construct targeting FGFR4 (shFGFR4). C Relative growth (as percentage of cell proliferation) of HPAF-II cells stably transduced with either the control vector (NTC) or the vector targeting FGFR4 (#884). Data presented are means ± SD of three biological replicates. D Proliferation (as total luminescence) of HPAF-II cell line stably transduced with NTC or two different short hairpin RNA against FGFR4 (#419 and #884) following 48 h stimulation with FGF2 (25 ng/mL), FGF10 (100 ng/mL), and FGF19 (100 ng/mL). Data presented as means ± SD (n = 3 biological replicates). *p < 0.05; and **p < 0.01 as determined by Student’s t test. E Scatter dot plot showing differences in tumour volumes between tumour-bearing mice transplanted with HPAF-II/NTC (n = 6 mice) and HPAF-II/shFGFR4 (n = 10 mice). Tumour volumes were measured by ultrasound 4 weeks after transplantation. *p < 0.05 as determined by Student’s t test. F Stacked bar plot showing the distribution of mice with or without metastatic lesions in the two different cohort of mice from (E) (upper panel). Lower panel, stacked bar plot showing the number of mice with individual or multiple metastatic lesions in the two different cohort from (E). On the right, representative Hematoxylin and Eosin staining of metastatic lesions at the liver (top and middle) and at the lungs (bottom) from a mouse bearing multiple metastases. The black arrows indicate the areas shown in the insets. Scale bar, 200 µm. G Line graph showing tumour volumes (mm3) of pancreatic masses detected upon the orthotopic injection of 1 × 106 cells from hT3 PDO into immunodeficient mice (n = 7). Means ± SD are shown. Mice were screened at 15, 21, 27, 32 days following transplantation. ****p < 0.001 by two-way ANOVA with Sidak’s test for multiple comparison. H Scatter plot showing the difference in tumour volumes between mice transplanted with PDA9 organoids either transduced with control vector (n = 10 mice) or with shFGFR4 (n = 7 mice). Tumour volumes were measured by ultrasound 4 weeks after transplantation. ***p < 0.01 as determined by Student’s t test. I Representative in situ hybridisation staining of FGFR4 in PDA9-O in mice from (H) and showing loss of FGFR4 in vivo. Scale bar, 50 μm.
Loss of FGFR4 is associated with increased basal-like/squamous features in PDAC
We next sought to explore whether loss of FGFR4 in a classical background could lead to changes in PDAC cell identity. FGFR4 knockdown in HPAF-II cells led to substantial changes in the transcriptome of this classical cell line (Supplementary Table 6 and Fig. 5A). Similarly, FGFR4 knockdown led to profound changes in the expression of genes that drive the signatures of molecular subtypes of PDAC (Fig. 5B) and deficient cells were classified as basal-like (Fig. 5C) suggesting that, in this cell line, FGFR4 is a superior barrier to the expression of ectopic gene programs (i.e. basal-like) compared to HNF1A. We then looked at changes in the expression of markers/drivers of subtype following FGFR4 silencing and found reduced expression of CDH1 (Fig. 5D), which is consistent with the reduction of E-cadherin (protein level) following stable knockdown (Fig. 4B). Of transcription factors previously associated with the basal-like/squamous lineage, ZBED2 showed the most prominent upregulation upon FGFR4 silencing (Fig. 5D). ZBED2 has been recently demonstrated to antagonise the action of interferon regulatory factor 1 (IRF1), to be selectively enriched in squamous tumours, and to prevent the growth arrest induced by Interferon in PDAC cells [41]. Accordingly, silencing of FGFR4 led to downregulation of gene sets related to interferon pathway in HPAF-II (Fig. 5E, Supplementary Table 7) while increasing for the expression of gene programs related to c-Myc activity and EMT. To corroborate our findings, we interrogated RNA-seq from patients’ tissues and found that ZBED2 is the gene whose expression is more prominent in FGFR4 low tumours of the ICGC cohort (Supplementary Fig. 5A). Furthermore, GSEA comparing the transcriptomes of FGFR4low versus FGFR4high tumours in both the ICGC and PanCuRx cohorts revealed the significant downregulation of terms related to “Interferon” in FGFR4low PDAC (Fig. 5F and Supplementary Fig. 5B). To further generalise our results