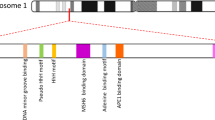
Abstract
POLE, POLD1, and NTHL1 are involved in DNA replication and have recently been recognized as hereditary cancer-predisposing genes, because their alterations are associated with colorectal cancer and other tumors. POLE/POLD1-associated syndrome shows an autosomal dominant inheritance, whereas NTHL1-associated syndrome follows an autosomal recessive pattern. Although the prevalence of germline monoallelic POLE/POLD1 and biallelic NTHL1 pathogenic variants is low, they determine different phenotypes with a broad tumor spectrum overlapping that of other hereditary conditions like Lynch Syndrome or Familial Adenomatous Polyposis. Endometrial and breast cancers, and probably ovarian and brain tumors are also associated with POLE/POLD1 alterations, while breast cancer and other unusual tumors are correlated with NTHL1 pathogenic variants. POLE-mutated colorectal and endometrial cancers are associated with better prognosis and may show favorable responses to immunotherapy. Since POLE/POLD1-mutated tumors show a high tumor mutational burden producing an increase in neoantigens, the identification of POLE/POLD1 alterations could help select patients suitable for immunotherapy treatment. In this review, we will investigate the role of POLE, POLD1, and NTHL1 genetic variants in cancer predisposition, discussing the potential future therapeutic applications and assessing the utility of performing a routine genetic testing for these genes, in order to implement prevention and surveillance strategies in mutation carriers.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer J Clin. 2018;68:394–424.
Short E, Thomas LE, Hurley J, Jose S, Sampson JR. Inherited predisposition to colorectal cancer: towards a more complete picture. J Med Genet. 2015;52:791–6.
van Wezel T, de Miranda NFCC, Morreau H, Schubert SA. The missing heritability of familial colorectal cancer. Mutagenesis. 2020;35:221–31.
Stjepanovic N, Moreira L, Carneiro F, Balaguer F, Cervantes A, Balmaña J, et al. Hereditary gastrointestinal cancers: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up†. Ann Oncol. 2019;30:1558–71.
Colas C, Coulet F, Svrcek M, Collura A, Fléjou J-F, Duval A, et al. Lynch or Not Lynch? Is that Always a Quest? 2012;113:121–66.
Vasen H, Watson P, Mecklin J, Lynch H. New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative Group on HNPCC☆. Gastroenterology. 1999;116:1453–6.
Umar A, Boland CR, Terdiman JP, Syngal S, Chapelle ADL, Ruschoff J, et al. Revised bethesda guidelines for hereditary nonpolyposis colorectal cancer (Lynch Syndrome) and microsatellite instability. JNCI J Natl Cancer Inst. 2004;96:261–8.
Nieuwenhuis MH, Vogt S, Jones N, Nielsen M, Hes FJ, Sampson JR, et al. Evidence for accelerated colorectal adenoma–carcinoma progression inMUTYH-associated polyposis? Gut. 2012;61:734–8.
Urso EDL, Ponz de Leon M, Vitellaro M, Piozzi GN, Bao QR, Martayan A, et al. Definition and management of colorectal polyposis not associated with APC/MUTYH germline pathogenic variants: AIFEG consensus statement. Dig Liver Dis. 2021;53:409–17.
Lindor NM, Rabe K, Petersen GM, Haile R, Casey G, Baron J, et al. Lower cancer incidence in Amsterdam-I criteria families without mismatch repair deficiency. JAMA. 2005;293:1979.
Palles C, Cazier J-B, Howarth KM, Domingo E, Jones AM, Broderick P, et al. Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat Genet. 2012;45:136–44.
Pursell ZF, Isoz I, Lundstrom EB, Johansson E, Kunkel TA. Yeast DNA polymerase participates in leading-strand DNA replication. Science. 2007;317:127–30.
Mertz TM, Baranovskiy AG, Wang J, Tahirov TH, Shcherbakova PV. Nucleotide selectivity defect and mutator phenotype conferred by a colon cancer-associated DNA polymerase δ mutation in human cells. Oncogene. 2017;36:4427–33.
Alani E, Thresher R, Griffith JD, Kolodner RD. Characterization of DNA-binding and strand-exchange stimulation properties of y-RPA, a yeast single-strand-DNA-binding protein. J Mol Biol. 1992;227:54–71.
Rayner E, van Gool IC, Palles C, Kearsey SE, Bosse T, Tomlinson I, et al. A panoply of errors: polymerase proofreading domain mutations in cancer. Nat Rev Cancer. 2016;16:71–81.
