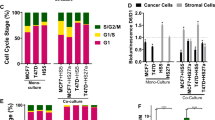
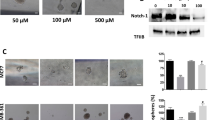
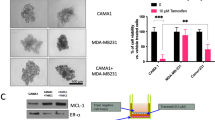
Abstract
Patients with estrogen receptor-positive (ER+) breast cancer, the most common subtype, remain at risk for lethal metastatic disease years after diagnosis. Recurrence arises partly because tumor cells in bone marrow become resistant to estrogen-targeted therapy. Here, we utilized a co-culture model of bone marrow mesenchymal stem cells (MSCs) and ER+ breast cancer cells to recapitulate interactions of cancer cells in bone marrow niches. ER+ breast cancer cells in direct contact with MSCs acquire cancer stem-like (CSC) phenotypes with increased resistance to standard antiestrogenic drugs. We confirmed that co-culture with MSCs increased labile iron in breast cancer cells, a phenotype associated with CSCs and disease progression. Clinically approved iron chelators and in-house lysosomal iron-targeting compounds restored sensitivity to antiestrogenic therapy. These findings establish iron modulation as a mechanism to reverse MSC-induced drug resistance and suggest iron modulation in combination with estrogen-targeted therapy as a promising, translatable strategy to treat ER+ breast cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
RNA sequencing data have been deposited in the Gene Expression Omnibus database (GEO accession number GSE196312).
Code availability
All custom MATLAB codes are available upon request through a material transfer agreement.
References
Savci-Heijink CD, Halfwerk H, Hooijer GKJ, Horlings HM, Wesseling J, van de Vijver MJ. Retrospective analysis of metastatic behaviour of breast cancer subtypes. Breast Cancer Res Treat. 2015;150:547–57.
Patel SA, Dave MA, Murthy RG, Helmy KY, Rameshwar P. Metastatic breast cancer cells in the bone marrow microenvironment: novel insights into oncoprotection. Oncol Rev. 2011;5:93–102.
Pantel K, Alix-Panabieres C. Bone marrow as a reservoir for disseminated tumor cells: a special source for liquid biopsy in cancer patients. Bonekey Rep. 2014;3:584.
Amanatullah DF, Tamaresis JS, Chu P, Bachmann MH, Hoang NM, Collyar D, et al. Local estrogen axis in the human bone microenvironment regulates estrogen receptor-positive breast cancer cells. Breast Cancer Res. 2017;19:121.
Jeselsohn R, Buchwalter G, De Angelis C, Brown M, Schiff R. ESR1 mutations—a mechanism for acquired endocrine resistance in breast cancer. Nat Rev Clin Oncol. 2015;12:573–83.
Katzenellenbogen JA, Mayne CG, Katzenellenbogen BS, Greene GL, Chandarlapaty S. Structural underpinnings of oestrogen receptor mutations in endocrine therapy resistance. Nat Rev Cancer. 2018;18:377–88.
Lavinsky RM, Jepsen K, Heinzel T, Torchia J, Mullen TM, Schiff R, et al. Diverse signaling pathways modulate nuclear receptor recruitment of N-CoR and SMRT complexes. Proc Natl Acad Sci USA. 1998;95:2920–5.
Johnston SR, Lu B, Scott GK, Kushner PJ, Smith IE, Dowsett M, et al. Increased activator protein-1 DNA binding and c-Jun NH2-terminal kinase activity in human breast tumors with acquired tamoxifen resistance. Clin Cancer Res. 1999;5:251–6.
Magnani L, Stoeck A, Zhang X, Lánczky A, Mirabella AC, Wang TL, et al. Genome-wide reprogramming of the chromatin landscape underlies endocrine therapy resistance in breast cancer. Proc Natl Acad Sci USA. 2013;110:E1490–9.
Muluhngwi P, Klinge CM. Roles for miRNAs in endocrine resistance in breast cancer. Endocr Relat Cancer. 2015;22:R279–300.
Plava J, Cihova M, Burikova M, Matuskova M, Kucerova L, Miklikova S. Recent advances in understanding tumor stroma-mediated chemoresistance in breast cancer. Mol Cancer. 2019;18:67.
Shiozawa Y, Pienta KJ, Taichman RS. Hematopoietic stem cell niche is a potential therapeutic target for bone metastatic tumors. Clin Cancer Res. 2011;17:5553–8.
Shi Z, Yang WM, Chen LP, Yang DH, Zhou Q, Zhu J, et al. Enhanced chemosensitization in multidrug-resistant human breast cancer cells by inhibition of IL-6 and IL-8 production. Breast Cancer Res Treat. 2012;135:737–47.