and understand whether the effect of FGFR4 downregulation on PDAC cell lineages is context dependent, we targeted the receptor by RNAi in two additional models of classical/progenitor (i.e. epithelial) PDAC: AsPC1 and SUIT2. These two cell lines have been extensively used to identify the molecular barriers to the expression of basal-like/squamous programs in PDAC [9, 38] and display high expression of FGFR4 (Supplementary Fig. 5C). Differently from HPAF-II, the forced downregulation of FGFR4 in these two models did not lead to a full switch of phenotype (Supplementary Fig. 5C). Nevertheless, the loss of FGFR4 led to the reduction of classical/progenitor programs and of the epithelial marker E-cadherin, the enrichment of transcriptional signatures of basal-like/squamous PDAC and of genesets related to increased proliferation and c-Myc activity (Fig. 5G, Supplementary Fig. 5D–H). While suggesting that the effect on “full subtype switch” is cell-context dependent, our results strongly suggest that FGFR4 limits the expression of gene programs of aggressive (i.e. basal-like/squamous) PDAC. To further ascertain whether FGFR4 expression is associated with changes in the expression of gene and gene programs of the two PDAC subtypes in human tissues, we interrogated the scRNA-Seq data from Peng et al. [27]. First, we confirmed the enrichment for FGFR4high cells in the epithelial compartment of patients displaying the classical phenotype (Supplementary Fig. 5I). Then, we separated cases with lowest and highest expression of FGFR4 (Supplementary Fig. 5J) to find that those with elevated epithelial expression of FGFR4 accordingly displayed elevated expression of classical markers (Supplementary Fig. 5K). Furthermore, FGFR4low cells were characterised by gene expression programs of basal-like/squamous subtypes (Fig. 5H), which included EMT related gene sets. Finally, we sorted out basal-like and classical cells from those cases and compared the transcriptome of cells displaying high and low expression of FGFR4. In line with the suggested role for FGFR4 in sustaining the classical phenotype, classical cells with the highest expression of FGFR4 displayed elevated expression of classical genes (Fig. 5I). Similarly, even within basal-like cells those with the highest expression of FGFR4 displayed elevated level of classical/epithelial PDAC while the ones with reduced expression of the gene showed increased expression of basal-like/squamous markers (Fig. 5I).
A Heatmap showing changes in the expression pattern of the 30 most differentially expressed genes in the comparison between: control and FGFR4 knock-down (siFGFR4). Z-scores derived from DESeq2-VST transformed counts. See also Supplementary Table 6. B GSEA plot evaluating the Basal-like signature upon FGFR4 knockdown in HPAF-II cell line. C Boxplots of GSVA score (based on the ssGSEA method) evaluating the Basal-like and Classical signatures [7] upon FGFR4 knockdown. D Expression of classical and basal-like genes in FGFR4-deficient HPAF-II cells (compared to parental cells) from RNA-Seq data. Data are presented as mean ± SD.*p < 0.05; **p < 0.01; and ****p < 0.0001 by Student t test. E Enrichment of selected pathways (GSEA) when comparing HPAF-II proficient (NTC) and deficient (siFGFR4) cells. GSEA was performed using gene sets from Hallmark database in MsigDB library. Displayed gene sets that passed false-discovery rate < 0.05. F Enrichment of Interferon related pathways when comparing FGFR4 low versus FGFR4 high tumours of the PanCuRx cohort [5]. GSEA was performed using gene sets from databases in MsigDB library. Displayed gene sets that passed false-discovery rate < 0.05. G GSEA plot evaluating the Bailey_Squamous (left panel) and the Bailey_Progenitor (right panel) signatures upon FGFR4 knockdown in AsPC1 and in SUIT2 cell lines, respectively. H Enrichment of selected pathways (GSEA) when comparing FGFR4 high versus FGFR4 cells from the scRNA-Seq dataset of Peng et al. [27]. See also Supplementary Fig. 5I. GSEA was performed using gene sets from Hallmark database in MsigDB library. I Expression of classical and basal-like/squamous genes in FGFR4 high cells (compared to FGFR4 low cells) displaying either a basal-like (left panel) or a classical (right panel) phenotype.
