Abstract
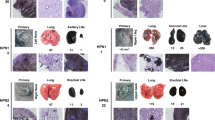
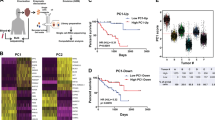
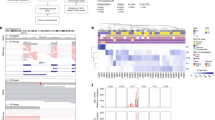
Hepatocyte growth factor-overexpressing mice that harbor a deletion of the Ink4a/p16 locus (HP mice) form melanomas with low metastatic potential in response to UV irradiation. Here we report that these tumors become highly metastatic following hemizygous deletion of the Nme1 and Nme2 metastasis suppressor genes (HPN mice). Whole-genome sequencing of melanomas from HPN mice revealed a striking increase in lung metastatic activity that is associated with missense mutations in eight signature genes (Arhgap35, Atp8b4, Brca1, Ift172, Kif21b, Nckap5, Pcdha2, and Zfp869). RNA-seq analysis of transcriptomes from HP and HPN primary melanomas identified a 32-gene signature (HPN lung metastasis signature) for which decreased expression is strongly associated with lung metastatic potential. Analysis of transcriptome data from The Cancer Genome Atlas revealed expression profiles of these genes that predict improved survival of patients with cutaneous or uveal melanoma. Silencing of three representative HPN lung metastasis signature genes (ARRDC3, NYNRIN, RND3) in human melanoma cells resulted in increased invasive activity, consistent with roles for these genes as mediators of the metastasis suppressor function of NME1 and NME2. In conclusion, our studies have identified a family of genes that mediate suppression of melanoma lung metastasis, and which may serve as prognostic markers and/or therapeutic targets for clinical management of metastatic melanoma.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Change history
11 October 2021
A Correction to this paper has been published: https://doi.org/10.1038/s41388-021-02045-4
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34.
Smith SC, Theodorescu D. Learning therapeutic lessons from metastasis suppressor proteins. Nat Rev Cancer. 2009;9:253–64.
Steeg PS, Bevilacqua G, Kopper L, Thorgerisson UR, Talmadge JE, Liotta LA, et al. Evidence for a novel gene associated with low tumor metastatic potential. J Natl Cancer Inst. 1988;80:200–5.
Zhang Q, McCorkle JR, Novak M, Yang M, Kaetzel DM. Metastasis suppressor function of NM23-H1 requires its 3’-5’ exonuclease activity. Int J Cancer. 2011;128:40–50.
Leone A, Flatow U, VanHoutte K, Steeg PS. Transfection of human nm23-H1 into the human MDA-MB-435 breast carcinoma cell line: effects on tumor metastatic potential, colonization and enzymatic activity. Oncogene. 1993;8:2325–33.
Hartsough MT, Steeg PS. Nm23/nucleoside diphosphate kinase in human cancers. J Bioenerg Biomembr. 2000;32:301–8.
Boissan M, Wendum D, Arnaud-Dabernat S, Munier A, Debray M, Lascu I, et al. Increased lung metastasis in transgenic NM23-Null/SV40 mice with hepatocellular carcinoma. J Natl Cancer Inst. 2005;97:836–45.
Pamidimukkala N, Puts GS, Leonard MK, Snyder D, Dabernat S, De Fabo EC, Noonan FP, Slominski A, Merlino G, Kaetzel DM. Nme1 and Nme2 genes exert metastasis suppressor activities in a genetically-engineered mouse model of UV-induced melanoma. Br J Cancer. 2021;124:161–5.
Marino N, Nakayama J, Collins JW, Steeg PS. Insights into the biology and prevention of tumor metastasis provided by the Nm23 metastasis suppressor gene. Cancer Metastasis Rev. 2012;31:593–603.
Horak CE, Lee JH, Elkahloun AG, Boissan M, Dumont S, Maga TK, et al. Nm23-H1 suppresses tumor cell motility by down-regulating the lysophosphatidic acid receptor EDG2. Cancer Res. 2007;67:7238–46.
Leonard MK, McCorkle JR, Snyder DE, Novak M, Zhang Q, Shetty AC, et al. Identification of a gene expression signature associated with the metastasis suppressor function of NME1: prognostic value in human melanoma. Lab Investig. 2017;98:327–38.
Horak CE, Mendoza A, Vega-Valle E, Albaugh M, Graff-Cherry C, McDermott WG, et al. Nm23-H1 suppresses metastasis by inhibiting expression of the lysophosphatidic acid receptor EDG2. Cancer Res. 2007;67:11751–9.
Leonard MK, Novak M, Snyder D, Snow G, Pamidimukkala N, McCorkle JR, et al. The metastasis suppressor NME1 inhibits melanoma cell motility via direct transcriptional induction of the integrin beta-3 gene. Exp Cell Res. 2019;374:85–93.
