Abstract
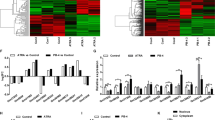
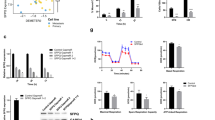
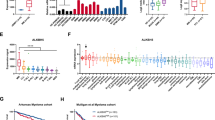
N6-methyladenosine (m6A) is the most universal internal RNA modification on messenger RNAs and regulates the fate and functions of m6A-modified transcripts through m6A-specific binding proteins. Nevertheless, the functional role and potential mechanism of the m6A reading proteins in ocular melanoma tumorigenicity, especially cancer stem-like cell (CSC) properties, remain to be elucidated. Herein, we demonstrated that the m6A reading protein YTHDF3 promotes the translation of the target transcript CTNNB1, contributing to ocular melanoma propagation and migration through m6A methylation. YTHDF3 is highly expressed in ocular melanoma stem-like cells and abundantly enriched in ocular melanoma tissues, which is related to poor clinical prognosis. Moreover, YTHDF3 is required for the maintenance of CSC properties and tumor initiation capacity in ocular melanoma both in vitro and in vivo. Ocular melanoma cells with targeted YTHDF3 knockdown exhibited inhibitory tumor proliferation and migration abilities. Transcriptome-wide mapping of m6A peaks and YTHDF3 binding peaks on mRNAs revealed a key target gene candidate, CTNNB1. Mechanistically, YTHDF3 enhances CTNNB1 translation through recognizing and binding the m6A peaks on CTNNB1 mRNA.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The raw sequencing data supporting the conclusions of this paper, including RNA-seq, miCLIP-seq, and iCLIP-seq data, have been deposited in the Genome Sequence Archive [65] in the BIG Data Center [66], Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under accession number CRA002956 (http://bigd.big.ac.cn/gsa/s/n110138p) and are publicly accessible at http://bigd.big.ac.cn/gsa.
References
Shi H, Zhang X, Weng YL, Lu Z, Liu Y, Lu Z, et al. m(6)A facilitates hippocampus-dependent learning and memory through YTHDF1. Nature. 2018;563:249–53.
Yang Y, Hsu PJ, Chen YS, Yang YG. Dynamic transcriptomic m(6)A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018;28:616–24.
Vu LP, Cheng Y, Kharas MG. The Biology of m(6)A RNA Methylation in Normal and Malignant Hematopoiesis. Cancer Discov. 2019;9:25–33.
Hsu PJ, Shi H, He C. Epitranscriptomic influences on development and disease. Genome Biol. 2017;18:197.
Yue Y, Liu J, He C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 2015;29:1343–55.
Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods. 2015;12:767–72.
Meyer KD. DART-seq: an antibody-free method for global m(6)A detection. Nat Methods. 2019;16:1275–80.
Jia R, Chai P, Wang S, Sun B, Xu Y, Yang Y, et al. m(6)A modification suppresses ocular melanoma through modulating HINT2 mRNA translation. Mol Cancer. 2019;18:161.
Yang S, Wei J, Cui YH, Park G, Shah P, Deng Y, et al. m(6)A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat Commun. 2019;10:2782.
Paris J, Morgan M, Campos J, Spencer GJ, Shmakova A, Ivanova I, et al. Targeting the RNA m(6)A Reader YTHDF2 Selectively Compromises Cancer Stem Cells in Acute Myeloid Leukemia. Cell Stem Cell. 2019;25:137–48. e136
Vu LP, Pickering BF, Cheng Y, Zaccara S, Nguyen D, Minuesa G, et al. The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med. 2017;23:1369–76.
Weng H, Huang H, Wu H, Qin X, Zhao BS, Dong L, et al. METTL14 Inhibits Hematopoietic Stem/Progenitor Differentiation and Promotes Leukemogenesis via mRNA m(6)A Modification. Cell Stem Cell. 2018;22:191–205. e199
Li Z, Qian P, Shao W, Shi H, He XC, Gogol M, et al. Suppression of m(6)A reader Ythdf2 promotes hematopoietic stem cell expansion. Cell Res. 2018;28:904–17.
Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, et al. m(6)A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell. 2017;31:591–606. e596
Yang J, Manson DK, Marr BP, Carvajal RD. Treatment of uveal melanoma: where are we now? Therapeutic Adv Med Oncol. 2018;10:1758834018757175.