Loss of FGFR4 leads to increased fluxes through the mTORC1 pathway in PDAC cells
Next, we used Enrichr [42] on gene expression data to infer signalling pathways’ dysregulation that might explain the increased aggressiveness observed in PDAC cells upon depletion of FGFR4 (Supplementary Table 8). We found the significant enrichment of gene sets related to the PI3K/AKT, ERBB, Axon guidance, and RAS signalling pathways in PDAC cells depleted for FGFR4 (Fig. 6A, Supplementary Table 8) as well as in the FGFR4low tumours of both the ICGC [3] and PanCuRx [5] cohorts (Fig. 6B, Supplementary Table 8). First, we evaluated the effect of FGFR4 downregulation on the main signaling activated through the stimulation of FGFRs, the MAPK pathway. Serum-starved HPAF-II and SUIT2 cell lines were stimulated for 20 min with FGF ligands or 2% FBS, which induced the phosphorylation of ERK in both parental and FGFR4-depleted cells (Supplementary Fig. 6A, B). This suggests that the loss of FGFR4 does not compromise signaling through this pathway. Furthermore, the baseline level of phospho-ERK were elevated in both HPAF-II and SUIT2 cell lines upon FGFR4 knockdown, which is in line with the pathway analysis from RNA-seq data. A common single nucleotide polymorphism in the exon 9 of the FGFR4 gene results in a nucleotide change (G388R), which has been shown to enhance STAT3 activation without altering MAPK/ERK pathway [43]. In our cohort, 7 out of 12 PDAC models carry the variant (see material and methods section). In line with a previous report [43], the polymorphic allele does not explain differences in FGFR4 transcript levels between cell lines (data not shown). Then, we tested the effect of FGFR4 knockdown on the activation of STAT3 in cell lines either wild-type (AsPC1) or carrying the polymorphic allele (HPAF-II and SUIT2). In line with the literature [43], the depletion of FGFR4 in SUIT2 and HPAF-II cell lines reduced the baseline and stimulated activation of STAT3 (Supplementary Fig. 6C). Conversely, the downregulation of FGFR4 in AsPC1 cell line did not dramatically affected the activation of STAT3, while consistently driving increased fluxes through MAPK at baseline (Supplementary Fig. 6C). Next, we sought to validate the inferred activation of the PI3K/AKT/mTOR pathway, which resulted the most dysregulated pathway in both cell lines and tissues at transcriptomic level (Fig. 6A, B).
A Enrichr pathway analysis of significantly over-represented genes in FGFR4 deficient HPAF-II cells. See also Supplementary Table 8. B Enrichr pathway analysis of significantly over-represented genes in FGFR4low tumours of the ICGC [3] (top) and PanCuRx [5] (bottom) cohorts. See also Supplementary Table 8. C Immunoblot analysis in whole-cell lysates of HPAF-II cell line transfected with either control (NTC) or siRNA targeting FGFR4. GAPDH was used as loading control. Quantification of changes in the phosphorylated levels of selected proteins (p-AKT, p-S6, and p-4E-BP1) between NTC and siFGFR4 at baseline is provided in the bar plots on the right (data presented as means ± SD of four biological replicates). *p < 0.05; **p < 0.01; ***p < 0.001 by Student’s t test. See also Supplementary Fig. 6D. D Scatter plot showing changes in the phosphorylated levels of 4E-BP1 in four different cell cultures (HPAF-II, PDA2, PDA9, PDA23) following transient knockdown of FGFR4. *p < 0.05 by Student’s t test. The quantification refers to the immunoblots of (C) and Supplementary Fig. 6E–G. E Representative images of the immunohistochemical staining for phosho-4E-BP1 in tissues from mice transplanted with either mock (NTC) or shRNA targeting FGFR4. Areas with different histologies were included in the analyses. White arrowhead refers to islet of Langerhans, which stained negative and used as internal control. The quantification is provided in the scatter dot plot (bottom panel) as percentage of brown pixel (mean intensity/nuclei). A minimum of five field of visualisation (FOVs, 20X areas) per mouse (5 mice/cohort) was analysed. Scale bars, 200 µm. F Immunodetection of DAPI (blue), and newly synthesised protein (green) in cycloheximide-treated HPAF-II cells (top), and HPAF-II transfected with control (NTC, middle panel) and siRNA targeting FGFR4 (bottom panel). Scale bar, 20 μm. G Quantitative Click-iT HPG Alexa Fluor 488 immunofluorescence labeling showed a significant increase in nascent protein synthesis following the silencing of FGFR4 in different monolayer cultures. Data presented as means ± SD (n = 3 biological replicates per cell culture). **p < 0.01; and ***p < 0.001 as determined by Student’s t test. H Cell viability of PDAC monolayer cultures transfected with either mock or siRNA targeting FGFR4 for 48 h and then challenged with 1 µM of the mTORC1 inhibitor Everolimus for 48 h (see methods). Data are displayed as heatmap of the percentage of inhibition relative to the untreated control and presented as mean of three independent experiments.