Briggs S, Tomlinson I. Germline and somatic polymerase ϵ and δ mutations define a new class of hypermutated colorectal and endometrial cancers. J Pathol. 2013;230:148–53.
Bellido F, Pineda M, Aiza G, Valdés-Mas R, Navarro M, Puente DA, et al. POLE and POLD1 mutations in 529 kindred with familial colorectal cancer and/or polyposis: review of reported cases and recommendations for genetic testing and surveillance. Genet Med. 2015;18:325–32.
Belhadj S, Quintana I, Mur P, Munoz-Torres PM, Alonso MH, Navarro M. NTHL1 biallelic mutations seldom cause colorectal cancer, serrated polyposis or a multi-tumor phenotype, in absence of colorectal adenomas. Sci Rep. 2019;9:1–5.
Limpose KL, Trego KS, Li Z, Leung SW, Sarker AH, Shah JA, et al. Overexpression of the base excision repair NTHL1 glycosylase causes genomic instability and early cellular hallmarks of cancer. Nucleic Acids Res. 2018;46:4515–32.
Mur P, García-Mulero S, del Valle J, Magraner-Pardo L, Vidal A, Pineda M, et al. Role of POLE and POLD1 in familial cancer. Genet Med. 2020;22:2089–2100.
Fang H, Barbour JA, Poulos RC, Katainen R, Aaltonen LA, Wong JWH. Mutational processes of distinct POLE exonuclease domain mutants drive an enrichment of a specific TP53 mutation in colorectal cancer. PLOS Genet. 2020;16:e1008572.
Yao J, Gong Y, Zhao W, Han Z, Guo S, Liu H. et al. Comprehensive analysis of POLE and POLD1 gene variations identifies cancer patients potentially benefit from immunotherapy in Chinese population. Sci Rep. 2019;9:1–14.
Haradhvala NJ, Kim J, Maruvka YE, Polak P, Rosebrock D, Livitz D. et al. Distinct mutational signatures characterize concurrent loss of polymerase proofreading and mismatch repair. Nat Commun. 2018;9:1–9.
He J, Ouyang W, Zhao W, Shao L, Li B, Liu B, et al. Distinctive genomic characteristics in POLE/POLD1-mutant cancers can potentially predict beneficial clinical outcomes in patients who receive immune checkpoint inhibitor. Ann Transl Med. 2021;9:129–129.
Valle L, Hernández-Illán E, Bellido F, Aiza G, Castillejo A, Castillejo M-I, et al. New insights into POLE and POLD1 germline mutations in familial colorectal cancer and polyposis. Hum Mol Genet. 2014;23:3506–12.
Spier I, Holzapfel S, Altmüller J, Zhao B, Horpaopan S, Vogt S, et al. Frequency and phenotypic spectrum of germline mutations inPOLEand seven other polymerase genes in 266 patients with colorectal adenomas and carcinomas. Int J Cancer. 2015;137:320–31.
Palles C, Martin L, Domingo E, Chegwidden L, McGuire J, Cuthill V, et al. The clinical features of polymerase proof-reading associated polyposis (PPAP) and recommendations for patient management. Familial Cancer. 2021; 1–13.
Chubb D, Broderick P, Dobbins SE, Frampton M, Kinnersley B, Penegar S. Rare disruptive mutations and their contribution to the heritable risk of colorectal cancer. Nat Commun. 2016;7:1–7.
Dong H, Bai Y, Cao X, Wang Y, Shi L, Li F, et al. Abstract 5137: comprehensive analysis of POLE and POLD1 mutation in 9322 Chinese cancer patients 2019: 5137–5137.
Rosner G, Gluck N, Carmi S, Bercovich D, Fliss-Issakov N, Ben-Yehoyada M, et al. POLD1 and POLE gene mutations in Jewish cohorts of early-onset colorectal cancer and of multiple colorectal adenomas. Dis Colon Rectum. 2018;61:1073–9.
Siraj AK, Bu R, Iqbal K, Parvathareddy SK, Masoodi T, Siraj N. et al. POLE and POLD1 germline exonuclease domain pathogenic variants, a rare event in colorectal cancer from the Middle East. Mol Genet Genom Med. 2020;8:1–11.
Djursby M, Madsen MB, Frederiksen JH, Berchtold LA, Therkildsen C, Willemoe GL, et al. New pathogenic germline variants in very early onset and familial colorectal cancer patients. Front Genet. 2020;11:566266.
Esteban-Jurado C, Giménez-Zaragoza D, Muñoz J, Franch-Expósito S, Álvarez-Barona M, Ocaña T, et al. POLE and POLD1 screening in 155 patients with multiple polyps and early-onset colorectal cancer. Oncotarget. 2017;8:26732–43.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–23.