Kaetzel DM, Leonard MK, Cook GS, Novak M, Jarrett SG, Yang X, et al. Dual functions of NME1 in suppression of cell motility and enhancement of genomic stability in melanoma. Naunyn Schmiedebergs Arch Pharmacol. 2015;388:199–206.
Yang M, Jarrett SG, Craven R, Kaetzel DM. YNK1, the yeast homolog of human metastasis suppressor NM23, is required for repair of UV radiation- and etoposide-induced DNA damage. Mutat Res. 2009;660:74–8.
Sheng Y, Xu M, Li C, Xiong Y, Yang Y, Kuang X, et al. Nm23-H1 is involved in the repair of ionizing radiation-induced DNA double-strand breaks in the A549 lung cancer cell line. BMC Cancer. 2018;18:710.
Xue R, Peng Y, Han B, Li X, Chen Y, Pei H. Metastasis suppressor NME1 promotes non-homologous end joining of DNA double-strand breaks. DNA Repair. 2019;77:27–35.
Recio JA, Noonan FP, Takayama H, Anver MR, Duray P, Rush WL, et al. Ink4a/arf deficiency promotes ultraviolet radiation-induced melanomagenesis. Cancer Res. 2002;62:6724–30.
Noonan FP, Recio JA, Takayama H, Duray P, Anver MR, Rush WL, et al. Neonatal sunburn and melanoma in mice. Nature. 2001;413:271–2.
Garibyan L, Fisher DE. How sunlight causes melanoma. Curr Oncol Rep. 2010;12:319–26.
Postel EH, Wohlman I, Zou X, Juan T, Sun N, D’Agostin D, et al. Targeted deletion of Nm23/nucleoside diphosphate kinase A and B reveals their requirement for definitive erythropoiesis in the mouse embryo. Dev Dyn. 2009;238:775–87.
Pérez-Guijarro E, Yang HH, Araya RE, El Meskini R, Michael HT, Vodnala SK, et al. Multimodel preclinical platform predicts clinical response of melanoma to immunotherapy. Nat Med. 2020;26:781–91.
Van Raamsdonk CD, Bezrookove V, Green G, Bauer J, Gaugler L, O’Brien JM, et al. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2009;457:599–602.
Van Raamsdonk CD, Griewank KG, Crosby MB, Garrido MC, Vemula S, Wiesner T, et al. Mutations in GNA11 in uveal melanoma. N Engl J Med. 2010;363:2191–99.
Diskin SJ, Eck T, Greshock J, Mosse YP, Naylor T, Stoeckert CJ Jr., et al. STAC: A method for testing the significance of DNA copy number aberrations across multiple array-CGH experiments. Genome Res. 2006;16:1149–58.
Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D, et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci USA. 2007;104:20007–12.
Ahn A, Chatterjee A, Eccles MR. The slow cycling phenotype: a growing problem for treatment resistance in melanoma. Mol Cancer Ther. 2017;16:1002–9.
Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan S, et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell. 2016;165:35–44.
Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350:207–11.
Boissan M, Lacombe M-L. NM23, an example of a metastasis suppressor gene. Bull Cancer. 2012;99:431–40.
Snyder D, Wang Y, Kaetzel DM. A rare subpopulation of melanoma cells with low expression of metastasis suppressor NME1 is highly metastatic in vivo. Sci Rep. 2020;10:1971.
Abkevich V, Timms KM, Hennessy BT, Potter J, Carey MS, Meyer LA, et al. Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. Br J Cancer. 2012;107:1776–82.
Popova T, Manié E, Rieunier G, Caux-Moncoutier V, Tirapo C, Dubois T, et al. Ploidy and large-scale genomic instability consistently identify basal-like breast carcinomas with BRCA1/2 inactivation. Cancer Res. 2012;72:5454–62.
Molina-Ortiz I, Bartolome RA, Hernandez-Varas P, Colo GP, Teixido J. Overexpression of E-cadherin on melanoma cells inhibits chemokine-promoted invasion involving p190RhoGAP/p120ctn-dependent inactivation of RhoA. J Biol Chem. 2009;284:15147–57.
Chen D, Li Y, Zhang X, Wu H, Wang Q, Cai J, et al. Ubiquitin ligase TRIM65 promotes colorectal cancer metastasis by targeting ARHGAP35 for protein degradation. Oncogene. 2019;38:6429–44.
Markby DW, Onrust R, Bourne HR. Separate GTP binding and GTPase activating domains of a G alpha subunit. Science. 1993;262:1895–901.
Spivak G. Transcription-coupled repair: an update. Arch Toxicol. 2016;90:2583–94.
Heraud C, Pinault M, Lagree V, Moreau V. p190RhoGAPs, the ARHGAP35- and ARHGAP5-encoded proteins, in health and disease. Cells. 2019;8:351.