We assessed the baseline and stimulated activation of the PI3K/AKT/mTOR pathway through the evaluation of the phosphorylated levels of AKT and of the downstream mTORC1 effectors (the inhibitory eIF4E-binding protein 4E-BP1, and the substrate of the S6 Kinase). Cells were serum-starved and then stimulated with FGF2, FGF19, and 2% of Fetal-bovine serum (FBS). In parental HPAF-II cell line, 20 min stimulation with either FGF ligands or FBS led to activation of AKT (Ser473), to increased phosphorylation of 4E-BP1, but not to increased phosphorylation of the S6K1 substrate S6 (Fig. 6C and Supplementary Fig. 6D). Strikingly, loss of FGFR4 in serum-starved condition was associated to increased activation of AKT, of S6K and phosphorylation of 4E-BP1 (Fig. 6C). As compared to the parental cell line, HPAF-II lacking FGFR4 showed higher induction of phosphorylated level of AKT, but no further increase in the phosphorylation of 4E-BP1, following stimulation with either FGFs or serum (Fig. 6C and Supplementary Fig. 6D). Overall, these data suggest that loss of FGFR4 in HPAF-II cell line increases the flux through the mTORC1 pathway in particular through the inhibition of the translational repressor 4E-BP1, which is known to be required for mTORC1-dependent regulation of proliferation in mammalian cells [44]. We replicated these results in five additional cell lines (PDA2-9-23, SUIT2, and AsPC1) by showing that the loss of FGFR4 primarily increases the phosphorylation of 4E-BP1 at baseline (Fig. 6D and Supplementary Fig. 6E–H). In keeping with this, we found increased levels of phosphorylated 4E-BP1 in pancreatic cancer tissues from mice transplanted with HPAF-II stably expressing shFGFR4 (Fig. 6E). Given that mTORC1 is the master regulator of protein synthesis [45], we measured changes in protein synthesis following FGFR4 knockdown in four different PDAC cell lines (see methods). Loss of FGFR4 significantly increased total protein synthesis (Fig. 6F, G) and coherently gene expression terms related to translational initiation were significantly enriched following FGFR4 knockdown (Supplementary Fig. 6I). Finally, we reasoned that if elevation of mTORC1 pathway activity was primarily responsible for the increased proliferation of PDAC cells upon FGFR4 depletion, then pharmacological inhibition of the pathway could represent a viable strategy to reverse this phenotype. Indeed, we found that, compared to the vehicle-treated cells, the treatment with the mTORC1 inhibitor (Everolimus) resulted in a larger inhibition of cell proliferation in FGFR4 depleted cells as opposed to parental cells (Fig. 6H). Overall, our results indicate that loss of FGFR4 in PDAC is associated to increased mTORC1-driven cell proliferation regardless of the genetic and transcriptomic background of the cells.
Discussion
Here, we show that the loss of FGFR4 expression in PDAC is invariably associated with the acquisition of a more aggressive phenotype by cancer cells driven by hyperactivation of the mTORC1 pathway. Conversely, we found that FGFR1 is a functional marker of the EMT. Receptor-Tyrosine Kinases (RTKs) initiated signalling is critical to cell fate determination [20, 21]. Therefore, we reasoned to explore the involvement of RTKs into the definition of PDAC subtypes which display distinct biological and clinical behaviours. The FGF-FGFR axis has been reportedly involved in pancreatic cell specification [22,23,24,25] and, while an aberrant FGFR signalling is reported in several malignancies [46, 47], its involvement in PDAC has been largely neglected so far. Amplification and overexpression of FGFR1 are described in up to 10% of patients with advanced PDAC [48] but rarer in resectable cohorts [3, 4]. Expression of FGFR4 is reported in a substantial fraction of PDAC but its role in mediating tumorigenesis and therapy resistance has been seldomly explored [49, 50]. Here, we provide multiple lines of evidence that FGFR4 expression is restricted to epithelial cells in both normal and diseased pancreata, selectively elevated in classical tumours, and correlated with better outcomes. As opposed to FGFR4, FGFR1 showed more promiscuous expression in both normal and diseased pancreata, was strongly associated with the EMT phenotype but not with epithelial (i.e. classical or basal-like) PDAC subtypes. Accordingly, genetic manipulation of FGFR1 in cells displaying mesenchymal traits and classified as basal-like based on transcriptomic profiling resulted in a reduced expression of markers/drivers of EMT and of the EMT gene program but did not lead to epithelial phenotype switch. Increased FGFR1 expression could be observed in PDAC cells following prolonged exposure to TGFβ1 and coherently reduced upon transient knockdown of the master-regulator ZEB1. Activation of FGFR1 is linked to EMT in prostate cancer [51], and both EMT and upregulation of FGFR1 has been linked to resistance to targeted therapies in lung cancers [52, 53]. In our study, selective inhibition of FGFR1 with BGJ398 did not dramatically affect proliferation of PDAC cells. Differently from FGFR1, we found a defined relationship between FGFR4 expression and the classical subtype of PDAC. Loss of FGFR4 in basal-like/squamous tumours was concordant with hypermethylation of the gene and reduced levels of active chromatin in its regulatory regions. Our results strongly suggest an epigenetic dysregulation of FGFR4 expression in basal-like/squamous tumours which is similar to that observed for several endodermal transcription factors. We also demonstrated that FGFR4 is a direct target of the classical transcription factor HNF1A. In line with recent reports, the silencing of HNF1A in the classical PDAC cell line HPAF-II was not sufficient to drive a subtype switch. In the same model, however, downregulation of FGFR4 was sufficient to drive a full subtype switch thus suggesting that FGFR4 has stronger “antibasal” function than HNF1A in this cell line. In other two well-established models of classical/progenitor PDAC, the forced downregulation of FGFR4 was not sufficient to drive a full switch of phenotype towards the basal-like subtype but was invariably associated with the reduced expression of classical genes and the enrichment for transcriptional programs of basal-like/squamous PDAC. This suggests that FGFR4 acts to maintain the classical phenotype and therefore to limit the expression of ectopic gene programs (i.e. basal-like/squamous) in PDAC. In keeping with that, the analysis of scRNA-Seq data demonstrated that, at individual cell level, FGFR4 expression is elevated in cells with a more classical/epithelial phenotype.