Campbell BB, Light N, Fabrizio D, Zatzman M, Fuligni F, de Borja R, et al. Comprehensive analysis of hypermutation in human cancer. Cell. 2017;171:1042–56. e1010
Strickler JH, Hanks BA, Khasraw M. Tumor mutational burden as a predictor of immunotherapy response: is more always better? Clin Cancer Res. 2021;27:1236–41.
Gelsomino F, Barbolini M, Spallanzani A, Pugliese G, Cascinu S. The evolving role of microsatellite instability in colorectal cancer: a review. Cancer Treat Rev. 2016;51:19–26.
Fabrizio DA, George TJ Jr, Dunne RF, Frampton G, Sun J, Gowen K, et al. Beyond microsatellite testing: assessment of tumor mutational burden identifies subsets of colorectal cancer who may respond to immune checkpoint inhibition. J Gastrointest Oncol. 2018;9:610–7.
Zhao P, Li L, Jiang X, Li Q. Mismatch repair deficiency/microsatellite instability-high as a predictor for anti-PD-1/PD-L1 immunotherapy efficacy. J Hematol Oncol. 2019;12:1–14.
Gong J, Wang C, Lee PP, Chu P, Fakih M. Response to PD-1 blockade in microsatellite stable metastatic colorectal cancer harboring aPOLEMutation. J Natl Compr Cancer Netw. 2017;15:142–7.
Jansen AML, van Wezel T, van den Akker BEWM, Ventayol Garcia M, Ruano D, Tops CMJ, et al. Combined mismatch repair and POLE/POLD1 defects explain unresolved suspected Lynch syndrome cancers. Eur J Hum Genet. 2015;24:1089–92.
Domingo E, Freeman-Mills L, Rayner E, Glaire M, Briggs S, Vermeulen L, et al. Somatic POLE proofreading domain mutation, immune response, and prognosis in colorectal cancer: a retrospective, pooled biomarker study. Lancet Gastroenterol Hepatol. 2016;1:207–16.
Fancello L, Gandini S, Pelicci PG, Mazzarella L. Tumor mutational burden quantification from targeted gene panels: major advancements and challenges. J Immun Ther Cancer. 2019;1–13.
Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl J Med. 2015;372:2509–20.
Silberman R, Steiner DF, Lo AA, Gomez A, Zehnder JL, Chu G. et al. Complete and prolonged response to immune checkpoint blockade in POLE-mutated colorectal cancer. JCO Precision Oncol. 2019;3:1–5.
Wang F, Zhao Q, Wang Y-N, Jin Y, He M-M, Liu Z-X, et al. Evaluation of POLE and POLD1 mutations as biomarkers for immunotherapy outcomes across multiple cancer types. JAMA Oncol. 2019;5:1504.
Forgó E, Gomez AJ, Steiner D, Zehnder J, Longacre TA. Morphological, immunophenotypical and molecular features of hypermutation in colorectal carcinomas with mutations in DNA polymerase ε (POLE). Histopathology. 2019;76:366–74.
Bourdais R, Rousseau B, Pujals A, Boussion H, Joly C, Guillemin A, et al. Polymerase proofreading domain mutations: New opportunities for immunotherapy in hypermutated colorectal cancer beyond MMR deficiency. Crit Rev Oncol/Hematol. 2017;113:242–8.
Rousseau B, Foote MB, Maron SB, Diplas BH, Lu S, Argilés G, et al. The spectrum of benefit from checkpoint blockade in hypermutated tumors. N. Engl J Med. 2021;384:1168–70.
Hu H, Cai W, Wu D, Hu W, Dong Wang L, Mao J, et al. Ultra‐mutated colorectal cancer patients with POLE driver mutations exhibit distinct clinical patterns. Cancer Med. 2020;10:135–42.
Nijman SMB. Synthetic lethality: general principles, utility and detection using genetic screens in human cells. FEBS Lett. 2011;585:1–6.
Kaelin WG. The concept of synthetic lethality in the context of anticancer therapy. Nat Rev Cancer. 2005;5:689–98.
Hocke S, Guo Y, Job A, Orth M, Ziesch A, Lauber K, et al. A synthetic lethal screen identifies ATR-inhibition as a novel therapeutic approach for POLD1-deficient cancers. Oncotarget. 2016;7:7080–95.
Job A, Tatura M, Schäfer C, Lutz V, Schneider H, Lankat-Buttgereit B. The POLD1R689W variant increases the sensitivity of colorectal cancer cells to ATR and CHK1 inhibitors. Sci Rep. 2020;10:1–12.
