Abstract
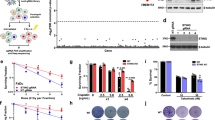
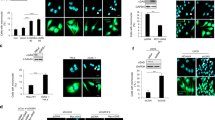
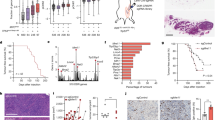
STING (Stimulator of Interferon Genes) is an endoplasmic reticulum-anchored adaptor of the innate immunity best known to trigger pro-inflammatory cytokine expression in response to pathogen infection. In cancer, this canonical pathway can be activated by intrinsic or drug-induced genomic instability, potentiating antitumor immune responses. Here we report that STING downregulation decreases cell survival and increases sensitivity to genotoxic treatment in a panel of breast cancer cell lines in a cell-autonomous manner. STING silencing impaired DNA Damage Response (53BP1) foci formation and increased DNA break accumulation. These newly identified properties were found to be independent of STING partner cGAS and of its canonical pro-inflammatory pathway. STING was shown to partially localize at the inner nuclear membrane in a variety of breast cancer cell models and clinical tumor samples. Interactomics analysis of nuclear STING identified several proteins of the DNA Damage Response, including the three proteins of the DNA-PK complex, further supporting a role of STING in the regulation of genomic stability. In breast and ovarian cancer patients that received adjuvant chemotherapy, high STING expression is associated with increased risk of relapse. In summary, this study highlights an alternative, non-canonical tumor-promoting role of STING that opposes its well-documented function in tumor immunosurveillance.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–92.
Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–8.
Liu S, Cai X, Wu J, Cong Q, Chen X, Li T, et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 2015;347:aaa2630.
Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Rohl I, et al. cGAS produces a 2’-5’-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498:380–4.
Abe T, Barber GN. Cytosolic-DNA-mediated, STING-dependent proinflammatory gene induction necessitates canonical NF-kappaB activation through TBK1. J Virol. 2014;88:5328–41.
Ranoa DRE, Widau RC, Mallon S, Parekh AD, Nicolae CM, Huang X, et al. STING promotes homeostasis via regulation of cell proliferation and chromosomal stability. Cancer Res. 2019;79:1465–79.
Bakhoum SF, Ngo B, Laughney AM, Cavallo JA, Murphy CJ, Ly P, et al. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature. 2018;553:467–72.
Dunphy G, Flannery SM, Almine JF, Connolly DJ, Paulus C, Jonsson KL, et al. Non-canonical activation of the DNA sensing adaptor STING by ATM and IFI16 mediates NF-kappaB signaling after nuclear DNA damage. Mol Cell. 2018;71:745–60 e5.
Barber GN. STING-dependent cytosolic DNA sensing pathways. Trends Immunol. 2014;35:88–93.
Deng L, Liang H, Xu M, Yang X, Burnette B, Arina A, et al. STING-dependent cytosolic DNA sensing promotes radiation-induced type I interferon-dependent antitumor immunity in immunogenic tumors. Immunity. 2014;41:843–52.
Vanpouille-Box C, Alard A, Aryankalayil MJ, Sarfraz Y, Diamond JM, Schneider RJ, et al. DNA exonuclease Trex1 regulates radiotherapy-induced tumour immunogenicity. Nat Commun. 2017;8:15618.
Wang H, Hu S, Chen X, Shi H, Chen C, Sun L, et al. cGAS is essential for the antitumor effect of immune checkpoint blockade. Proc Natl Acad Sci USA. 2017;114:1637–42.
Harding SM, Benci JL, Irianto J, Discher DE, Minn AJ, Greenberg RA. Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature. 2017;548:466–70.
Brown JS, Sundar R, Lopez J. Combining DNA damaging therapeutics with immunotherapy: more haste, less speed. Br J Cancer. 2018;118:312–24.
Ahn J, Xia T, Konno H, Konno K, Ruiz P, Barber GN. Inflammation-driven carcinogenesis is mediated through STING. Nat Commun. 2014;5:5166.