We additionally showed that FGFR4 loss is associated with enhanced malignancy of PDAC cells by increasing in vitro and in vivo proliferation as well as enhancing metastatization of a cell line of the classical background. This finding further corroborates the evidence that basal-like cells tend to accumulate in advanced stages of the disease. While repression of “basalness/squamousness” seems an intrinsic feature of FGFR4, we also showed that its silencing in FGFR4low basal-like models increases malignant behaviour suggesting a bona fide tumour suppressive role for endogenous levels of FGFR4 in PDAC. Others have recently suggested that RTKs in PDAC might exert an oncogenic role only when overexpressed [54]. In keeping with this, acceleration of cell proliferation could be observed in established and primary cell lines regardless of their background and upon either transient or stable knockdown of FGFR4. Accordingly, gene programs related to cell cycle and proliferation were highly enriched in the comparison between low versus high FGFR4 tumours of the ICGC and PanCuRx cohorts [3, 5]. Pharmacological inhibition of FGFR4 had little to no effect on the short-term proliferation of several PDAC cell lines. Interestingly, stimulation of FGFR4high classical cells with FGF2 and FGF19 led to reduced cell proliferation, which partially agrees with results from Motoda et al. [50] that suggested a tumour suppressive role of FGF19 stimulation on cells expressing FGFR4 in PDAC. Conversely, FGF ligands did not exert inhibitor effects on the proliferation of cells deficient for FGFR4. Mechanistically, downregulation of FGFR4 was associated with increased basal fluxes through the MAPK and the mTORC1 signaling pathways regardless of the genomic and transcriptomic background of the cells. Accordingly, PI3K/Akt/mTOR transcriptional signatures as well as the inhibitory phosphorylation of one of the best characterised mTORC1 substrate, 4E-BP1, were significantly enriched in both cell lines and tissues displaying reduced expression of FGFR4. Phosphorylation of 4E-BP1 by mTORC1 has been demonstrated to regulate mTORC1-driven proliferation in mammalian cells [44, 45] and accordingly its levels were higher in all cell lines tested following silencing of FGFR4 and were concordant with increased cell proliferation and protein synthesis. In keeping with this, pharmacological inhibition of mTORC1 with Everolimus showed superior effect in reducing the proliferation of cells deficient for FGFR4 as opposed to proficient cells. In summary, we show that endogenous levels of FGFR4 limit the malignant phenotype of PDAC. In particular, the loss of FGFR4 was associated with increased activity of the mTORC1 pathway in PDAC cells. Finally, we propose FGFR4 as a valuable marker for the prognostic stratification of PDAC patients.
Data availability
The primary RNA-Seq data generated for this study will be available in a public, open access repository. All other relevant data are already available as Supplementary material.
References
Michl P, Lohr M, Neoptolemos JP, Capurso G, Rebours V, Malats N et al. UEG position paper on pancreatic cancer. Bringing pancreatic cancer to the 21st century: Prevent, detect, and treat the disease earlier and better. United Eur Gastroenterol J. 2021;9:860–71.
Rahib L, Wehner MR, Matrisian LM, Nead KT. Estimated Projection of US Cancer Incidence and Death to 2040. JAMA Netw Open. 2021;4:e214708.
Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52.