McIlwain DR, Berger T, Mak TW. Caspase functions in cell death and disease. Cold Spring Harbor Perspectives. Cold Spring Harbor Perspect Biol. 2013;5:a008656.
Hamidi H, Ivaska J. Every step of the way: integrins in cancer progression and metastasis. Nat Rev Cancer. 2018;18:533–48.
Balint K, Xiao M, Pinnix CC, Soma A, Veres I, Juhasz I, et al. Activation of Notch1 signaling is required for beta-catenin-mediated human primary melanoma progression. J Clin Investig. 2005;115:3166–76.
Zhang K, Wong P, Salvaggio C, Salhi A, Osman I, Bedogni B. Synchronized targeting of notch and ERBB signaling suppresses melanoma tumor growth through inhibition of Notch1 and ERBB3. J Investig Dermatol. 2016;136:464–72.
Fei F, Qu J, Zhang M, Li Y, Zhang S. S100A4 in cancer progression and metastasis: a systematic review. Oncotarget. 2017;8:73219–39.
Bektic J, Pfeil K, Berger AP, Ramoner R, Pelzer A, Schafer G, et al. Small G-protein RhoE is underexpressed in prostate cancer and induces cell cycle arrest and apoptosis. Prostate. 2005;64:332–40.
Rubenstein NM, Chan JF, Kim JY, Hansen SH, Firestone GL. Rnd3/RhoE induces tight junction formation in mammary epithelial tumor cells. Exp Cell Res. 2005;305:74–82.
Chen LH, Kuo WH, Tsai MH, Chen PC, Hsiao CK, Chuang EY, et al. Identification of prognostic genes for recurrent risk prediction in triple negative breast cancer patients in Taiwan. PLoS ONE. 2011;6:e28222.
Ekizoglu S, Seven D, Ulutin T, Guliyev J, Buyru N. Investigation of the SLC22A23 gene in laryngeal squamous cell carcinoma. BMC Cancer. 2018;18:477.
Tamura K, Makino A, Hullin-Matsuda F, Kobayashi T, Furihata M, Chung S, et al. Novel lipogenic enzyme ELOVL7 is involved in prostate cancer growth through saturated long-chain fatty acid metabolism. Cancer Res. 2009;69:8133–40.
Arakaki AKS, Pan WA, Trejo J. GPCRs in cancer: protease-activated receptors, endocytic adaptors and signaling. Int J Mol Sci. 2018;19:1886.
Draheim KM, Chen HB, Tao Q, Moore N, Roche M, Lyle S. ARRDC3 suppresses breast cancer progression by negatively regulating integrin beta4. Oncogene. 2010;29:5032–47.
Soung YH, Pruitt K, Chung J. Epigenetic silencing of ARRDC3 expression in basal-like breast cancer cells. Sci Rep. 2014;4:3846.
Zheng Y, Lin ZY, Xie JJ, Jiang FN, Chen CJ, Li JX, et al. ARRDC3 inhibits the progression of human prostate cancer through ARRDC3-ITGβ4 pathway. Curr Mol Med. 2017;17:221–9.
Xiao J, Shi Q, Li W, Mu X, Peng J, Li M, et al. ARRDC1 and ARRDC3 act as tumor suppressors in renal cell carcinoma by facilitating YAP1 degradation. Am J Cancer Res. 2018;8:132–43.
Yang Z, Shang J, Li N, Zhang L, Tang T, Tian G, et al. Development and validation of a 10-gene prognostic signature for acute myeloid leukaemia. J Cell Mol Med. 2020;24:4510–23.
Mahamdallie S, Yost S, Poyastro-Pearson E, Holt E, Zachariou A, Seal S, et al. Identification of new Wilms tumour predisposition genes: an exome sequencing study. Lancet Child Adolesc Health. 2019;3:322–31.
Paysan L, Piquet L, Saltel F, Moreau V. Rnd3 in cancer: a review of the evidence for tumor promoter or suppressor. Mol Cancer Res. 2016;14:1033–44.
Klein RM, Aplin AE. Rnd3 regulation of the actin cytoskeleton promotes melanoma migration and invasive outgrowth in three dimensions. Cancer Res. 2009;69:2224–33.
Tomayko MM, Reynolds CP. Determination of subcutaneous tumor size in athymic (nude) mice. Cancer Chemother Pharmacol. 1989;24:148–54.
Aaron KA, Manojlovic Z, Tu N, Xu Y, Jin Y, Chang S, et al. What genes can tell: a closer look at vestibular schwannoma. Otol Neurotol. 2020;41:522–9.
Manojlovic Z, Christofferson A, Liang WS, Aldrich J, Washington M, Wong S, et al. Comprehensive molecular profiling of 718 Multiple Myelomas reveals significant differences in mutation frequencies between African and European descent cases. PLoS Genet. 2017;13:e1007087.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550.