Wu Z, Oeck S, West AP, Mangalhara KC, Sainz AG, Newman LE, et al. Mitochondrial DNA stress signalling protects the nuclear genome. Nat Metab. 2019;1:1209–18.
Chen Q, Boire A, Jin X, Valiente M, Er EE, Lopez-Soto A, et al. Carcinoma-astrocyte gap junctions promote brain metastasis by cGAMP transfer. Nature. 2016;533:493–8.
Gaston J, Cheradame L, Yvonnet V, Deas O, Poupon MF, Judde JG, et al. Intracellular STING inactivation sensitizes breast cancer cells to genotoxic agents. Oncotarget. 2016;7:77205–24.
Li T, Chen ZJ. The cGAS-cGAMP-STING pathway connects DNA damage to inflammation, senescence, and cancer. J Exp Med. 2018;215:1287–99.
Erdal E, Haider S, Rehwinkel J, Harris AL, McHugh PJ. A prosurvival DNA damage-induced cytoplasmic interferon response is mediated by end resection factors and is limited by Trex1. Genes Dev. 2017;31:353–69.
Parkes EE, Walker SM, Taggart LE, McCabe N, Knight LA, Wilkinson R, et al. Activation of STING-dependent innate immune signaling by S-phase-specific DNA damage in breast cancer. J Natl Cancer Inst. 2017;109:djw199.
Mackenzie KJ, Carroll P, Martin CA, Murina O, Fluteau A, Simpson DJ, et al. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 2017;548:461–5.
Lan YY, Londono D, Bouley R, Rooney MS, Hacohen N. Dnase2a deficiency uncovers lysosomal clearance of damaged nuclear DNA via autophagy. Cell Rep. 2014;9:180–92.
Hartlova A, Erttmann SF, Raffi FA, Schmalz AM, Resch U, Anugula S, et al. DNA damage primes the type I interferon system via the cytosolic DNA sensor STING to promote anti-microbial innate immunity. Immunity. 2015;42:332–43.
Mazur L, Opydo-Chanek M, Stojak M, Wojcieszek K. Mafosfamide as a new anticancer agent: preclinical investigations and clinical trials. Anticancer Res. 2012;32:2783–9.
Basit A, Cho MG, Kim EY, Kwon D, Kang SJ, Lee JH. The cGAS/STING/TBK1/IRF3 innate immunity pathway maintains chromosomal stability through regulation of p21 levels. Exp Mol Med. 2020;52:643–57.
Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C. Clonogenic assay of cells in vitro. Nat Protoc. 2006;1:2315–9.
Gaston J, Cheradame L, Yvonnet V, Deas O, Poupon MF, Judde JG, et al. Correction: intracellular STING inactivation sensitizes breast cancer cells to genotoxic agents. Oncotarget. 2019;10:4249–51.
Schultz LB, Chehab NH, Malikzay A, Halazonetis TD. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J Cell Biol. 2000;151:1381–90.
Lukas C, Savic V, Bekker-Jensen S, Doil C, Neumann B, Pedersen RS, et al. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat Cell Biol. 2011;13:243–53.
Chen H, Pei R, Zhu W, Zeng R, Wang Y, Wang Y, et al. An alternative splicing isoform of MITA antagonizes MITA-mediated induction of type I IFNs. J Immunol. 2014;192:1162–70.
Liu H, Zhang H, Wu X, Ma D, Wu J, Wang L, et al. Nuclear cGAS suppresses DNA repair and promotes tumorigenesis. Nature. 2018;563:131–6.
Burleigh K, Maltbaek JH, Cambier S, Green R, Gale M, Jr, James RC, et al. Human DNA-PK activates a STING-independent DNA sensing pathway. Sci Immunol. 2020;5:eaba4219.
Ferguson BJ, Mansur DS, Peters NE, Ren H, Smith GL. DNA-PK is a DNA sensor for IRF-3-dependent innate immunity. Elife 2012;1:e00047.
Kondo T, Kobayashi J, Saitoh T, Maruyama K, Ishii KJ, Barber GN, et al. DNA damage sensor MRE11 recognizes cytosolic double-stranded DNA and induces type I interferon by regulating STING trafficking. Proc Natl Acad Sci USA. 2013;110:2969–74.