Cancer Genome Atlas Research Network. Electronic address aadhe, Cancer Genome Atlas Research N. Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell. 2017;32:185–203 e113.
Chan-Seng-Yue M, Kim JC, Wilson GW, Ng K, Figueroa EF, O’Kane GM, et al. Transcription phenotypes of pancreatic cancer are driven by genomic events during tumor evolution. Nat Genet. 2020;52:231–40.
Collisson EA, Bailey P, Chang DK, Biankin AV. Molecular subtypes of pancreatic cancer. Nat Rev Gastroenterol Hepatol. 2019;16:207–20.
Moffitt RA, Marayati R, Flate EL, Volmar KE, Loeza SG, Hoadley KA, et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet. 2015;47:1168–78.
Hayashi A, Fan J, Chen R, Ho Y-j, Makohon-Moore AP, Lecomte N, et al. A unifying paradigm for transcriptional heterogeneity and squamous features in pancreatic ductal adenocarcinoma. Nat Cancer. 2020;1:59–74.
Kloesch B, Ionasz V, Paliwal S, Hruschka N, Martinez De Villarreal J, Öllinger R, et al. A GATA6-centred gene regulatory network involving HNFs and ΔNp63 controls plasticity and immune escape in pancreatic cancer. Gut. 2021;0:1–12.
Martinelli P, Carrillo-De Santa Pau E, Cox T, Sainz B, Dusetti N, Greenhalf W, et al. GATA6 regulates EMT and tumour dissemination, and is a marker of response to adjuvant chemotherapy in pancreatic cancer. Gut. 2017;66:1665–76.
Somerville TD, Biffi G, Dassler-Plenker J, Hur SK, He XY, Vance KE, et al. Squamous trans-differentiation of pancreatic cancer cells promotes stromal inflammation. Elife 2020;9:e53381.
Andricovich J, Perkail S, Kai Y, Casasanta N, Peng W, Tzatsos A. Loss of KDM6A Activates Super-Enhancers to Induce Gender-Specific Squamous-like Pancreatic Cancer and Confers Sensitivity to BET Inhibitors. Cancer Cell. 2018;33:512–526.e518.
Porter RL, Magnus NKC, Thapar V, Morris R, Szabolcs A, Neyaz A, et al. Epithelial to mesenchymal plasticity and differential response to therapies in pancreatic ductal adenocarcinoma. Proc Natl Acad Sci USA. 2019;116:26835–45.
Dreyer SB, Upstill-Goddard R, Paulus-Hock V, Paris C, Lampraki EM, Dray E, et al. Targeting DNA Damage Response and Replication Stress in Pancreatic Cancer. Gastroenterology. 2021;160:362–377.e313.
Brunton H, Caligiuri G, Cunningham R, Upstill-Goddard R, Bailey UM, Garner IM, et al. HNF4A and GATA6 Loss Reveals Therapeutically Actionable Subtypes in Pancreatic Cancer. Cell Rep. 2020;31:107625.
Aung KL, Fischer SE, Denroche RE, Jang GH, Dodd A, Creighton S, et al. Genomics-Driven Precision Medicine for Advanced Pancreatic Cancer: Early Results from the COMPASS Trial. Clin Cancer Res. 2018;24:1344–54.
Nicolle R, Blum Y, Duconseil P, Vanbrugghe C, Brandone N, Poizat F, et al. Establishment of a pancreatic adenocarcinoma molecular gradient (PAMG) that predicts the clinical outcome of pancreatic cancer. EBioMedicine. 2020;57:102858.
Kloesch B, Ionasz V, Paliwal S, Hruschka N, Martinez de Villarreal J, Ollinger R, et al. A GATA6-centred gene regulatory network involving HNFs and DeltaNp63 controls plasticity and immune escape in pancreatic cancer. Gut. 2021;71:766–77.
Kalisz M, Bernardo E, Beucher A, Maestro MA, Del Pozo N, Millan I, et al. HNF1A recruits KDM6A to activate differentiated acinar cell programs that suppress pancreatic cancer. EMBO J. 2020;39:e102808.
Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–34.
Neben CL, Lo M, Jura N, Klein OD. Feedback regulation of RTK signaling in development. Dev Biol. 2019;447:71–89.
Bhushan A, Itoh N, Kato S, Thiery JP, Czernichow P, Bellusci S, et al. Fgf10 is essential for maintaining the proliferative capacity of epithelial progenitor cells during early pancreatic organogenesis. Development. 2001;128:5109–17.