Missotten GS, Keijser S, De Keizer RJ, De, Wolff-Rouendaal D. Conjunctival melanoma in the Netherlands: a nationwide study. Investig Ophthalmol Vis Sci. 2005;46:75–82.
Shields CL, Shields JA, Gunduz K, Cater J, Mercado GV, Gross N et al. Conjunctival melanoma: risk factors for recurrence, exenteration, metastasis, and death in 150 consecutive patients. Arch Ophthalmol. (Chicago, Ill: 1960) 2000;118:1497–507.
Tuomaala S, Eskelin S, Tarkkanen A, Kivela T. Population-based assessment of clinical characteristics predicting outcome of conjunctival melanoma in whites. Investig Ophthalmol Vis Sci. 2002;43:3399–408.
Smit KN, Jager MJ, de Klein A, Kili E. Uveal melanoma: towards a molecular understanding. Prog Retin Eye Res. 2019;75:100800
Feng X, Degese MS, Iglesias-Bartolome R, Vaque JP, Molinolo AA, Rodrigues M, et al. Hippo-independent activation of YAP by the GNAQ uveal melanoma oncogene through a trio-regulated rho GTPase signaling circuitry. Cancer Cell. 2014;25:831–45.
Yu FX, Luo J, Mo JS, Liu G, Kim YC, Meng Z, et al. Mutant Gq/11 promote uveal melanoma tumorigenesis by activating YAP. Cancer Cell. 2014;25:822–30.
Moore AR, Ran L, Guan Y, Sher JJ, Hitchman TD, Zhang JQ, et al. GNA11 Q209L Mouse Model Reveals RasGRP3 as an Essential Signaling Node in Uveal Melanoma. Cell Rep. 2018;22:2455–68.
Larsen AC. Conjunctival malignant melanoma in Denmark: epidemiology, treatment and prognosis with special emphasis on tumorigenesis and genetic profile. Acta Ophthalmol. 2016;1:1–27. 94 Thesis
Li P, He J, Yang Z, Ge S, Zhang H, Zhong Q, et al. ZNNT1 long noncoding RNA induces autophagy to inhibit tumorigenesis of uveal melanoma by regulating key autophagy gene expression. Autophagy. 2019;16:1186–99.
Shang Q, Li Y, Wang H, Ge S, Jia R. Altered expression profile of circular RNAs in conjunctival melanoma. Epigenomics. 2019;11:787–804.
Clarke MF, Dick JE, Dirks PB, Eaves CJ, Jamieson CH, Jones DL, et al. Cancer stem cells-perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 2006;66:9339–44.
Braun KM. Cutaneous cancer stem cells: beta-catenin strikes again. Cell Stem Cell. 2008;2:406–8.
Angeloni V, Tiberio P, Appierto V, Daidone MG. Implications of stemness-related signaling pathways in breast cancer response to therapy. Semin Cancer Biol. 2015;31:43–51.
Murphy GF, Wilson BJ, Girouard SD, Frank NY, Frank MH. Stem cells and targeted approaches to melanoma cure. Mol Asp Med. 2014;39:33–49.
de Waard NE, Kolovou PE, McGuire SP, Cao J, Frank NY, Frank MH, et al. Expression of Multidrug Resistance Transporter ABCB5 in a Murine Model of Human Conjunctival Melanoma. Ocul Oncol Pathol. 2015;1:182–9.
Zhang J, Liu S, Ye Q, Pan J. Transcriptional inhibition by CDK7/9 inhibitor SNS-032 abrogates oncogene addiction and reduces liver metastasis in uveal melanoma. Mol Cancer. 2019;18:140.
Lai CY, Schwartz BE, Hsu MY. CD133+ melanoma subpopulations contribute to perivascular niche morphogenesis and tumorigenicity through vasculogenic mimicry. Cancer Res. 2012;72:5111–8.
Batlle E, Clevers H. Cancer stem cells revisited. Nat Med. 2017;23:1124–34.
Giraud J, Failla LM, Pascussi JM, Lagerqvist EL, Ollier J, Finetti P, et al. Autocrine Secretion of Progastrin Promotes the Survival and Self-Renewal of Colon Cancer Stem-like Cells. Cancer Res. 2016;76:3618–28.