Church DN, Briggs SEW, Palles C, Domingo E, Kearsey SJ, Grimes JM, et al. DNA polymerase ɛ and δ exonuclease domain mutations in endometrial cancer. Hum Mol Genet. 2013;22:2820–8.
McConechy MK, Talhouk A, Leung S, Chiu D, Yang W, Senz J, et al. Endometrial carcinomas with POLE exonuclease domain mutations have a favorable prognosis. Clin Cancer Res. 2016;22:2865–73.
León‐Castillo A, Britton H, McConechy MK, McAlpine JN, Nout R, Kommoss S, et al. Interpretation of somatic POLE mutations in endometrial carcinoma. J Pathol. 2020;250:323–35.
Howitt BE, Shukla SA, Sholl LM, Ritterhouse LL, Watkins JC, Rodig S, et al. Association of polymerase e–mutated and microsatellite-instable endometrial cancers with neoantigen load, number of tumor-infiltrating lymphocytes, and expression of PD-1 and PD-L1. JAMA Oncol. 2015;1:1319.
Mehnert JM, Panda A, Zhong H, Hirshfield K, Damare S, Lane K, et al. Immune activation and response to pembrolizumab in POLE-mutant endometrial cancer. J Clin Investig. 2016;126:2334–40.
Imboden S, Nastic D, Ghaderi M, Rydberg F, Rau TT, Mueller MD, et al. Phenotype of POLE-mutated endometrial cancer. Plos One. 2019;14:e0214318.
Siraj AK, Parvathareddy SK, Bu R, Iqbal K, Siraj S, Masoodi T. Germline POLE and POLD1 proofreading domain mutations in endometrial carcinoma from Middle Eastern region. Cancer Cell Int. 2019;19:1–9.
Wong A, Kuick CH, Wong WL, Tham JM, Mansor S, Loh E, et al. Mutation spectrum of POLE and POLD1 mutations in South East Asian women presenting with grade 3 endometrioid endometrial carcinomas. Gynecologic Oncol. 2016;141:113–20.
He Y, Wang T, Li N, Yang B, Hu Y. Clinicopathological characteristics and prognostic value of POLE mutations in endometrial cancer. Medicine. 2020;99:e19281.
Hansen MF, Johansen J, Bjørnevoll I, Sylvander AE, Steinsbekk KS, Sætrom P, et al. A novel POLE mutation associated with cancers of colon, pancreas, ovaries and small intestine. Fam Cancer. 2015;14:437–48.
Aoude LG, Heitzer E, Johansson P, Gartside M, Wadt K, Pritchard AL, et al. POLE mutations in families predisposed to cutaneous melanoma. Fam Cancer. 2015;14:621–8.
Siraj AK, Bu R, Arshad M, Iqbal K, Parvathareddy SK, Masoodi T, et al. POLE and POLD1 pathogenic variants in the proofreading domain in papillary thyroid cancer. Endocr Connect. 2020;9:923–32.
Vande Perre P, Siegfried A, Corsini C, Bonnet D, Toulas C, Hamzaoui N, et al. Germline mutation p.N363K in POLE is associated with an increased risk of colorectal cancer and giant cell glioblastoma. Fam Cancer. 2018;18:173–8.
Hamzaoui N, Alarcon F, Leulliot N, Guimbaud R, Buecher B, Colas C, et al. Genetic, structural, and functional characterization of POLE polymerase proofreading variants allows cancer risk prediction. Genet Med. 2020;22:1533–41.
Gao S, Zhang X, Song Q, Liu J, Ji X, Wang P. POLD1 deficiency is involved in cognitive function impairment in AD patients and SAMP8 mice. Biomed Pharmacother. 2019;114:108833.
Liu J, Liu Y, Fu J, Liu C, Yang T, Zhang X. Preliminary study on the function of the POLD1 (CDC2) EXON2 c.56G>A mutation. Mol Genet Genom Med. 2020;8:1–6.
Weren RDA, Ligtenberg MJL, Kets CM, de Voer RM, Verwiel ETP, Spruijt L, et al. A germline homozygous mutation in the base-excision repair gene NTHL1 causes adenomatous polyposis and colorectal cancer. Nat Genet. 2015;47:668–71.
Grolleman JE, de Voer RM, Elsayed FA, Nielsen M, Weren RDA, Palles C, et al. Mutational signature analysis reveals NTHL1 deficiency to cause a multi-tumor phenotype. Cancer Cell. 2019;35:256–66. e255
Li N, Zethoven M, McInerny S, Devereux L, Huang Y-K, Thio N. et al. Evaluation of the association of heterozygous germline variants in NTHL1 with breast cancer predisposition: an international multi-center study of 47,180 subjects. npj Breast Cancer. 2021;7:1–12.