Elghazi L, Cras-Meneur C, Czernichow P, Scharfmann R. Role for FGFR2IIIb-mediated signals in controlling pancreatic endocrine progenitor cell proliferation. Proc Natl Acad Sci USA. 2002;99:3884–9.
Goncalves CA, Larsen M, Jung S, Stratmann J, Nakamura A, Leuschner M, et al. A 3D system to model human pancreas development and its reference single-cell transcriptome atlas identify signaling pathways required for progenitor expansion. Nat Commun. 2021;12:3144.
Hebrok M, Kim SK, Melton DA. Notochord repression of endodermal Sonic hedgehog permits pancreas development. Genes Dev. 1998;12:1705–13.
Lin W, Noel P, Borazanci EH, Lee J, Amini A, Han IW, et al. Single-cell transcriptome analysis of tumor and stromal compartments of pancreatic ductal adenocarcinoma primary tumors and metastatic lesions. Genome Med. 2020;12:80.
Peng J, Sun BF, Chen CY, Zhou JY, Chen YS, Chen H, et al. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res. 2019;29:725–38.
Steele NG, Carpenter ES, Kemp SB, Sirihorachai V, The S, Delrosario L, et al. Multimodal Mapping of the Tumor and Peripheral Blood Immune Landscape in Human Pancreatic Cancer. Nat Cancer. 2020;1:1097–112.
Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods. 2019;16:1289–96.
Muraro MJ, Dharmadhikari G, Grun D, Groen N, Dielen T, Jansen E, et al. A Single-Cell Transcriptome Atlas of the Human Pancreas. Cell Syst. 2016;3:385–394 e383.
Grun D, Muraro MJ, Boisset JC, Wiebrands K, Lyubimova A, Dharmadhikari G, et al. De Novo Prediction of Stem Cell Identity using Single-Cell Transcriptome Data. Cell Stem Cell. 2016;19:266–77.
Segerstolpe A, Palasantza A, Eliasson P, Andersson EM, Andreasson AC, Sun X, et al. Single-Cell Transcriptome Profiling of Human Pancreatic Islets in Health and Type 2 Diabetes. Cell Metab. 2016;24:593–607.
Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinforma. 2013;14:7.
Balli D, Rech AJ, Stanger BZ, Vonderheide RH. Immune Cytolytic Activity Stratifies Molecular Subsets of Human Pancreatic Cancer. Clin Cancer Res. 2017;23:3129–38.
Guagnano V, Furet P, Spanka C, Bordas V, Le Douget M, Stamm C, et al. Discovery of 3-(2,6-dichloro-3,5-dimethoxyphenyl)-1-{6-[4-(4-ethyl-piperazin-1-yl)-phenylamin o]-pyrimidin-4-yl}-1-methyl-urea (NVP-BGJ398), a potent and selective inhibitor of the fibroblast growth factor receptor family of receptor tyrosine kinase. J Med Chem. 2011;54:7066–83..
Diaferia GR, Balestrieri C, Prosperini E, Nicoli P, Spaggiari P, Zerbi A, et al. Dissection of transcriptional and cis-regulatory control of differentiation in human pancreatic cancer. EMBO J. 2016;35:595–617.
Shah P, Ding Y, Niemczyk M, Kudla G, Plotkin JB. Rate-limiting steps in yeast protein translation. Cell. 2013;153:1589–601.
Somerville TDD, Xu Y, Miyabayashi K, Tiriac H, Cleary CR, Maia-Silva D, et al. TP63-Mediated Enhancer Reprogramming Drives the Squamous Subtype of Pancreatic Ductal Adenocarcinoma. Cell Rep. 2018;25:1741–1755.e1747.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50.
Hagel M, Miduturu C, Sheets M, Rubin N, Weng W, Stransky N, et al. First Selective Small Molecule Inhibitor of FGFR4 for the Treatment of Hepatocellular Carcinomas with an Activated FGFR4 Signaling Pathway. Cancer Disco. 2015;5:424–37.
Somerville TDD, Xu Y, Wu XS, Maia-Silva D, Hur SK, de Almeida LMN, et al. ZBED2 is an antagonist of interferon regulatory factor 1 and modifies cell identity in pancreatic cancer. Proc Natl Acad Sci USA. 2020;117:11471–82.
Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–97.
Ulaganathan VK, Sperl B, Rapp UR, Ullrich A. Germline variant FGFR4 p.G388R exposes a membrane-proximal STAT3 binding site. Nature. 2015;528:570–4.
Dowling RJ, Topisirovic I, Alain T, Bidinosti M, Fonseca BD, Petroulakis E, et al. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science. 2010;328:1172–6.
Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–13.