Chen DR, Lu DY, Lin HY, Yeh WL. Mesenchymal stem cell-induced doxorubicin resistance in triple negative breast cancer. Biomed Res Int. 2014;2014:532161.
Yeh WL, Tsai CF, Chen DR. Peri-foci adipose-derived stem cells promote chemoresistance in breast cancer. Stem Cell Res Ther. 2017;8:177.
Houthuijzen JM, Daenen LGM, Roodhart JML, Voest EE. The role of mesenchymal stem cells in anti-cancer drug resistance and tumour progression. Br J cancer. 2012;106:1901–6.
Graham N, Qian B-Z. Mesenchymal stromal cells: emerging roles in bone metastasis. Int J Mol Sci. 2018;19:1121.
Kucerova L, Skolekova S, Matuskova M, Bohac M, Kozovska Z. Altered features and increased chemosensitivity of human breast cancer cells mediated by adipose tissue-derived mesenchymal stromal cells. BMC Cancer. 2013;13:535.
Luo M, Brooks M, Wicha M. Epithelial-mesenchymal plasticity of breast cancer stem cells: implications for metastasis and therapeutic resistance. Curr Pharm Des. 2015;21:1301–10.
Pastushenko I, Brisebarre A, Sifrim A, Fioramonti M, Revenco T, Boumahdi S, et al. Identification of the tumour transition states occurring during EMT. Nature. 2018;556:463–8.
Sarmiento-Castro A, Caamaño-Gutiérrez E, Sims AH, Hull NJ, James MI, Santiago-Gómez A, et al. Increased expression of interleukin-1 receptor characterizes anti-estrogen-resistant ALDH+ breast cancer stem cells. Stem Cell Rep. 2020;15:307–16.
Pogribny IP, Tryndyak VP, Pogribna M, Shpyleva S, Surratt G, Gamboa da Costa G, et al. Modulation of intracellular iron metabolism by iron chelation affects chromatin remodeling proteins and corresponding epigenetic modifications in breast cancer cells and increases their sensitivity to chemotherapeutic agents. Int J Oncol. 2013;42:1822–32.
Recalcati S, Gammella E, Cairo G. Dysregulation of iron metabolism in cancer stem cells. Free Radic Biol Med. 2019;133:216–20.
Brown RAM, Richardson KL, Kabir TD, Trinder D, Ganss R, Leedman PJ. Altered iron metabolism and impact in cancer biology, metastasis, and immunology. Front Oncol. 2020;10:476.
Forciniti S, Greco LA-O, Grizzi FA-OX, Malesci A, Laghi LA-O. Iron metabolism in cancer progression. Int J Mol Sci. 2020;21:2257.
Tury SA-OX, Assayag F, Bonin F, Chateau-Joubert S, Servely JL, Vacher S, et al. The iron chelator deferasirox synergises with chemotherapy to treat triple-negative breast cancers. J Pathol. 2018;246:103–14.
Goto W, Kashiwagi S, Asano Y, Takada K, Morisaki T, Takahashi K, et al. Inhibitory effects of iron depletion plus eribulin on the breast cancer microenvironment. BMC Cancer. 2020;20:1215.
Chanvorachote P, Luanpitpong SA-O. Iron induces cancer stem cells and aggressive phenotypes in human lung cancer cells. Am J Physiol Cell Physiol. 2016;310:C728–39.
Huang J, Woods P, Normolle D, Goff JP, Benos PV, Stehle CJ, et al. Downregulation of estrogen receptor and modulation of growth of breast cancer cell lines mediated by paracrine stromal cell signals. Breast Cancer Res Treat. 2017;161:229–43.
Colacino JA, Azizi E, Brooks MD, Harouaka R, Fouladdel S, McDermott SP, et al. Heterogeneity of human breast stem and progenitor cells as revealed by transcriptional profiling. Stem Cell Rep. 2018;10:1596–609.
Hu J, Lei H, Fei X, Liang S, Xu H, Qin D, et al. NES1/KLK10 gene represses proliferation, enhances apoptosis and down-regulates glucose metabolism of PC3 prostate cancer cells. Sci Rep. 2015;5:17426.
Yousef GM, Yacoub GM, Polymeris ME, Popalis C, Soosaipillai A, Diamandis EP. Kallikrein gene downregulation in breast cancer. Br J Cancer. 2004;90:167–72.
Adamo A, Delfino P, Gatti A, Bonato A, Takam Kamga P, Bazzoni R, et al. HS-5 and HS-27A stromal cell lines to study bone marrow mesenchymal stromal cell-mediated support to cancer development. Front Cell Dev Biol. 2020;8:584232.