Sui H, Zhou M, Imamichi H, Jiao X, Sherman BT, Lane HC, et al. STING is an essential mediator of the Ku70-mediated production of IFN-lambda1 in response to exogenous DNA. Sci Signal. 2017;10:eaah5054.
Xia T, Konno H, Ahn J, Barber GN. Deregulation of STING signaling in colorectal carcinoma constrains DNA damage responses and correlates with tumorigenesis. Cell Rep. 2016;14:282–97.
Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci USA. 2009;106:20842–6.
Dobrzynska A, Gonzalo S, Shanahan C, Askjaer P. The nuclear lamina in health and disease. Nucleus 2016;7:233–48.
Britton S, Coates J, Jackson SP. A new method for high-resolution imaging of Ku foci to decipher mechanisms of DNA double-strand break repair. J Cell Biol. 2013;202:579–95.
Sawasdichai A, Chen HT, Abdul Hamid N, Jayaraman PS, Gaston K In situ subcellular fractionation of adherent and non-adherent mammalian cells. J Vis Exp. 2010;41:1958.
Malik P, Korfali N, Srsen V, Lazou V, Batrakou DG, Zuleger N, et al. Cell-specific and lamin-dependent targeting of novel transmembrane proteins in the nuclear envelope. Cell Mol Life Sci. 2010;67:1353–69.
Shang G, Zhang C, Chen ZJ, Bai XC, Zhang X Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP. Nature. 2019;389–93.
Gonugunta VK, Sakai T, Pokatayev V, Yang K, Wu J, Dobbs N, et al. Trafficking-mediated STING degradation requires sorting to acidified endolysosomes and can be targeted to enhance anti-tumor response. Cell Rep. 2017;21:3234–42.
Chen H, Sun H, You F, Sun W, Zhou X, Chen L, et al. Activation of STAT6 by STING is critical for antiviral innate immunity. Cell. 2011;147:436–46.
Smeenk G, Wiegant WW, Marteijn JA, Luijsterburg MS, Sroczynski N, Costelloe T, et al. Poly(ADP-ribosyl)ation links the chromatin remodeler SMARCA5/SNF2H to RNF168-dependent DNA damage signaling. J Cell Sci. 2013;126:889–903. Pt 4
Lans H, Marteijn JA, Vermeulen W. ATP-dependent chromatin remodeling in the DNA-damage response. Epigenetics Chromatin. 2012;5:4.
Oliveira DV, Kato A, Nakamura K, Ikura T, Okada M, Kobayashi J, et al. Histone chaperone FACT regulates homologous recombination by chromatin remodeling through interaction with RNF20. J Cell Sci. 2014;127:763–72. Pt 4
Clarke TL, Sanchez-Bailon MP, Chiang K, Reynolds JJ, Herrero-Ruiz J, Bandeiras TM, et al. PRMT5-dependent methylation of the TIP60 coactivator RUVBL1 is a key regulator of homologous recombination. Mol Cell. 2017;65:900–16 e7.
Potts PR, Porteus MH, Yu H. Human SMC5/6 complex promotes sister chromatid homologous recombination by recruiting the SMC1/3 cohesin complex to double-strand breaks. EMBO J. 2006;25:3377–88.
Shin YJ, Kim MS, Kim MS, Lee J, Kang M, Jeong JH. High-mobility group box 2 (HMGB2) modulates radioresponse and is downregulated by p53 in colorectal cancer cell. Cancer Biol Ther. 2013;14:213–21.
Blackford AN, Jackson SP. ATM, ATR, and DNA-PK: The Trinity at the Heart of the DNA Damage Response. Mol Cell. 2017;66:801–17.
Ochi T, Blackford AN, Coates J, Jhujh S, Mehmood S, Tamura N, et al. DNA repair. PAXX, a paralog of XRCC4 and XLF, interacts with Ku to promote DNA double-strand break repair. Science. 2015;347:185–8.
Xia T, Konno H, Barber GN. Recurrent loss of STING signaling in melanoma correlates with susceptibility to viral oncolysis. Cancer Res. 2016;76:6747–59.