Groves A, Gleeson M, Spigelman AD. NTHL1-associate polyposis: first Australian case report. Fam Cancer. 2019;18:179–82.
Rivera B, Castellsagué E, Bah I, van Kempen LC, Foulkes WD. Biallelic NTHL1 mutations in a woman with multiple primary tumors. N. Engl J Med. 2015;373:1985–6.
Weren RDA, Ligtenberg MJL, Geurts van Kessel A, De Voer RM, Hoogerbrugge N, Kuiper RP. NTHL1 and MUTYH polyposis syndromes: two sides of the same coin? J Pathol. 2018;244:135–42.
Elsayed FA, Grolleman JE, Ragunathan A, Buchanan DD, van Wezel T, de Voer RM, et al. Monoallelic NTHL1 loss-of-function variants and risk of polyposis and colorectal cancer. Gastroenterology. 2020;159:2241–3. e2246
Das L, Quintana VG, Sweasy JB. NTHL1 in genomic integrity, aging and cancer. DNA Repair. 2020;93:102920.
Drost J, van Boxtel R, Blokzijl F, Mizutani T, Sasaki N, Sasselli V, et al. Use of CRISPR-modified human stem cell organoids to study the origin of mutational signatures in cancer. Science. 2017;358:234–8.
Pilati C, Shinde J, Alexandrov LB, Assié G, André T, Hélias‐Rodzewicz Z, et al. Mutational signature analysis identifies MUTYH deficiency in colorectal cancers and adrenocortical carcinomas. J Pathol. 2017;242:10–15.
Marie Lewis K. Identifying hereditary cancer: genetic counseling and cancer risk assessment. Curr Probl Cancer. 2014;38:216–25.
Fanale D, Incorvaia L, Filorizzo C, Bono M, Fiorino A, Calò V, et al. Detection of germline mutations in a cohort of 139 patients with bilateral breast cancer by multi-gene panel testing: impact of pathogenic variants in other genes beyond BRCA1/2. Cancers. 2020;12:2415.
Hegde M, Ferber M, Mao R, Samowitz W, Ganguly A. ACMG technical standards and guidelines for genetic testing for inherited colorectal cancer (Lynch syndrome, familial adenomatous polyposis, and MYH-associated polyposis). Genet Med. 2013;16:101–16.
Dhooge M, Baert-Desurmont S, Corsini C, Caron O, Andrieu N, Berthet P, et al. National recommendations of the French Genetics and Cancer Group - Unicancer on the modalities of multi-genes panel analyses in hereditary predispositions to tumors of the digestive tract. Eur J Med Genet. 2020;63:104080.
Mao R, Krautscheid P, Graham RP, Ganguly A, Shankar S, Ferber M et al. Genetic testing for inherited colorectal cancer and polyposis, 2021 revision: a technical standard of the American College of Medical Genetics and Genomics (ACMG). Genet Med. 2021; 1–11.
Acknowledgements
The authors thank Dr. Chiara Drago for the English language revision.
Author information
Authors and Affiliations
Contributions
LM, DF, CB, AR, and VB conceived, wrote, and critically revised the manuscript with the contribution of AF, LRC, RS, CF, and AD. Literature data were acquired and analyzed by LM, DF, CB, AF, LRC, RS, CF, and AD. The figures of the manuscript were conceived and designed by AF, CB, and LRC. The tables were conceived and designed by LM, DF, and CB. AF, LRC, RS, CF, and AD participated to the critical revision of the manuscript. All authors have read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Magrin, L., Fanale, D., Brando, C. et al. POLE, POLD1, and NTHL1: the last but not the least hereditary cancer-predisposing genes. Oncogene 40, 5893–5901 (2021). https://doi.org/10.1038/s41388-021-01984-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-021-01984-2
This article is cited by
-
Multiple duodenal epithelial tumors in a patient with polymerase proofreading-associated polyposis in POLE variant
Clinical Journal of Gastroenterology (2024)
-
Evaluation of inherited germline mutations in cancer susceptibility genes among pancreatic cancer patients: a single-center study
Molecular Medicine (2023)
-
Personalisierte Prävention und Früherkennung am Beispiel des Lynch-Syndroms
Die Onkologie (2023)
-
The clinicopathological characteristics of POLE-mutated/ultramutated endometrial carcinoma and prognostic value of POLE status: a meta-analysis based on 49 articles incorporating 12,120 patients
BMC Cancer (2022)
-
MUTYH-associated tumor syndrome: The other face of MAP
Oncogene (2022)