Bystrom LM, Guzman ML, Rivella S. Iron and reactive oxygen species: friends or foes of cancer cells? Antioxid Redox Signal. 2014;20:1917–24.
Chirillo R, Aversa I, Di Vito A, Salatino A, Battaglia AM, Sacco A, et al. FtH-mediated ROS dysregulation promotes CXCL12/CXCR4 axis activation and EMT-like trans-differentiation in erythroleukemia K562 cells. Front Oncol. 2020;10:698.
Yang J, Bielenberg DR, Rodig SJ, Doiron R, Clifton MC, Kung AL, Strong RK, et al. Lipocalin 2 promotes breast cancer progression. Proc Natl Acad Sci USA. 2009;106:3913–8.
Müller SA-O, Sindikubwabo F, Cañeque TA-O, Lafon A, Versini A, Lombard B, et al. CD44 regulates epigenetic plasticity by mediating iron endocytosis. Nat Chem. 2020;12:929–38.
Mai TT, Hamaï A, Hienzsch A, Cañeque T, Müller S, Wicinski J, et al. Salinomycin kills cancer stem cells by sequestering iron in lysosomes. Nat Chem. 2017;9:1025–33.
Liang W, Ferrara N. Iron metabolism in the tumor microenvironment: contributions of innate immune cells. Front Immunol. 2020;11:626812.
Leimberg MJ, Prus E, Konijn AM, Fibach E. Macrophages function as a ferritin iron source for cultured human erythroid precursors. J Cell Biochem. 2008;103:1211–8.
Ören B, Urosevic J, Mertens C, Mora J, Guiu M, Gomis RR, et al. Tumour stroma-derived lipocalin-2 promotes breast cancer metastasis. J Pathol. 2016;239:274–85.
Blanchette-Farra N, Kita D, Konstorum A, Tesfay L, Lemler D, Hegde P, et al. Contribution of three-dimensional architecture and tumor-associated fibroblasts to hepcidin regulation in breast cancer. Oncogene. 2018;37:4013–32.
El Hout M, Dos Santos L, Hamaï A, Mehrpour M. A promising new approach to cancer therapy: targeting iron metabolism in cancer stem cells. Semin Cancer Biol. 2018;53:125–38.
Schonberg DL, Miller TE, Wu Q, Flavahan WA, Das NK, Hale JS, et al. Preferential iron trafficking characterizes glioblastoma stem-like cells. Cancer Cell. 2015;28:441–55.
Rodriguez R, Schreiber SL, Conrad M. Persister cancer cells: Iron addiction and vulnerability to ferroptosis. Mol Cell. 2021;82:728–40.
Huczyński A, Janczak J, Antoszczak M, Wietrzyk J, Maj E, Brzezinski B. Antiproliferative activity of salinomycin and its derivatives. Bioorg Med Chem Lett. 2012;22:7146–50.
Katsura Y, Ohara T, Noma K, Ninomiya T, Kashima H, Kato T, et al. A novel combination cancer therapy with iron chelator targeting cancer stem cells via suppressing stemness. Cancers. 2019;11:177.
Saeki I, Yamamoto N, Yamasaki T, Takami T, Maeda M, Fujisawa K, et al. Effects of an oral iron chelator, deferasirox, on advanced hepatocellular carcinoma. World J Gastroenterol. 2016;22:8967–77.
Mody K, Mansfield AS, Vemireddy L, Nygren P, Gulbo J, Borad M. A phase I study of the safety and tolerability of VLX600, an Iron Chelator, in patients with refractory advanced solid tumors. Invest N Drugs. 2019;37:684–92.
Raggi C, Gammella E, Correnti M, Buratti P, Forti E, Andersen JB, et al. Dysregulation of iron metabolism in cholangiocarcinoma stem-like cells. Sci Rep. 2017;7:17667.
Buschhaus JM, Humphries BA, Eckley SS, Robison TH, Cutter AC, Rajendran S, et al. Targeting disseminated estrogen-receptor-positive breast cancer cells in bone marrow.
Buschhaus JM, Luker KE, Luker GD. A facile, in vitro 384-well plate system to model disseminated tumor cells in the bone marrow microenvironment. In: Lacorazza HD, editor. Cellular quiescence: methods and protocols. New York, NY: Springer New York; 2018. p. 201–13.
Humphries BA, Buschhaus JM, Chen YC, Haley HR, Qyli T, Chiang B, et al. Plasminogen Activator Inhibitor 1 (PAI1) promotes actin cytoskeleton reorganization and glycolytic metabolism in triple-negative breast cancer. Mol Cancer Res. 2019;17:1142–54.