Frattini V, Pagnotta SM, Tala, Fan JJ, Russo MV, Lee SB, et al. A metabolic function of FGFR3-TACC3 gene fusions in cancer. Nature. 2018;553:222–7.
Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer. 2010;10:116–29.
Aguirre AJ, Nowak JA, Camarda ND, Moffitt RA, Ghazani AA, Hazar-Rethinam M, et al. Real-time Genomic Characterization of Advanced Pancreatic Cancer to Enable Precision Medicine. Cancer Disco. 2018;8:1096–111.
Sasaki N, Gomi F, Yoshimura H, Yamamoto M, Matsuda Y, Michishita M et al. FGFR4 Inhibitor BLU9931 Attenuates Pancreatic Cancer Cell Proliferation and Invasion While Inducing Senescence: Evidence for Senolytic Therapy Potential in Pancreatic Cancer. Cancers (Basel). 2020;12:2976.
Motoda N, Matsuda Y, Onda M, Ishiwata T, Uchida E, Naito Z. Overexpression of fibroblast growth factor receptor 4 in high-grade pancreatic intraepithelial neoplasia and pancreatic ductal adenocarcinoma. Int J Oncol. 2011;38:133–43.
Acevedo VD, Gangula RD, Freeman KW, Li R, Zhang Y, Wang F, et al. Inducible FGFR-1 activation leads to irreversible prostate adenocarcinoma and an epithelial-to-mesenchymal transition. Cancer Cell. 2007;12:559–71.
Raoof S, Mulford IJ, Frisco-Cabanos H, Nangia V, Timonina D, Labrot E, et al. Targeting FGFR overcomes EMT-mediated resistance in EGFR mutant non-small cell lung cancer. Oncogene. 2019;38:6399–413.
Kitai H, Ebi H, Tomida S, Floros KV, Kotani H, Adachi Y, et al. Epithelial-to-Mesenchymal Transition Defines Feedback Activation of Receptor Tyrosine Kinase Signaling Induced by MEK Inhibition in KRAS-Mutant Lung Cancer. Cancer Disco. 2016;6:754–69.
Meyers N, Gerard C, Lemaigre FP, Jacquemin P. Differential impact of the ERBB receptors EGFR and ERBB2 on the initiation of precursor lesions of pancreatic ductal adenocarcinoma. Sci Rep. 2020;10:5241.
Acknowledgements
We gratefully acknowledge the Centro Piattaforme Tecnologiche (CPT - University of Verona, Verona, Italy) for granting access to the genomic facility of University of Verona. In addition, we acknowledge the assistance provided by Serena Pedron, Valerio Crestani, Claudia Parolini, e Paola Perfetti for the execution of immunohistochemical staining. VC is supported by Associazione Italiana Ricerca sul Cancro (AIRC; Grant No. 18178). VC is also supported by the EU (MSCA project PRECODE, grant No: 861196) and the National Cancer Institute (NCI, HHSN26100008). EB is supported by the AIRC (Grant No. IG20583), and by Institutional funds of Università Cattolica del Sacro Cuore (UCSC-project D1). AS is supported by AIRC 5 × 1000, grant n 12182. SDa is supported by AIRC (24043). CF was supported for by the Fondazione Umberto Veronesi. AdC was supported by AIRC (19404, 20872). PD is supported by Fondazione Nadia Valsecchi Onlus. EF is supported by AIRC (25286). This study was conducted with the support of the Ontario Institute for Cancer Research through funding provided by the Government of Ontario. The funding agencies had no role in the collection, analysis, and interpretation of data or in the writing of the paper.
Author information
Authors and Affiliations
Contributions
SDa, EB and VC designed the research; SDa, LV, CF, GTT, AdC, EF, CV, GF, FL, SA, GL and AM performed experiments; FP, PD, DP and PB analysed omics data and generated displays; RTL, AP, ES and CB collected samples and clinicopathological information; CL and AS performed histopathological evaluation of human tissues and xenografts; MM and EB analysed treatment data; VC, EB and AS acquired funding; SDa, FP, LV and VC wrote the paper; VC and EB supervised the study. All authors approved the final version of the paper.
Corresponding author
Ethics declarations
Competing interests
EB received speakers’ and travels’ fee from MSD, Astra-Zeneca, Pfizer, Eli-Lilly, BMS, Novartis and Roche. EB received institutional research grants from Astra-Zeneca, Roche. The other authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
D’Agosto, S., Pezzini, F., Veghini, L. et al. Loss of FGFR4 promotes the malignant phenotype of PDAC. Oncogene 41, 4371–4384 (2022). https://doi.org/10.1038/s41388-022-02432-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-022-02432-5