Morrison GJ, Cunha AT, Jojo N, Xu Y, Xu Y, Kwok E, et al. Cancer transcriptomic profiling from rapidly enriched circulating tumor cells. Int J Cancer. 2020;146:2845–54.
Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14:417–9.
Roberts A, Trapnell C, Donaghey J, Rinn JL, Pachter L. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 2011;12:r22.
Roberts A, Pachter L. Streaming fragment assignment for real-time analysis of sequencing experiments. Nat Methods. 2013;10:71–3.
Kim D, Salzberg SL. TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol. 2011;12:r72.
Fisher ML, Ciavattone N, Grun D, Adhikary G, Eckert RL. Sulforaphane reduces YAP/∆Np63α signaling to reduce cancer stem cell survival and tumor formation. Oncotarget. 2017;8:73407–18.
Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet. 2012;44:1006–14.
Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–63.
Van Allen EM, Wagle N, Sucker A, Treacy DJ, Johannessen CM, Goetz EM, et al. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov. 2014;4:94–109.
Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371:2189–99.
Berger MF, Hodis E, Heffernan TP, Deribe YL, Lawrence MS, Protopopov A, et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012;485:502–6.
Liang WS, Hendricks W, Kiefer J, Schmidt J, Sekar S, Carpten J, et al. Integrated genomic analyses reveal frequent TERT aberrations in acral melanoma. Genome Res. 2017;27:524–32.
Shain AH, Garrido M, Botton T, Talevich E, Yeh I, Sanborn JZ, et al. Exome sequencing of desmoplastic melanoma identifies recurrent NFKBIE promoter mutations and diverse activating mutations in the MAPK pathway. Nat Genet. 2015;47:1194–9.
Johansson P, Aoude LG, Wadt K, Glasson WJ, Warrier SK, Hewitt AW, et al. Deep sequencing of uveal melanoma identifies a recurrent mutation in PLCB4. Oncotarget. 2016;7:4624–31.
Kandasamy S, Adhikary G, Rorke EA, Friedberg JS, Mickle MB, Alexander HR, et al. The YAP1 signaling inhibitors, verteporfin and CA3, suppress the mesothelioma cancer stem cell phenotype. Mol. Cancer Res. 2020;18:343–51.
Acknowledgements
The authors gratefully acknowledge the expert technical assistance of A. Greenawalt and B. Hazzard, and to I. Violich for providing guidance in development of the mouse computational pipeline. This work was supported by the National Institutes of Health/National Cancer Institute through research grants R01 CA83237, R01 CA159871 and R01 CA159871-S1 (DMK), R01 CA211909 (RLE), center core grant P30 CA134274 and training grant T32 CA154274, by research grant R01 AR071189 (ATS) from the National Institutes of Health/National Institute of Arthritis and Musculoskeletal and Skin Diseases, and by education grant R25 GM055036 from the National Institutes of Health/National Institute of General Medical Sciences. The study was also supported by funding from the Maryland Stem Cell Research Foundation (MSCRFI-1638, DMK), the Veterans Administration (VA Merit Grant 1I01BX004293, ATS), and the Baltimore Research and Education Foundation, VA Maryland Health Care System.
Author information
Authors and Affiliations
Contributions
Conceptualization: JDC and DMK. Methodology: MKL, ECD-F, FPN, MGW, DWC, ZM, GM, RLE, and DMK. Formal analysis: MKL, ATS, ZM, and DMK. Investigation: MKL, GSP, NP, DS, GA, YX, EK, YJ, NM, MN, RMS, ACS, C-PD, MR, ATS, MGW, and DMK. Data curation: MKL, AM, ACS, and DMK. Writing (original draft): DMK; Writing (review and editing): MKL, RLE, GM, JDC, ZM, and DMK. Visualization: ATS, ZM, and DMK. Supervision: JDC, ZM, ATS, RLE, and DMK. Project administration: RLE, JDC, ZM, and DMK. Funding acquisition: RLE and DMK.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Leonard, M.K., Puts, G.S., Pamidimukkala, N. et al. Comprehensive molecular profiling of UV-induced metastatic melanoma in Nme1/Nme2-deficient mice reveals novel markers of survival in human patients. Oncogene 40, 6329–6342 (2021). https://doi.org/10.1038/s41388-021-01998-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-021-01998-w
This article is cited by
-
The m6A demethylases FTO and ALKBH5 aggravate the malignant progression of nasopharyngeal carcinoma by coregulating ARHGAP35
Cell Death Discovery (2024)
-
Genome-wide CRISPR screening identifies a role for ARRDC3 in TRP53-mediated responses
Cell Death & Differentiation (2024)