Konno H, Yamauchi S, Berglund A, Putney RM, Mule JJ, Barber GN. Suppression of STING signaling through epigenetic silencing and missense mutation impedes DNA damage mediated cytokine production. Oncogene. 2018;37:2037–51.
Liang D, Xiao-Feng H, Guan-Jun D, Er-Ling H, Sheng C, Ting-Ting W, et al. Activated STING enhances Tregs infiltration in the HPV-related carcinogenesis of tongue squamous cells via the c-jun/CCL22 signal. Biochim Biophys Acta. 2015;1852:2494–503.
Baird JR, Friedman D, Cottam B, Dubensky TW Jr, Kanne DB, Bambina S, et al. Radiotherapy combined with novel STING-targeting oligonucleotides results in regression of established tumors. Cancer Res. 2016;76:50–61.
Bu Y, Liu F, Jia QA, Yu SN. Decreased expression of TMEM173 predicts poor prognosis in patients with hepatocellular carcinoma. PLoS One. 2016;11:e0165681.
Song S, Peng P, Tang Z, Zhao J, Wu W, Li H, et al. Decreased expression of STING predicts poor prognosis in patients with gastric cancer. Sci Rep. 2017;7:39858.
Bakhoum SF, Cantley LC. The multifaceted role of chromosomal instability in cancer and its microenvironment. Cell. 2018;174:1347–60.
Litvin O, Schwartz S, Wan Z, Schild T, Rocco M, Oh NL, et al. Interferon alpha/beta enhances the cytotoxic response of MEK inhibition in melanoma. Mol Cell. 2015;57:784–96.
Linsley PS, Speake C, Whalen E, Chaussabel D. Copy number loss of the interferon gene cluster in melanomas is linked to reduced T cell infiltrate and poor patient prognosis. PLoS One. 2014;9:e109760.
Chen Y, Wang L, Jin J, Luan Y, Chen C, Li Y, et al. p38 inhibition provides anti-DNA virus immunity by regulation of USP21 phosphorylation and STING activation. J Exp Med. 2017;214:991–1010.
Jiang H, Xue X, Panda S, Kawale A, Hooy RM, Liang F, et al. Chromatin-bound cGAS is an inhibitor of DNA repair and hence accelerates genome destabilization and cell death. EMBO J. 2019;38:e102718.
Chen H, Chen H, Zhang J, Wang Y, Simoneau A, Yang H, et al. cGAS suppresses genomic instability as a decelerator of replication forks. Sci Adv. 2020;6:eabb8941.
Goodwin JF, Knudsen KE. Beyond DNA repair: DNA-PK function in cancer. Cancer Discov. 2014;4:1126–39.
Mohiuddin IS, Kang MH. DNA-PK as an emerging therapeutic target in cancer. Front Oncol. 2019;9:635.
Neal JA, Meek K. Choosing the right path: does DNA-PK help make the decision? Mutat Res. 2011;711:73–86.
Schirmer EC, Florens L, Guan T, Yates JR 3rd, Gerace L. Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science. 2003;301:1380–2.
Malik P, Zuleger N, las Heras JI, Saiz-Ros N, Makarov AA, Lazou V, et al. NET23/STING promotes chromatin compaction from the nuclear envelope. PLoS ONE. 2014;9:e111851.
Katta SS, Smoyer CJ, Jaspersen SL. Destination: inner nuclear membrane. Trends Cell Biol. 2014;24:221–9.
Sullivan T, Escalante-Alcalde D, Bhatt H, Anver M, Bhat N, Nagashima K, et al. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J Cell Biol. 1999;147:913–20.
Marnef A, Legube G. Organizing DNA repair in the nucleus: DSBs hit the road. Curr Opin Cell Biol. 2017;46:1–8.
Oza P, Jaspersen SL, Miele A, Dekker J, Peterson CL. Mechanisms that regulate localization of a DNA double-strand break to the nuclear periphery. Genes Dev. 2009;23:912–27.