Eckley SSBJ, Humphries BA, Robison TH, Luker KE, Luker GD. Short-term environmental conditioning generates cellular memory that enhances tumorigenic potential of triple-negative breast cancer cells. Tomography. 2019;5:346–57.
Luker G, Pica C, Song J, Luker K, Piwnica-Worms D. Imaging 26S proteasome activity and inhibition in living mice. Nat Med. 2003;9:969–73.
Cavnar S, Xiao A, Gibbons A, Rickelmann A, Neely T, Luker K, et al. Imaging sensitivity of quiescent cancer cells to metabolic perturbations in bone marrow spheroids. Tomography. 2016;2:146–57.
Luo M, Shang L, Brooks MD, Jiagge E, Zhu Y, Buschhaus JM, et al. Targeting breast cancer stem cell state equilibrium through modulation of redox signaling. Cell Metab. 2018;28:69–86.
Ge SX, Son EW, Yao R. iDEP: an integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinforma. 2018;19:534.
Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehar J, et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–73.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50.
Ray P, Stacer A, Fenner J, Cavnar S, Mequiar K, Brown M, et al. CXCL12-γ in primary tumors drives breast cancer metastasis. Oncogene. 2015;34:2043–51.
Acknowledgements
We thank Sean Linkes, Ann Marie Deslauriers-Cox, and Michael Pihalja for assistance with Flow cytometry. We acknowledge support from the Advanced Genomics Core, the Flow Cytometry Core, and the Center for Molecular Imaging Core of the University of Michigan Medical School’s Biomedical Research Core Facilities.
Funding
We acknowledge support to the University of Michigan Rogel Cancer Center through National Institutes of Health grant P30CA046592 for flow cytometry and animal imaging studies. The authors acknowledge funding from NIH grants R01CA100768, R01CA238042, R01GM138385, R01CA148828, R01CA245546, R01DK095201, R01CA248160, R35CA197585, R01CA2449310, U01CA210152, R01CA238023, R33CA225549, R50CA221807, and R37CA222563 and funding from the Breast Cancer Research Foundation grant BCRF-18-173. RR lab is funded by the European Research Council under the European Union’s Horizon 2020 research and innovation programme grant agreement No 647973 (R.R.), the Foundation Charles Defforey-Institut de France, and Ligue Contre le Cancer Equipe Labellisée. This material is based upon work supported by the National Science Foundation Graduate Research Fellowship under Grant No. DGE 1256260 (JMB and AJM). BAH, Ph.D., was supported by an American Cancer Society - Michigan Cancer Research Fund Postdoctoral Fellowship, PF-18-236-01-CCG. ZCN is supported by the Michigan Postdoctoral Pioneer Program at the University of Michigan Medical School.
Author information
Authors and Affiliations
Contributions
JMB conceptualized and designed the study, performed experiments, wrote MATLAB code, acquired funding, analyzed data, performed bioinformatics analyses, and wrote the paper. SR performed experiments, analyzed data, and wrote the paper. BAH provided reagents, performed experiments, acquired funding, and analyzed data. ACC, NGC, AMB, and ASB performed experiments. AJM performed experiments and acquired funding. TC provided reagents. YMS, MSW, and RR provided reagents and acquired funding. ZCN, DS, and PG acquired funding and performed bioinformatics analyses. CAL provided reagents, acquired funding, and performed bioinformatics analyses. GDL conceptualized and designed the study, acquired funding, and wrote the paper. All authors reviewed the paper before submission.
Corresponding author
Ethics declarations
Competing interests
CAL has received consulting fees from Astellas Pharmaceuticals, Odyssey Therapeutics, and T-Knife Therapuetics, and is an inventor on patents pertaining to Kras regulated metabolic pathways, redox control pathways in pancreatic cancer, and targeting the GOT1-pathway as a therapeutic approach (US Patent No: 2015126580-A1, 05/07/2015; US Patent No: 20190136238, 05/09/2019; International Patent No: WO2013177426-A2, 04/23/2015). GDL has received research funding from InterAx Biotech AG and Polyphor (now part of Spexis). The remaining authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Buschhaus, J.M., Rajendran, S., Humphries, B.A. et al. Effects of iron modulation on mesenchymal stem cell-induced drug resistance in estrogen receptor-positive breast cancer. Oncogene 41, 3705–3718 (2022). https://doi.org/10.1038/s41388-022-02385-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-022-02385-9
This article is cited by
-
Characterizing heterogeneous single-cell dose responses computationally and experimentally using threshold inhibition surfaces and dose-titration assays
npj Systems Biology and Applications (2024)
-
Bone marrow mesenchymal stem cells combined with estrogen synergistically promote endometrial regeneration and reverse EMT via Wnt/β-catenin signaling pathway
Reproductive Biology and Endocrinology (2022)