Li A, Chen YS, Ping XL, Yang X, Xiao W, Yang Y, et al. Cytoplasmic m(6)A reader YTHDF3 promotes mRNA translation. Cell Res. 2017;27:444–7.
Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, et al. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 2017;27:315–28.
Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017;27:626–41.
Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature. 2015;523:231–5.
Rappa G, Mercapide J, Anzanello F, Le TT, Johlfs MG, Fiscus RR, et al. Wnt interaction and extracellular release of prominin-1/CD133 in human malignant melanoma cells. Exp Cell Res. 2013;319:810–9.
Newell F, Kong Y, Wilmott JS, Johansson PA, Ferguson PM, Cui C, et al. Whole-genome landscape of mucosal melanoma reveals diverse drivers and therapeutic targets. Nat Commun. 2019;10:3163.
Jin B, Zhang P, Zou H, Ye H, Wang Y, Zhang J, et al. Verification of EZH2 as a druggable target in metastatic uveal melanoma. Mol Cancer. 2020;19:52.
Mackiewicz J, Burzykowski T, Izycki D, Mackiewicz A. Re-induction using whole cell melanoma vaccine genetically modified to melanoma stem cells-like beyond recurrence extends long term survival of high risk resected patients - updated results. J Immunother Cancer. 2018;6:134.
Zhou J, Jin B, Jin Y, Liu Y, Pan J. The antihelminthic drug niclosamide effectively inhibits the malignant phenotypes of uveal melanoma in vitro and in vivo. Theranostics. 2017;7:1447–62.
Kalirai H, Damato BE, Coupland SE. Uveal melanoma cell lines contain stem-like cells that self-renew, produce differentiated progeny, and survive chemotherapy. Investig Ophthalmol Vis Sci. 2011;52:8458–66.
Shidal C, Singh NP, Nagarkatti P, Nagarkatti M. MicroRNA-92 Expression in CD133(+) Melanoma Stem Cells Regulates Immunosuppression in the Tumor Microenvironment via Integrin-Dependent Activation of TGFbeta. Cancer Res. 2019;79:3622–35.
Terai K, Bi D, Liu Z, Kimura K, Sanaat Z, Dolatkhah R, et al. A Novel Oncolytic Herpes Capable of Cell-Specific Transcriptional Targeting of CD133+/- Cancer Cells Induces Significant Tumor Regression. Stem Cells (Dayt, Ohio). 2018;36:1154–69.
Liu L, Liu X, Dong Z, Li J, Yu Y, Chen X, et al. N6-methyladenosine-related Genomic Targets are Altered in Breast Cancer Tissue and Associated with Poor Survival. J Cancer. 2019;10:5447–59.
Ni W, Yao S, Zhou Y, Liu Y, Huang P, Zhou A, et al. Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m(6)A reader YTHDF3. Mol Cancer. 2019;18:143.
Zhang Y, Wang X, Zhang X, Wang J, Ma Y, Zhang L, et al. RNA-binding protein YTHDF3 suppresses interferon-dependent antiviral responses by promoting FOXO3 translation. Proc Natl Acad Sci USA. 2019;116:976–81.
Chang G, Shi L, Ye Y, Shi H, Zeng L, Tiwary S, et al. YTHDF3 Induces the Translation of m(6)A-Enriched Gene Transcripts to Promote Breast Cancer Brain Metastasis. Cancer Cell. 2020;38:857–71.
Gao Y, Pei G, Li D, Li R, Shao Y, Zhang QC, et al. Multivalent m(6)A motifs promote phase separation of YTHDF proteins. Cell Res. 2019;29:767–9.
Katoh M. Canonical and non-canonical WNT signaling in cancer stem cells and their niches: Cellular heterogeneity, omics reprogramming, targeted therapy and tumor plasticity (Review). Int J Oncol. 2017;51:1357–69.
Wang Y, He L, Du Y, Zhu P, Huang G, Luo J, et al. The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell. 2015;16:413–25.
Sun Q, Lee W, Mohri Y, Takeo M, Lim CH, Xu X, et al. A novel mouse model demonstrates that oncogenic melanocyte stem cells engender melanoma resembling human disease. Nat Commun. 2019;10:5023.