Ryu T, Spatola B, Delabaere L, Bowlin K, Hopp H, Kunitake R, et al. Heterochromatic breaks move to the nuclear periphery to continue recombinational repair. Nat Cell Biol. 2015;17:1401–11.
Lemaitre C, Grabarz A, Tsouroula K, Andronov L, Furst A, Pankotai T, et al. Nuclear position dictates DNA repair pathway choice. Genes Dev. 2014;28:2450–63.
Chung DK, Chan JN, Strecker J, Zhang W, Ebrahimi-Ardebili S, Lu T, et al. Perinuclear tethers license telomeric DSBs for a broad kinesin- and NPC-dependent DNA repair process. Nat Commun. 2015;6:7742.
Legrier ME, Bieche I, Gaston J, Beurdeley A, Yvonnet V, Deas O, et al. Activation of IFN/STAT1 signalling predicts response to chemotherapy in oestrogen receptor-negative breast cancer. Br J Cancer. 2016;114:177–87.
Marangoni E, Vincent-Salomon A, Auger N, Degeorges A, Assayag F, de Cremoux P, et al. A new model of patient tumor-derived breast cancer xenografts for preclinical assays. Clin Cancer Res. 2007;13:3989–98.
Baghirova S, Hughes BG, Hendzel MJ, Schulz R. Sequential fractionation and isolation of subcellular proteins from tissue or cultured cells. MethodsX. 2015;2:440–5.
Bolte S, Cordelieres FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224:213–32. Pt 3
Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–82.
Schmidt U, Weigert M, Broaddus C, Myers G, editors. Cell Detection with Star-convex Polygons. In Medical Image Computing and Computer Assisted Intervention – MICCAI 2018 (eds. Frangi, A. F. et al.), Springer International Publishing, 2018;11071:265–73.
Guzman C, Bagga M, Kaur A, Westermarck J, Abankwa D. ColonyArea: an ImageJ plugin to automatically quantify colony formation in clonogenic assays. PLoS One. 2014;9:e92444.
Nagy A, Lanczky A, Menyhart O, Gyorffy B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep. 2018;8:9227.
Acknowledgements
This study was funded by XenTech, annual funding from Inserm and the University Paris Descartes, and ECLER non-profit association. JG and LC were recipient of a CIFRE fellowship from the Association Nationale de la Recherche et de la Technologie (ANRT). MM is supported by the Ligue Nationale contre le Cancer. We are grateful to Patrice Codogno, Etienne Morel, Ganna Panasyuk, Thierry Dubois and Frédéric Rieux-Laucat for helpful discussions, to Dr Orgunc and Pr S. Fillatreau for providing biological materials, and to Katheryn Meek for critical reading of the manuscript.
Author information
Authors and Affiliations
Contributions
Conception and design: LC, JG, SC, VG. Development of methodology: LC, ICG, NG, AS, SC, VG. Acquisition of data: LC, ICG, AS, VJ, MP. Analysis and interpretation of data: LC, ICG, NG, AS, NR-R, MM, SC, VG. Writing, review, and/or revision of the manuscript: LC, ICG, AS, MM, J-GJ, SC, VG. Administrative, technical, or material support: LC, ICG, AS, VJ, NR-R, J-GJ, SC, VG. Study supervision: SC, VG.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Cheradame, L., Guerrera, I.C., Gaston, J. et al. STING protects breast cancer cells from intrinsic and genotoxic-induced DNA instability via a non-canonical, cell-autonomous pathway. Oncogene 40, 6627–6640 (2021). https://doi.org/10.1038/s41388-021-02037-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-021-02037-4
This article is cited by
-
The tumor cell-intrinsic cGAS–STING pathway is associated with the high density of CD8+ T cells after chemotherapy in esophageal squamous cell carcinoma
Esophagus (2024)
-
Ligands stimulating antitumour immunity as the next G-quadruplex challenge
Molecular Cancer (2022)
-
Nuclear soluble cGAS senses double-stranded DNA virus infection
Communications Biology (2022)
-
Topoisomerase I poison-triggered immune gene activation is markedly reduced in human small-cell lung cancers by impairment of the cGAS/STING pathway
British Journal of Cancer (2022)