Liu L, Wang J, Sun G, Wu Q, Ma J, Zhang X, et al. m(6)A mRNA methylation regulates CTNNB1 to promote the proliferation of hepatoblastoma. Mol Cancer. 2019;18:188.
Han B, Yan S, Wei S, Xiang J, Liu K, Chen Z, et al. YTHDF1-mediated translation amplifies Wnt-driven intestinal stemness. EMBO Rep. 2020;21:e49229.
Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347:70–8.
Moore MJ, Zhang C, Gantman EC, Mele A, Darnell JC, Darnell RB. Mapping Argonaute and conventional RNA-binding protein interactions with RNA at single-nucleotide resolution using HITS-CLIP and CIMS analysis. Nat Protoc. 2014;9:263–93.
Shah A, Qian Y, Weyn-Vanhentenryck SM, Zhang C. CLIP Tool Kit (CTK): a flexible and robust pipeline to analyze CLIP sequencing data. Bioinforma (Oxf, Engl). 2017;33:566–7.
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinforma (Oxf, Engl). 2014;30:2114–20.
Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinforma (Oxf, Engl). 2010;26:589–95.
Weyn-Vanhentenryck SM, Mele A, Yan Q, Sun S, Farny N, Zhang Z, et al. HITS-CLIP and integrative modeling define the Rbfox splicing-regulatory network linked to brain development and autism. Cell Rep. 2014;6:1139–52.
Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinforma (Oxf, Engl). 2010;26:841–2.
Zhao L, Guo Z, Wang P, Zheng M, Yang X, Liu Y, et al. Proteomics of epicardial adipose tissue in patients with heart failure. J Cell Mol Med. 2020;24:511–20.
Zhang SS, Chen TT, Zhu JW, Zhou Q, Chen X, Wang YQ, et al. [GSA: Genome Sequence Archive]. Yi Chuan. 2018;40:1044–7.
Members BDC. The BIG Data Center: from deposition to integration to translation. Nucleic Acids Res. 2017;45:D18–D24.
Acknowledgements
We express our sincere thanks for all the patients participated in our study and wish them good health. This work was supported by the National Key Research and Development Plan (2017YFE0196300), the National Natural Science Foundation of China (81772875, U1932135, 82073889), the Clinical Research Plan of SHDC (SHDC2020CR1009A), the Science and Technology Commission of Shanghai (20DZ2270800 and 19JC1410200).
Author information
Authors and Affiliations
Contributions
XQF and YGY designed the research, supervised the experiments and approved the paper. YFX and XYH performed the experiments with assistance from RBJ, PWC, FL, and YY; SZW and BFS performed the bioinformatics analyses and contributed to the experimental candidate selection; YFX, XYH, and SZW analyzed the data, and drafted the paper. SFG and RBJ was responsible for sample collection. All of the authors read and approved this paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
Written informed consent was obtained from all patients enrolled in this study. The animal research was approved by the Shanghai Jiao Tong University School of Medicine Animal Care and Use Committee.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Xu, Y., He, X., Wang, S. et al. The m6A reading protein YTHDF3 potentiates tumorigenicity of cancer stem-like cells in ocular melanoma through facilitating CTNNB1 translation. Oncogene 41, 1281–1297 (2022). https://doi.org/10.1038/s41388-021-02146-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-021-02146-0
This article is cited by
-
RBM3 suppresses stemness remodeling of prostate cancer in bone microenvironment by modulating N6-methyladenosine on CTNNB1 mRNA
Cell Death & Disease (2023)
-
METTL14-dependent maturation of pri-miR-17 regulates mitochondrial homeostasis and induces chemoresistance in colorectal cancer
Cell Death & Disease (2023)
-
N6-methyladenosine reader YTHDF3 contributes to the aerobic glycolysis of osteosarcoma through stabilizing PGK1 stability
Journal of Cancer Research and Clinical Oncology (2023)
-
A novel MYCN-YTHDF1 cascade contributes to retinoblastoma tumor growth by eliciting m6A -dependent activation of multiple oncogenes
Science China Life Sciences (2023)
-
Crosstalk among m6A RNA methylation, hypoxia and metabolic reprogramming in TME: from immunosuppressive microenvironment to clinical application
Journal of Hematology & Oncology (2022)